Physiological and Proteomic Responses of Pitaya to PEG-Induced Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Stress Treatments
2.3. Physiological Measurements
2.4. Protein Extraction
2.5. Trypsin Digestion
2.6. LC-MS/MS Analysis
2.7. Database Search
2.8. Data Analysis
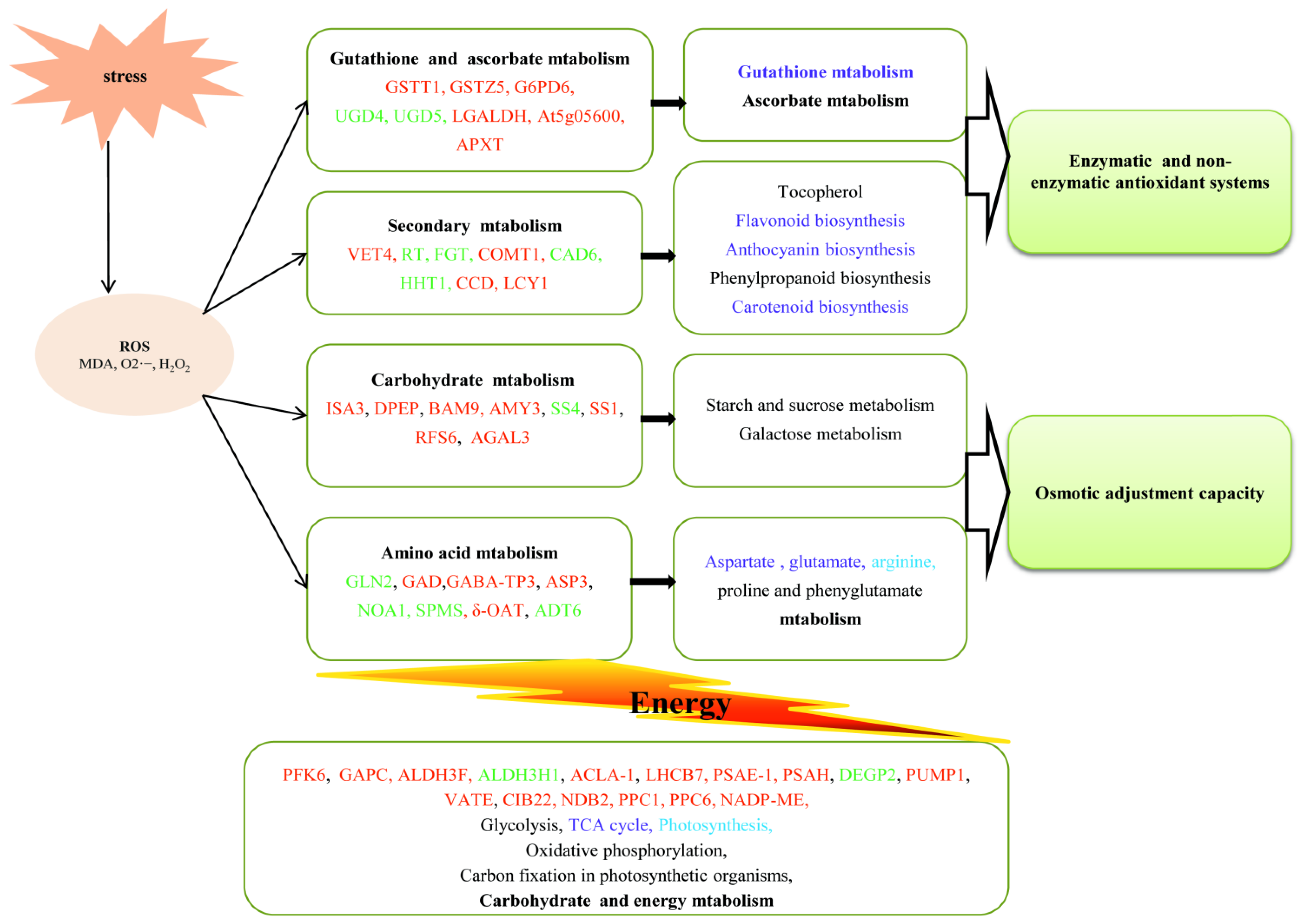
3. Results
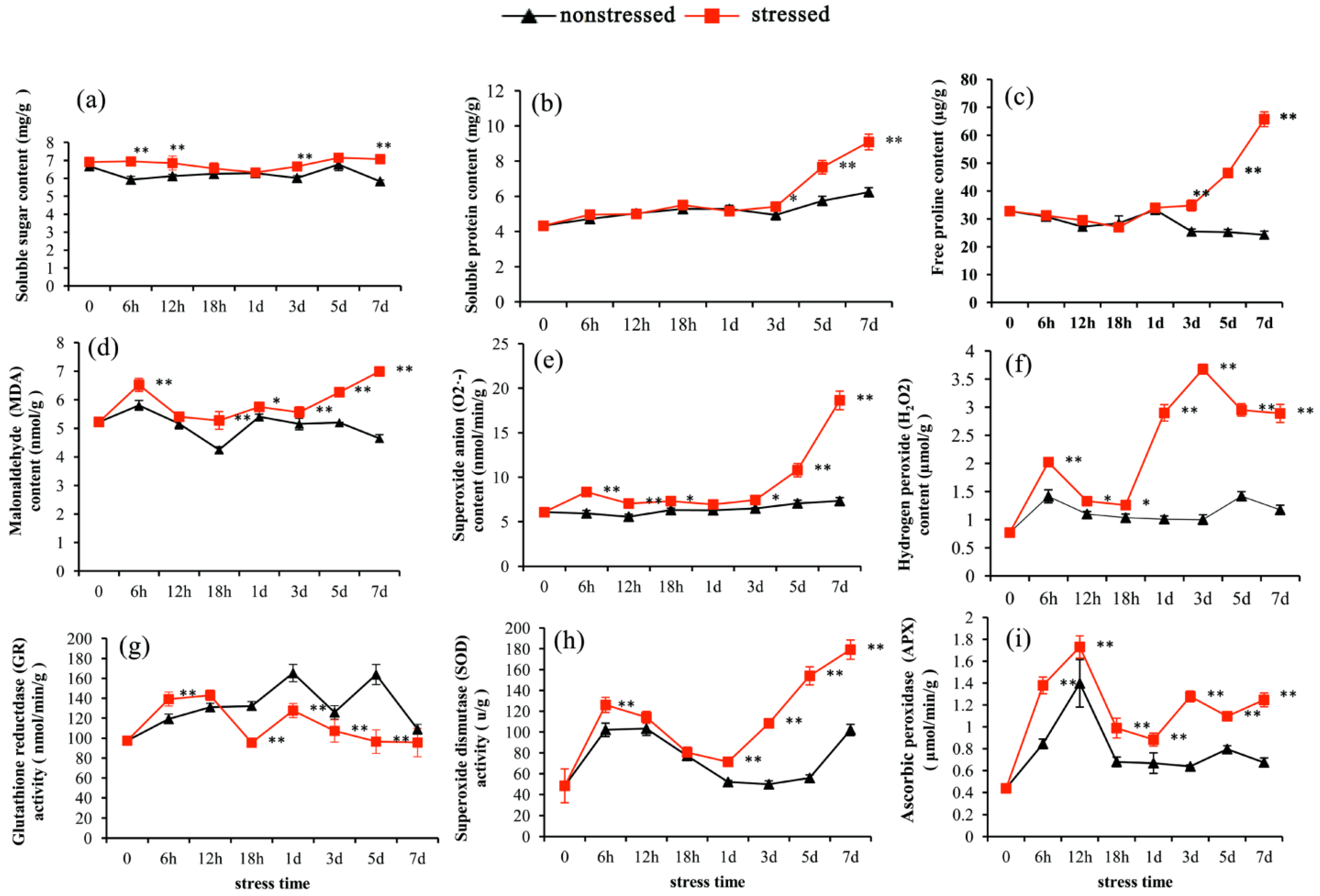
3.1. Physiological Responses of Pitaya to Drought Stress
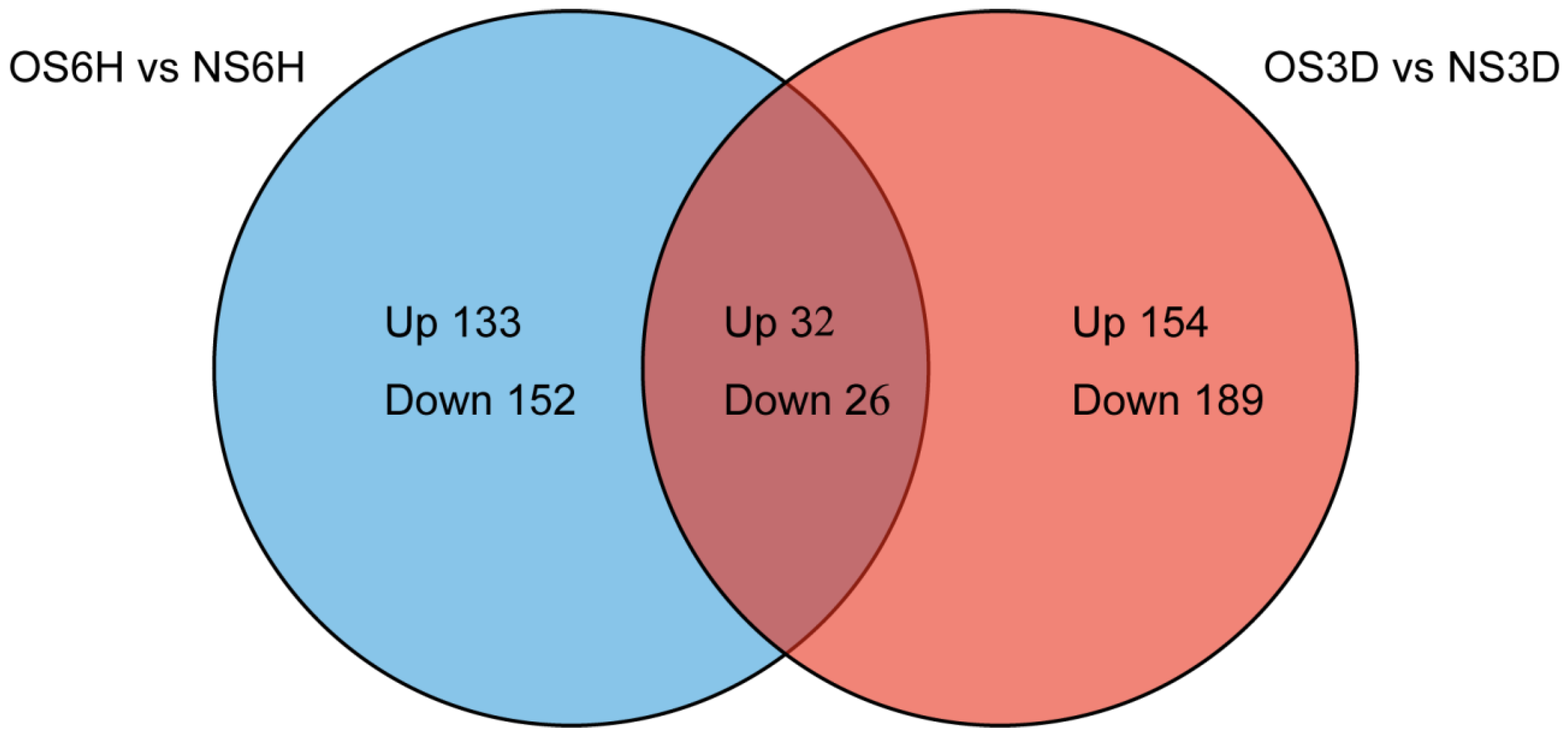
3.2. Primary Data Analysis and Protein Detection
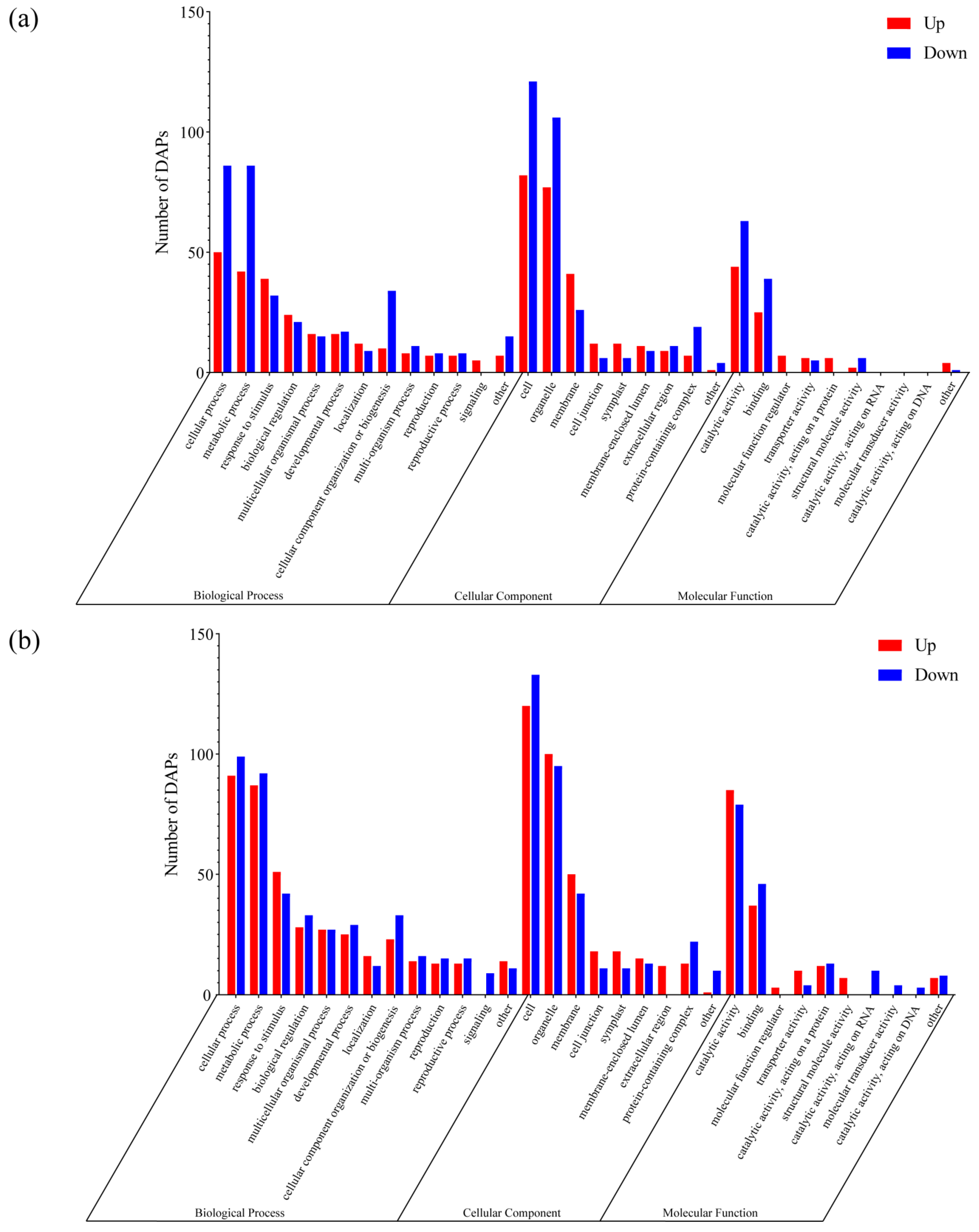
3.3. Identification and Functional Analysis of DAPs
3.4. DAPs Involved in Carbohydrate Metabolism
3.5. DAPs Involved in Energy Metabolism
3.6. DAPs Involved in Amino Acid Metabolism
3.7. DAPs Involved in GSH and AsA Metabolism
3.8. DAPs Involved in Secondary Metabolism
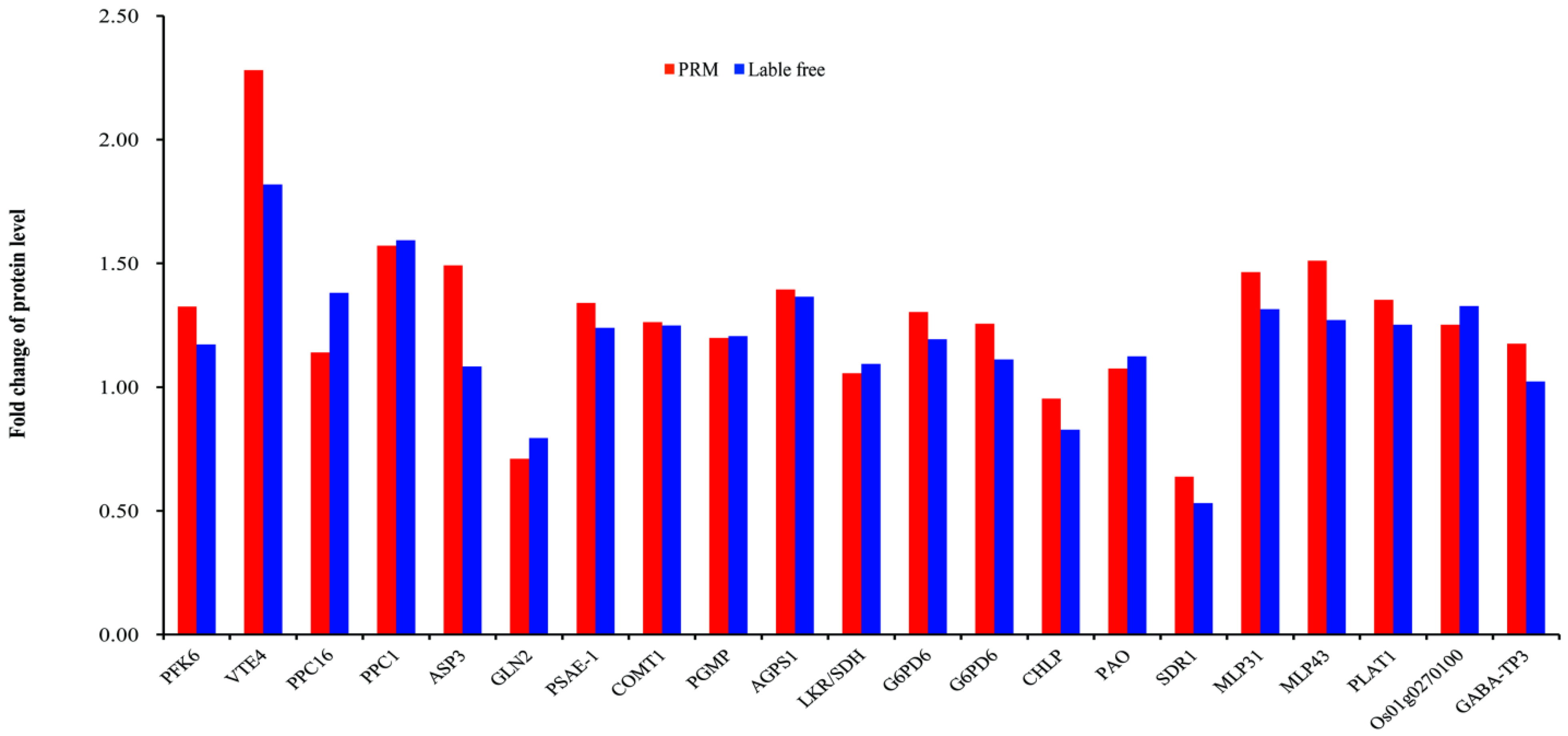
3.9. Validation of Proteomics Data by PRM
4. Discussion
4.1. Physiological Responses
4.2. Carbohydrate Metabolism
4.3. Energy Metabolism
4.4. Amino Acid Metabolism
4.5. GSH and AsA Metabolism
4.6. Secondary Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PEG | Polyethylene glycol |
| MDA | Malondialdehyde |
| O2- | Superoxide anion |
| H2O2 | Hydrogen peroxide |
| SOD | Superoxide dismutase |
| ASA | Ascorbate |
| GSH | Glutathione |
| APX | Ascorbic peroxidase |
| ROS | Reactive oxygen species |
| GO | Gene ontology |
| DAPs | Differentially accumulated proteins |
| PUMP1 | Mitochondrial uncoupling protein 1 |
| LHCB7 | Chlorophyll a-b binding protein 7 |
| PSAE-1 | Photosystem I reaction center subunit IV |
| PSAH | Photosystem I reaction center subunit VI |
| GAPC | Glyceraldehyde-3-phosphate dehydrogenase |
| SS | Starch synthase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GAD | Glutamate decarboxylase |
| ASP3 | Aspartate aminotransferase 3 |
| OAT | Ornithine aminotransferase |
| NOA1 | NO-associated protein 1 |
| SPMS | Spermine synthase |
| PRM | Parallel reaction monitoring |
| G6PD6 | Glucose-6-phosphate 1-dehydrogenase 6 |
| At5g05600 | Probable 2-oxoglutarate-dependent dioxygenase |
| GST | Glutathione S-transferase |
| LGALDH | L-galactose dehydrogenase |
| VET4 | Probable tocopherol O-methyltransferase |
| CCD | Probable carotenoid cleavage dioxygenase |
| RT | Anthocyanidin-3-O-glucoside rhamnosyltransferase |
| COMT1 | Caffeic acid 3-O-methyltransferase |
| HHT1 | Omega-hydroxypalmitate O-feruloyl transferase |
References
- Suleymanov, S.Y.; Aliyeva, D.R.; Mammadov, A.C.; Aliyev, J.A. Drought-induced changes in photosynthetic apparatus and antioxidant components of wheat (Triticum durum Desf.) varieties. Photosynth. Res. 2016, 130, 215–223. [Google Scholar]
- Anwar, A.; She, M.; Wang, K.; Riaz, B.; Ye, X. Biological roles of ornithine aminotransferase (OAT) in plant stress tolerance: Present progress and future perspectives. Int. J. Mol. Sci. 2018, 19, 3681. [Google Scholar] [CrossRef]
- Thirunavukkarasu, N.; Sharma, R.; Singh, N.; Shiriga, K.; Mohan, S.; Mittal, S.; Mittal, S.; Mallikarjuna, M.G.; Rao, A.R.; Dash, P.K.; et al. Genomewide expression and functional interactions of genes under drought stress in maize. Int. J. Genom. 2017, 1–14. [Google Scholar] [CrossRef]
- Claeys, H.; Inzé, D. The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol. 2013, 162, 1768–1779. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, P.; Shao, J.; Li, C.; Wang, B.; Guo, X.; Yan, B.; Xia, Y.; Peng, M. Analysis of different strategies adapted by two cassava cultivars in response to drought stress: Ensuring survival or continuing growth. J. Exp. Bot. 2015, 66, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shi, S. Physiological and proteomic responses of contrasting Alfalfa (Medicago sativa L.) varieties to PEG-Induced osmotic stress. Front. Plant Sci. 2018, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Gupta, S.K.; Majumder, B.; Maurya, V.K.; Deeba, F.; Alam, A.; Pandey, V. Salicylic acid mediated growth, physiological and proteomic responses in two wheat varieties under drought stress. J. Proteom. 2017, 163, 28–51. [Google Scholar] [CrossRef]
- Ma, X.; Wang, P.; Zhou, S.; Sun, Y.; Liu, N.; Li, X.; Hou, Y. De novo transcriptome sequencing and comprehensive analysis of the drought-responsive genes in the desert plant Cynanchum komarovii. BMC Genom. 2015, 16, 753. [Google Scholar] [CrossRef]
- Carmo, L.S.T.; Martins, A.C.Q.; Martins, C.C.C.; Passos, M.A.S.; Silva, L.P.; Araujo, A.C.G.; Brasileiro, A.C.M.; Miller, R.N.G.; Guimarães, P.M.; Mehta, A. Comparative proteomics and gene expression analysis in Arachis duranensis reveal stress response proteins associated to drought tolerance. J. Proteom. 2019, 192, 299–310. [Google Scholar] [CrossRef]
- Gharechahi, J.; Hajirezaei, M.R.; Salekdeh, G.H. Comparative proteomic analysis of tobacco expressing cyanobacterial flavodoxin and its wild type under drought stress. J. Plant Physiol. 2015, 175, 48–58. [Google Scholar] [CrossRef]
- Wang, X.; Oh, M.; Sakata, K.; Komatsu, S. Gel-free/label-free proteomic analysis of root tip of soybean over time under flooding and drought stresses. J. Proteom. 2016, 13, 42–55. [Google Scholar] [CrossRef]
- Lalit, A.; Swati, G.; Mishra, S.K.; Garima, P.; Susheel, K.; Chauhan, P.S.; Chakrabarty, D.; Nautiyal, C.S. Elucidation of complex nature of PEG induced drought-stress response in rice root using comparative proteomics approach. Front. Plant Sci. 2016, 7, 1466. [Google Scholar]
- Li, N.; Zhang, S.; Liang, Y.; Qi, Y.; Chen, J.; Zhu, W.; Zhang, L. Label-free quantitative proteomic analysis of drought stress-responsive late embryogenesis abundant proteins in the seedling leaves of two wheat (Triticum aestivum L.) genotypes. J. Proteom. 2018, 172, 122–142. [Google Scholar] [CrossRef]
- Dong, H.; Li, Y.; Fan, H.; Zhou, D.; Li, H. Quantitative proteomics analysis reveals resistance differences of banana cultivar ‘Brazilian’ to Fusarium oxysporum f. sp. cubense races 1 and 4. J. Proteom. 2019, 203, 103376. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, S.S.; Abdi, F.; Abdollahi, M.R.; Tahmasebi-Enferadi, S.; Maleki, M. Phenological, morpho-physiological and proteomic responses of Triticum boeoticum to drought stress. Plant Physiol. Biochem. 2020, 156, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, J.; Wang, F.; Wang, L.; Xu, Z. Morpho-physiological and proteomic responses to water stress in two contrasting tobacco varieties. Sci. Rep. 2019, 9, 18523. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, T.A.; Takahashi, L.S. Physical and chemical characteristics of pitaya fruits at physiological maturity. Genet. Mol. Res. 2015, 14, 14422–14439. [Google Scholar] [CrossRef] [PubMed]
- Hua, Q.; Chen, C.; Tel Zur, N.; Wang, H.; Wu, J.; Chen, J.; Zhang, Z.; Zhao, J.; Hu, G.; Qin, Y. Metabolomic characterization of pitaya fruit from three red-skinned cultivars with different pulp colors. Plant Physiol. Biochem. 2018, 126, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Nie, Q.; Qiao, G.; Peng, L.; Wen, X. Transcriptional activation of long terminal repeat retrotransposon sequences in the genome of pitaya under abiotic stress. Plant Physiol. Bioechem. 2019, 135, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.J.; Yan, F.X.; Qiao, G.; Zhang, B.X.; Wen, X.P. Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene 2014, 533, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.; Arnon, D.I. The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347, 32. [Google Scholar]
- Buysse, J.; Merckx, R. An improved colorimetric method to quantify sugar content of plant tissue. J. Exp. Bot. 1993, 44, 1627–1629. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil. 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Castrejón, S.E.; Yatsimirsky, A.K. Cyclodextrin enhanced fluorimetric determination of malonaldehyde by the thiobarbituric acid method. Talanta 1997, 44, 951–957. [Google Scholar] [CrossRef]
- Lin, Y.F.; Lin, Y.X.; Lin, H.T.; Zhang, S.; Chen, Y.H.; Shi, J. Inhibitory effects of propyl gallate on browning and its relationship to active oxygen metabolism in pericarp of harvested longan fruit. LWT Food Sci. Technol. 2015, 60, 1122–1128. [Google Scholar] [CrossRef]
- Sim, Y.H.; Yao, J.M.; Hou, Y.S.; Wang, L.; Zhao, L.C. Variations of hydrogen peroxide and catalase expression in Bombyx eggs during diapause initiation and termination. Arch. Insect Biochem. Physiol. 2011, 77, 72–80. [Google Scholar] [CrossRef] [PubMed]
- García-Triana, A.; Zenteno-Savín, T.; Peregrino-Uriarte, A.B.; Yepiz-Plascencia, G. Hypoxia, reoxygenation and cytosolic manganese superoxide dismutase (cMnSOD) silencing in Litopenaeus vannamei: Effects on cMnSOD transcripts, superoxide dismutase activity and superoxide anion production capacity. Dev. Comp. Immunol. 2010, 34, 1230–1235. [Google Scholar] [CrossRef]
- Foster, J.G.; Hess, J.L. Responses of superoxide dismutase and glutathione reductase activities in cotton leaf tissue exposed to an atmosphere enriched in oxygen. Plant Physiol. 1980, 66, 482–487. [Google Scholar] [CrossRef]
- Ullah, S.; Kolo, Z.; Egbichi, I.; Keyster, M.; Ludidi, N. Nitric oxide influences glycine betaine content and ascorbate peroxidase activity in maize. S. Afr. J. Bot. 2016, 105, 218–225. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Huang, X.; Liu, L.W.; Wang, P.Y.; Long, Q.S.; Tao, Q.Q.; Li, Z.; Yang, S. Identification of racemic and chiral carbazole derivatives containing an isopropanolamine linker as prospective surrogates against plant pathogenic bacteria: In vitro and in vivo assays and quantitative proteomics. J. Agric. Food Chem. 2019, 67, 7512–7525. [Google Scholar] [CrossRef]
- Cui, G.; Zhao, Y.; Zhang, J.; Chao, M.; Xie, K.; Zhang, C.; Sun, F.; Liu, S.; Xi, Y. Proteomic analysis of the similarities and differences of soil drought and polyethylene glycol stress responses in wheat (Triticum aestivum L.). Plant Mol. Biol. 2019, 100, 391–410. [Google Scholar] [CrossRef]
- Wang, A.H.; Yang, L.; Yao, X.Z.; Wen, X.P. Overexpression of the pitaya phosphoethanolamine N -methyltransferase gene (HpPEAMT) enhanced simulated drought stress in tobacco. Plant Cell Tissue Organ Cult. 2021, 146, 29–40. [Google Scholar] [CrossRef]
- You, J.; Hu, H.; Xiong, L. An ornithine δ-aminotransferase gene OsOAT confers drought and oxidative stress tolerance in rice. Plant Sci. 2012, 197, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Adamipour, N.; Khosh-Khui, M.; Salehi, H.; Razi, H.; Karami, A.; Moghadam, A. Metabolic and genes expression analyses involved in proline metabolism of two rose species under drought stress. Plant Physiol. Biochem. 2020, 155, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Faize, M.; Burgos, L.; Faize, L.; Piqueras, A.; Nicolas, E.; Barba-Espin, G.; Clemente-Moreno, M.J.; Alcobendas, R.; Artlip, T.; Hernandez, J.A. Involvement of cytosolic ascorbate peroxidase and Cu/Zn-superoxide dismutase for improved tolerance against drought stress. J. Exp. Bot. 2011, 62, 2599–2613. [Google Scholar] [CrossRef] [PubMed]
- Ansari, W.A.; Atri, N.; Ahmad, J.; Qureshi, M.I.; Singh, B.; Kumar, R.; Rai, V.; Pandey, S. Drought mediated physiological and molecular changes in muskmelon (Cucumis melo L.). PLoS ONE 2019, 14, e0222647. [Google Scholar] [CrossRef]
- Cao, Y.; Luo, Q.; Tian, Y.; Meng, F. Physiological and proteomic analyses of the drought stress response in Amygdalus Mira (Koehne) Yü et Lu roots. BMC Plant Biol. 2017, 17, 53. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation ofexcess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Iqbal, N.; Hussain, S.; Raza, M.A.; Yang, C.Q.; Safdar, M.E.; Brestic, M.; Aziz, A.; Hayyat, M.S.; Asghar, M.A.; Wang, X.C.; et al. Drought tolerance of soybean (Glycine max L. Merr.) by improved photosynthetic characteristics and an efficient antioxidant enzyme activities under a split-root system. Front. Physiol. 2019, 10, 786. [Google Scholar] [CrossRef]
- Hao, P.; Zhu, J.; Gu, A.; Lv, D.; Ge, P.; Chen, G.; Li, X.; Yan, Y. An integrative proteome analysis of different seedling organs in tolerant and sensitive wheat cultivars under drought stress and recovery. Proteomics 2015, 15, 1544–1563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, H.; Yang, S. Cytosolic TaGAPC2 enhances tolerance to drought stress in transgenic arabidopsis plants. Int. J. Mol. Sci. 2020, 21, 7499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Z.; Ji, H.; Zhou, Y.; Yang, S. TaWRKY40 transcription factor positively regulate the expression of TaGAPC1 to enhance drought tolerance. BMC Genom. 2019, 20, 795. [Google Scholar] [CrossRef]
- Huang, W.; Ma, X.; Wang, Q.; Gao, Y.; Xue, Y.; Niu, X.; Yu, G.; Liu, Y. Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant Mol. Biol. 2008, 68, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Kirch, H.H.; Schlingensiepen, S.; Kotchoni, S.; Sunkar, R.; Bartels, D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.J.; Senning, M.; Fischer-Stettler, M.; Streb, S.; Ast, M.; Neuhaus, H.E.; Zeeman, S.C.; Sonnewald, S.; Sonnewald, U. Simultaneous silencing of isoamylases ISA1, ISA2 and ISA3 by multi-target RNAi in potato tubers leads to decreased starch content and an early sprouting phenotype. PLoS ONE 2017, 12, e0181444. [Google Scholar] [CrossRef]
- Steichen, J.M.; Petty, R.V.; Sharke, T.D. Domain characterization of a 4-alpha-glucanotransferase essential for maltose metabolism in photosynthetic leaves. J. Biol. Chem. 2008, 283, 20797–20804. [Google Scholar] [CrossRef]
- Malinova, I.; Alseekh, S.; Feil, R.; Fernie, A.R.; Baumann, O.; Schöttler, M.A.; Lunn, J.E.; Fettke, J. Starch synthase 4 and plastidal phosphorylase differentially affect starch granule number and morphology. Plant Physiol. 2017, 174, 73–85. [Google Scholar] [CrossRef]
- Li, C.Y.; Weiss, D.; Goldschmidt, E.E. Effects of carbohydrate starvation on gene expression in citrus root. Planta 2003, 217, 11–20. [Google Scholar] [CrossRef]
- Griffiths, H.; Parry, M.A.J. Plant responses to water stress. Ann. Bot. 2002, 89, 801–802. [Google Scholar] [CrossRef]
- Szydlowski, N.; Ragel, P.; Hennen-Bierwagen, T.A.; Planchot, V.; Myers, A.M.; Mérida, A.; d’Hulst, C.; Wattebled, F. Integrated functions among multiple starch synthases determine both amylopectin chain length and branch linkage location in arabidopsis leaf starch. J. Exp. Bot. 2011, 62, 4547–4559. [Google Scholar] [CrossRef]
- Sweetman, C.S.; Waterman, C.D.; Rainbird, B.M.; Smith, P.M.C.; Jenkins, C.D.; Day, D.A.; Soole, K.L. AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress. Plant Physiol. 2019, 181, 774–788. [Google Scholar] [CrossRef]
- Sun-Wada, G.H.; Wada, Y. Role of vacuolar-type proton ATPase in signal transduction. Biochim. Biophys. Acta 2015, 1847, 1166–1172. [Google Scholar] [CrossRef]
- Barreto, P.; Okura, V.K.; Neshich, I.A.; Maia Ide, G.; Arruda, P. Overexpression of UCP1 in tobacco induces mitochondrial biogenesis and amplifies a broad stress response. BMC Plant Biol. 2014, 14, 144. [Google Scholar] [CrossRef]
- Barreto, P.; Okura, V.; Pena, I.A.; Maia, R.; Maia, I.G.; Arruda, P. Overexpression of mitochondrial uncoupling protein 1 (UCP1) induces a hypoxic response in Nicotiana tabacum leaves. J. Exp. Bot. 2016, 67, 301–313. [Google Scholar] [CrossRef]
- Wen, Z.; Zhang, M. Possible involvement of phosphoenolpyruvate carboxylase and NAD-malic enzyme in response to drought stress. A case study: A succulent nature of the C4-NAD-ME type desert plant, Salsola lanata (Chenopodiaceae). Funct. Plant Biol. 2017, 44, 1219–1228. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, B.; Ding, H.; Zhang, J.; Li, S. Review: The role of NADP-malic enzyme in plants under stress. Plant Sci. 2019, 281, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Kim, Y.J.; Sukweenadhi, J.; Rahimi, S.; Kwon, W.S.; Yang, D.C. Molecular characterization of 5 chlorophyll a/b-binding protein genes from panax ginseng meyer and their expression analysis during abiotic stresses. Photosynthetica 2016, 54, 446–458. [Google Scholar] [CrossRef]
- Hao, X.; Li, J.; Gao, S.; Tuerxun, Z.; Chang, X.; Hu, W.; Chen, G.; Huang, Q. SsPsaH, a H subunit of the photosystem I reaction center of Suaeda salsa, confers the capacity of osmotic adjustment in tobacco. Genes Genom. 2020, 42, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Haussühl, K.; Andersson, B.; Adamska, I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 2011, 20, 713–722. [Google Scholar]
- James, D.; Borphukan, B.; Fartyal, D.; Ram, B.; Singh, J.; Manna, M.; Sheri, V.; Panditi, V.; Yadav, R.; Achary, V.M.M.; et al. Concurrent overexpression of OsGS1;1 and OsGS2 genes in transgenic Rice (Oryza sativa L.): Impact on tolerance to abiotic stresses. Front. Plant Sci. 2018, 21, 786. [Google Scholar] [CrossRef]
- Cevik, S.; Akpinar, G.; Yildizli, A.; Kasap, M.; Karaosmanoglu, K.; Unyayar, S. Comparative physiological and leaf proteome analysis between drought-tolerant chickpea Cicer reticulatum and drought-sensitive chickpea C. arietinum. J. Biosci. 2019, 44, 20. [Google Scholar] [CrossRef]
- Cai, H.; Zhou, Y.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 2009, 28, 527–537. [Google Scholar] [CrossRef]
- Díaz, P.; Betti, M.; Sánchez, D.H.; Udvardi, M.K.; Monza, J.; Márquez, A.J. Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 2010, 188, 1001–1013. [Google Scholar] [CrossRef]
- Hyun, T.K.; Eom, S.H.; Han, X.; Kim, J.S. Evolution and expression analysis of the soybean glutamate decarboxylase gene family. J. Biosci. 2014, 39, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Chen, X.; Lv, S.; Jiang, P.; Feng, J.; Fan, P.; Nie, L.; Li, Y. Virus-induced gene silencing reveals control of reactive oxygen species accumulation and salt tolerance in tomato by γ-aminobutyric acid metabolic pathway. Plant Cell Environ. 2015, 38, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Xu, Y.; Zhang, X.; Peng, Y.; Ma, X.; Huang, L.; Yan, Y. The physiological and iTRAQ-based proteomic analyses reveal the function of spermidine on improving drought tolerance in white clover. J. Proteome Res. 2016, 15, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, Y.; Meng, Q.; Shi, F.; Liu, J.; Li, Y. Molecular cloning, identification of GSTs family in sunflower and their regulatory roles in biotic and abiotic stress. World J. Microbiol. Biotechnol. 2018, 34, 109. [Google Scholar] [CrossRef]
- Wang, X.; Ruan, M.; Wan, Q.; He, W.; Yang, L.; Liu, X.; He, L.; Yan, L.; Bi, Y. Nitric oxide and hydrogen peroxide increase glucose-6-phosphate dehydrogenase activities and expression upon drought stress in soybean roots. Plant Cell Rep. 2020, 39, 63–73. [Google Scholar] [CrossRef]
- Vítámvás, P.; Urban, M.O.; Škodáček, Z.; Kosová, K.; Pitelková, I.; Vítámvás, J.; Renaut, J.; Prášil, I.T. Quantitative analysis of proteome extracted from barley crowns grown under different drought conditions. Front. Plant Sci. 2015, 6, 479. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.; Sterky, F.; Amini, B.; Lundeberg, J.; Kleczkowski, L.A. Molecular cloning and characterization of a cDNA encoding poplar UDP-glucose dehydrogenase, a key gene of hemicellulose/pectin formation. Biochim. Biophys. Acta 2021, 1576, 53–58. [Google Scholar] [CrossRef]
- Gupta, S.; Mishra, S.K.; Misra, S.; Pandey, V.; Agrawal, L.; Nautiyal, C.S.; Chauhan, P.S. Revealing the complexity of protein abundance in chickpea root under drought-stress using a comparative proteomics approach. Plant Physiol. Biochem. 2020, 151, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.H.; Qi, B.X.; Wang, X.Q.; Shen, L.Y.; Luo, J.; Zhang, Y.X. Proteomic analysis of the key mechanism of exocarp russet pigmentation of semi-russet pear under rainwater condition. Sci. Hortic. 2019, 254, 178–186. [Google Scholar] [CrossRef]
- Li, W.; Lu, J.; Lu, K.; Yuan, J.; Huang, J.; Du, H.; Li, J. Cloning and phylogenetic analysis of brassica napus L. Caffeic acid O-Methyltransferase 1 gene family and its expression pattern under drought stress. PLoS ONE 2016, 11, e0165975. [Google Scholar] [CrossRef]
- Mato, M.; Ozeki, Y.; Itoh, Y.; Higeta, D.; Yoshitama, K.; Teramoto, S.; Aida, R.; Ishikura, N.; Shibata, M. Isolation and characterization of a cDNA clone of UDP-galactose: Flavonoid 3-O-galactosyltransferase (UF3GaT) expressed in Vigna mungo seedlings. Plant Cell Physiol. 1998, 39, 1145–1155. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rubio-Moraga, A.; Rambla, J.L.; Fernández-de-Carmen, A.; Trapero-Mozos, A.; Ahrazem, O.; Orzáez, D.; Granell, A.; Gómez-Gómez, L. New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol. Biol. 2014, 86, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Guo, J.; Zhang, W.; Jin, L.; Liu, P.; Chen, X.; Li, F.; Wei, P.; Li, Z.; Li, W.; et al. Cloning of the Lycopene β-cyclase Gene in Nicotiana tabacum and its overexpression confers salt and drought tolerance. Int. J. Mol. Sci. 2015, 16, 30438–30457. [Google Scholar] [CrossRef]
- Yang, M.; Fan, Z.; Xie, Y.; Fang, L.; Wang, X.; Yuan, Y.; Li, R. Transcriptome analysis of the effect of bisphenol A exposure on the growth, photosynthetic activity and risk of microcystin-LR release by Microcystis aeruginosa. J. Hazard. Mater. 2020, 397, 122746. [Google Scholar] [CrossRef]

| Protein ID | Description | Protein Abbreviation | OS6H/NS6H Ratio | OS6H/NS6H p-Value | OS3D/NS3D Ratio | OS3D/NS3D p-Value |
|---|---|---|---|---|---|---|
| Glycolysis | ||||||
| TRINITY_DN1285_c0_g1_m.2918 | Pyrophosphate—fructose 6-phosphate 1-phosphotransferase subunit beta | PFP-BETA | 1.2232 | 0.028736 | ns | 0.491619 |
| TRINITY_DN1345_c0_g1_m.3326 | Enolase | PGH1 | 1.2113 | 0.023062 | ns | 0.176205 |
| TRINITY_DN1541_c1_g1_m.4614 | Triosephosphate isomerase | -- | 1.2205 | 0.048792 | ns | 0.190237 |
| TRINITY_DN22699_c0_g1_m.8480 | Plastidial pyruvate kinase 2 | PKP2 | 0.7949 | 0.012965 | ns | 0.071468 |
| TRINITY_DN680_c2_g1_m.20738 | Fructose-1,6-bisphosphatase | -- | 0.6861 | 0.001059 | ns | 0.068059 |
| TRINITY_DN700_c0_g1_m.21125 | Phosphoglucomutase | PGMP | 1.2058 | 0.003547 | ns | 0.09038 |
| TRINITY_DN8402_c0_g1_m.23765 | Aldehyde dehydrogenase family 3 member F1 | ALDH3F1 | 1.4108 | 0.034248 | 1.4821 | 0.004249 |
| TRINITY_DN956_c0_g1_m.25499 | Glyceraldehyde-3-phosphate dehydrogenase | GAPC | 1.2105 | 0.00537 | ns | 0.23787 |
| TRINITY_DN956_c0_g1_m.25500 | Glyceraldehyde-3-phosphate dehydrogenase | GAPC | 1.2849 | 0.006127 | ns | 0.272781 |
| TRINITY_DN1378_c0_g1_m.3547 | ATP-dependent 6-phosphofructokinase 6 | PFK6 | ns | 0.052857 | 2.8748 | 0.008707 |
| TRINITY_DN188_c0_g2_m.6607 | Aldehyde dehydrogenase family 3 member F1 | ALDH3F1 | ns | 0.102474 | 0.7175 | 0.042614 |
| TRINITY_DN2017_c0_g1_m.7240 | Pyruvate decarboxylase 4 | PDC4 | ns | 0.091026 | 1.7692 | 0.002958 |
| TRINITY_DN2017_c0_g2_m.7243 | Pyruvate decarboxylase 1 | PDC1 | ns | 0.087881 | 1.2044 | 0.012331 |
| TRINITY_DN2514_c0_g1_m.9503 | Aldehyde dehydrogenase family 3 member H1 | ALDH3H1 | ns | 0.724813 | 0.6052 | 0.012778 |
| TRINITY_DN5539_c0_g1_m.18129 | Alcohol dehydrogenase class-3 | -- | ns | 0.526004 | 1.2465 | 0.00285 |
| TRINITY_DN785_c0_g1_m.22777 | Hexokinase-3 | At1g50460 | ns | 0.97966423 | 0.6069 | 0.002177 |
| Starch and sucrose metabolism | ||||||
| TRINITY_DN120_c1_g1_m.2258 | Beta-fructofuranosidase, soluble isoenzyme I | INV*DC4 | 1.2139 | 0.025473 | ns | 0.36858184 |
| TRINITY_DN1281_c0_g1_m.2874 | 4-alpha-glucanotransferase | DPEP | 1.3783 | 0.001518 | ns | 0.065598773 |
| TRINITY_DN2311_c0_g1_m.8701 | Alpha-1,4 glucan phosphorylase L isozyme | -- | 1.2988 | 0.015376 | 1.6666 | 0.033337 |
| TRINITY_DN243_c0_g1_m.9202 | Glucose-1-phosphate adenylyltransferase large subunit 1 | AGPS1 | 1.3654 | 0.002383 | ns | 0.065961158 |
| TRINITY_DN243_c0_g2_m.9203 | Glucose-1-phosphate adenylyltransferase large subunit 1 | AGPS1 | 1.4417 | 0.005241 | ns | 0.082784957 |
| TRINITY_DN3421_c0_g1_m.12709 | Probable starch synthase 4 | SS4 | 0.8081 | 0.000869 | ns | 0.436338494 |
| TRINITY_DN396_c0_g2_m.14182 | Isoamylase 3 | ISA3 | 1.2597 | 0.038832 | 2.2276 | 0.005774 |
| TRINITY_DN558_c0_g2_m.18286 | Glucan endo-1,3-beta-glucosidase 1 | At1g11820 | 0.7347 | 0.009098 | ns | 0.258131706 |
| TRINITY_DN6728_c0_g1_m.20594 | Inactive beta-amylase 9 | BAM9 | 2.8986 | 5.89E-05 | ns | 0.422387234 |
| TRINITY_DN8071_c0_g1_m.23172 | Glucose-1-phosphate adenylyltransferase small subunit | AGPB1 | 1.3814 | 0.013501 | ns | 0.152380603 |
| TRINITY_DN2107_c0_g1_m.7698 | Probable alpha,alpha-trehalose-phosphate synthase | TPS11 | ns | 0.107875285 | 0.6784 | 0.047779 |
| TRINITY_DN409_c0_g3_m.14564 | Alpha-glucosidase | -- | ns | 0.145618967 | 1.3875 | 0.047325 |
| TRINITY_DN483_c0_g2_m.16576 | Alpha-1,4 glucan phosphorylase L isozyme, | -- | ns | 0.241229313 | 1.2924 | 0.04298 |
| TRINITY_DN4852_c0_g1_m.16596 | Alpha-amylase 3 | AMY3 | ns | 0.629154053 | 1.4422 | 0.043718 |
| TRINITY_DN6640_c0_g1_m.20450 | Glucose-1-phosphate adenylyltransferase large subunit | AGPS1 | ns | 0.11514614 | 1.3167 | 0.030977 |
| TRINITY_DN7735_c0_g1_m.22545 | 4-alpha-glucanotransferase | DPEP | ns | 0.15889801 | 1.3818 | 0.01198 |
| TRINITY_DN8896_c0_g1_m.24535 | Starch synthase 1 | SS1 | ns | 0.999352844 | 1.4676 | 0.046376 |
| Citrate cycle (TCA cycle) | ||||||
| TRINITY_DN12082_c0_g1_m.2243 | Citrate synthase | -- | ns | 0.664702 | 1.2021 | 0.020504 |
| TRINITY_DN3909_c0_g2_m.14018 | ATP-citrate synthase alpha chain protein 1 | ACLA-1 | ns | 0.137801 | 1.2304 | 0.002227 |
| Galactose metabolism | ||||||
| TRINITY_DN455_c0_g1_m.15769 | Probable galactinol--sucrose galactosyltransferase 6 | RFS6 | 1.255 | 0.02696 | ns | |
| TRINITY_DN4902_c0_g1_m.16720 | Probable galactinol--sucrose galactosyltransferase 6 | RFS6 | 1.3 | 0.044023 | ns | |
| TRINITY_DN327_c0_g1_m.12252 | Alpha-galactosidase 3 | AGAL3 | ns | 1.4405 | 0.029031 |
| Protein ID | Description | Protein Abbreviation | OS6H/NS6H Ratio | OS6H/NS6H p-Value | OS3D/NS3D Ratio | Os3d/Ns3d p-Value |
|---|---|---|---|---|---|---|
| Oxidative phosphorylation | ||||||
| TRINITY_DN130_c0_g2_m.3091 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 9 | CIB22 | 1.215 | 0.006 | ns | 0.481 |
| TRINITY_DN284_c10_g1_m.10791 | V-type proton ATPase subunit G 1 | VATG1 | 2.006 | 0.022 | ns | 0.355 |
| TRINITY_DN5440_c0_g1_m.17954 | External alternative NAD(P)H-ubiquinone oxidoreductase B2 | NDB2 | 1.295 | 0.024 | ns | 0.406 |
| TRINITY_DN7109_c0_g2_m.21284 | Mitochondrial uncoupling protein 1 | PUMP1 | 1.285 | 0.006 | 1.2331 | 0.016 |
| TRINITY_DN1470_c0_g1_m.4137 | V-type proton ATPase subunit a3 | VHA-a3 | ns | 0.059 | 1.592 | 0.034 |
| TRINITY_DN30311_c0_g1_m.11465 | V-type proton ATPase subunit E | VATE | ns | 0.039 | 1.224 | 0.025 |
| TRINITY_DN5440_c0_g1_m.17953 | External alternative NAD(P)H-ubiquinone oxidoreductase B2 | NDB2 | ns | 0.084 | 1.202 | 0.014 |
| TRINITY_DN6767_c0_g2_m.20656 | Acyl carrier protein 2 | MTACP2 | 1.264 | 0.003 | ns | 0.131 |
| TRINITY_DN10196_c0_g1_m.265 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 | At3g12260 | ns | 0.293 | 0.826 | 0.014 |
| TRINITY_DN517_c0_g3_m.17409 | Plasma membrane ATPase 4 | PMA4 | ns | 0.15 | 0.726 | 0.022 |
| Carbon fixation in photosynthetic organisms | ||||||
| TRINITY_DN1003_c0_g1_m.71 | NAD-dependent malic enzyme 62 kDa isoform | -- | 1.2388 | 0.005 | ns | 0.196015 |
| TRINITY_DN1003_c0_g1_m.72 | NAD-dependent malic enzyme 62 kDa isoform | -- | 1.2197 | 0.004638 | 1.2602 | 0.018536 |
| TRINITY_DN1199_c0_g1_m.2179 | NAD-dependent malic enzyme 59 kDa isoform | -- | 1.2055 | 0.012349 | ns | 0.187842 |
| TRINITY_DN21851_c0_g1_m.8123 | NADP-dependent malic enzyme | -- | 1.4738 | 0.028002 | ns | 0.184188 |
| TRINITY_DN54_c0_g1_m.18065 | Phosphoenolpyruvate carboxylase | PPC16 | 1.3809 | 0.006254 | 2.0725 | 0.015201 |
| TRINITY_DN5722_c0_g1_m.18543 | Phosphoenolpyruvate carboxylase 1 | PPC1 | 1.5942 | 0.029038 | 2.5273 | 0.005758 |
| TRINITY_DN5722_c0_g1_m.18545 | Phosphoenolpyruvate carboxylase 1 | PPC1 | 1.7533 | 0.024815 | 1.471 | 0.019766 |
| Photosynthesis | ||||||
| TRINITY_DN3928_c2_g1_m.14056 | Chlorophyll a-b binding protein 7 | LHCB7 | 1.311 | 0.048 | ns | 0.216 |
| TRINITY_DN2207_c0_g1_m.8225 | Photosystem I reaction center subunit IV | PSAE-1 | 1.24 | 0.003 | ns | 0.244 |
| TRINITY_DN4734_c0_g1_m.16295 | Photosystem I reaction center subunit VI | PSAH | 1.422 | 0.048 | ns | 0.091 |
| TRINITY_DN2097_c1_g1_m.7621 | Protease Do-like 2 | DEGP2 | 0.746 | 0.007 | ns | 0.39 |
| Protein ID | Description | Protein Abbreviation | OS6H/NS6H Ratio | OS6H/NS6H p-Value | OS3D/NS3D Ratio | OS3D/NS3D p-Value |
|---|---|---|---|---|---|---|
| Aspartate and glutamate metabolism | ||||||
| TRINITY_DN1488_c1_g1_m.4241 | Glutamine synthetase | GLN2 | 0.7957 | 0.026414 | ns | 0.97597626 |
| TRINITY_DN1404_c3_g1_m.3689 | Glutamate decarboxylase | GAD | ns | 0.0013574 | 1.3966 | 0.027208 |
| TRINITY_DN294_c0_g1_m.11144 | Gamma aminobutyrate transaminase 3 | GABA-TP3 | ns | 0.683342268 | 1.2024 | 0.010061 |
| TRINITY_DN7757_c0_g1_m.22593 | Aspartate aminotransferase 3 | ASP3 | ns | 0.141381977 | 1.272 | 0.00091 |
| TRINITY_DN1488_c1_g1_m.4239 | Glutamine synthetase | GLN2 | ns | 0.064775788 | 0.8262 | 0.037899 |
| TRINITY_DN15906_c0_g1_m.4931 | Glutamine synthetase nodule isozyme | -- | ns | -- | 0.7607 | 0.027319 |
| Arginine and proline metabolism | ||||||
| TRINITY_DN1617_c1_g1_m.5135 | Ornithine aminotransferase | DELTA-OAT | 1.2603 | 0.008374713 | ns | 0.258228322 |
| TRINITY_DN809_c0_g2_m.23215 | Proline-rich receptor-like protein kinase | PERK1 | ns | 0.160140522 | 0.7234 | 0.001426311 |
| TRINITY_DN6277_c0_g2_m.19711 | NO-associated protein 1 | NOA1 | 0.8235 | 0.019199161 | ns | 0.055348656 |
| TRINITY_DN7062_c0_g1_m.21204 | Spermine synthase | SPMS | 0.7974 | 0.014501 | ns | 0.35623713 |
| Phenylalanine metabolism | ||||||
| TRINITY_DN10229_c1_g1_m.315 | Hydroxyphenylpyruvate reductase | HPPR | ns | 0.735219447 | 1.2086 | 0.003324 |
| TRINITY_DN3220_c0_g1_m.12107 | Probable enoyl-CoA hydratase 1 | ECHIA | 0.8204 | 0.031414137 | ns | 0.293016083 |
| TRINITY_DN185_c0_g1_m.6424 | Histidinol-phosphate aminotransferase | HPA | ns | 0.024259888 | 0.8195 | 0.015681 |
| TRINITY_DN2397_c0_g1_m.9007 | Arogenate dehydratase/prephenate dehydratase 6 | ADT6 | ns | 0.081337319 | 0.7012 | 0.012254 |
| Protein ID | Description | Protein Abbreviation | OS6H/NS6H Ratio | OS6H/NS6H p-Value | OS3D/NS3D Ratio | OS3D/NS3D p-Value |
|---|---|---|---|---|---|---|
| Ascorbate metabolism | ||||||
| TRINITY_DN1613_c0_g1_m.5115 | UDP-glucose 6-dehydrogenase 4 | UGD4 | 0.6064 | 0.006265 | ns | 0.151395367 |
| TRINITY_DN1613_c0_g2_m.5117 | UDP-glucose 6-dehydrogenase 5 | UGD5 | 0.8276 | 0.020249 | ns | 0.671819139 |
| TRINITY_DN1595_c0_g1_m.4962 | L-galactose dehydrogenase | LGALDH | ns | 0.701474521 | 1.291 | 0.036156 |
| TRINITY_DN3248_c0_g1_m.12163 | Probable 2-oxoglutarate-dependent dioxygenase | At5g05600 | 1.8511 | 0.006704 | 4.1204 | 0.0000578 |
| TRINITY_DN893_c0_g1_m.24609 | L-ascorbate peroxidase T | APXT | ns | 0.841331205 | 1.2801 | 0.0015628 |
| TRINITY_DN893_c0_g1_m.24610 | L-ascorbate peroxidase T | APXT | 1.2609 | 0.039256535 | ns | 0.056852039 |
| TRINITY_DN427_c0_g2_m.15091 | L-ascorbate oxidase homolog | Bp10 | ns | 0.562150148 | 1.2159 | 0.013967466 |
| Glutathione metabolism | ||||||
| TRINITY_DN4975_c0_g1_m.16910 | Glutathione S-transferase T1 | GSTT1 | ns | 0.117978 | 1.6817 | 0.0466 |
| TRINITY_DN178_c0_g1_m.6079 | Protein IN2-1 homolog B | GSTZ5 | ns | 0.262995961 | 1.3097 | 0.011018 |
| TRINITY_DN2046_c0_g1_m.7392 | Glucose-6-phosphate 1-dehydrogenase 6 | G6PD6 | ns | 0.019788019 | 1.2133 | 0.009388 |
| TRINITY_DN2046_c0_g1_m.7393 | Glucose-6-phosphate 1-dehydrogenase 6 | G6PD6 | ns | 0.321337379 | 1.255 | 0.04952 |
| Protein ID | Description | Protein Abbreviation | OS6H/NS6H Ratio | OS6H/NS6H p-Value | OS3D/NS3D Ratio | OS3D/NS3D p-Value |
|---|---|---|---|---|---|---|
| Tocopherol | ||||||
| TRINITY_DN1553_c0_g1_m.4682 | Probable tocopherol O-methyltransferase | VET4 | 1.82 | 0.014 | 2.725 | 0.014 |
| Flavone and flavonol biosynthesis | ||||||
| TRINITY_DN12687_c0_g1_m.2773 | Anthocyanidin-3-O-glucoside rhamnosyltransferase | RT | ns | 0.299 | 0.729 | 0.017 |
| Phenylpropanoid biosynthesis | ||||||
| TRINITY_DN6823_c0_g1_m.20755 | Caffeic acid 3-O-methyltransferase | COMT1 | 1.249 | 0.049 | 2.209 | 0.014 |
| TRINITY_DN10780_c0_g1_m.946 | Omega-hydroxypalmitate O-feruloyl transferase | HHT1 | ns | 0.748 | 0.374 | 0.000248 |
| TRINITY_DN8170_c0_g2_m.23343 | Probable cinnamyl alcohol dehydrogenase | CAD6 | ns | 0.178 | 0.72 | 0.048 |
| Anthocyanin biosynthesis | ||||||
| TRINITY_DN142_c0_g2_m.3870 | Anthocyanidin 3-O-glucosyltransferase 2 | FGT | ns | 0.522 | 0.792 | 0.049 |
| Carotenoid biosynthesis | ||||||
| TRINITY_DN2792_c0_g2_m.10610 | Probable carotenoid cleavage dioxygenase 4 | CCD4 | ns | 0.264956118 | 1.4016 | 0.038862 |
| TRINITY_DN92199_c0_g1_m.25001 | Lycopene beta cyclase | LCY1 | ns | 0.699025813 | 1.3876 | 0.014785649 |
| Antioxidation system | ||||||
| TRINITY_DN4793_c1_g1_m.16452 | Superoxide dismutase [Cu-Zn] | SODCP | 1.3609 | 0.013743234 | ns | 0.204307412 |
| TRINITY_DN13770_c0_g1_m.3537 | Peroxidase 4 | GSVIVT00023967001 | ns | 0.866 | 3.076 | 0.043 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Ma, C.; Ma, H.; Qiu, Z.; Wen, X. Physiological and Proteomic Responses of Pitaya to PEG-Induced Drought Stress. Agriculture 2021, 11, 632. https://doi.org/10.3390/agriculture11070632
Wang A, Ma C, Ma H, Qiu Z, Wen X. Physiological and Proteomic Responses of Pitaya to PEG-Induced Drought Stress. Agriculture. 2021; 11(7):632. https://doi.org/10.3390/agriculture11070632
Chicago/Turabian StyleWang, Aihua, Chao Ma, Hongye Ma, Zhilang Qiu, and Xiaopeng Wen. 2021. "Physiological and Proteomic Responses of Pitaya to PEG-Induced Drought Stress" Agriculture 11, no. 7: 632. https://doi.org/10.3390/agriculture11070632
APA StyleWang, A., Ma, C., Ma, H., Qiu, Z., & Wen, X. (2021). Physiological and Proteomic Responses of Pitaya to PEG-Induced Drought Stress. Agriculture, 11(7), 632. https://doi.org/10.3390/agriculture11070632






