Epistatic QTLsPlay a Major Role in Nitrogen Use Efficiency and Its Component Traits in Indian Spring Wheat
Abstract
:1. Introduction
2. Materials and Methods
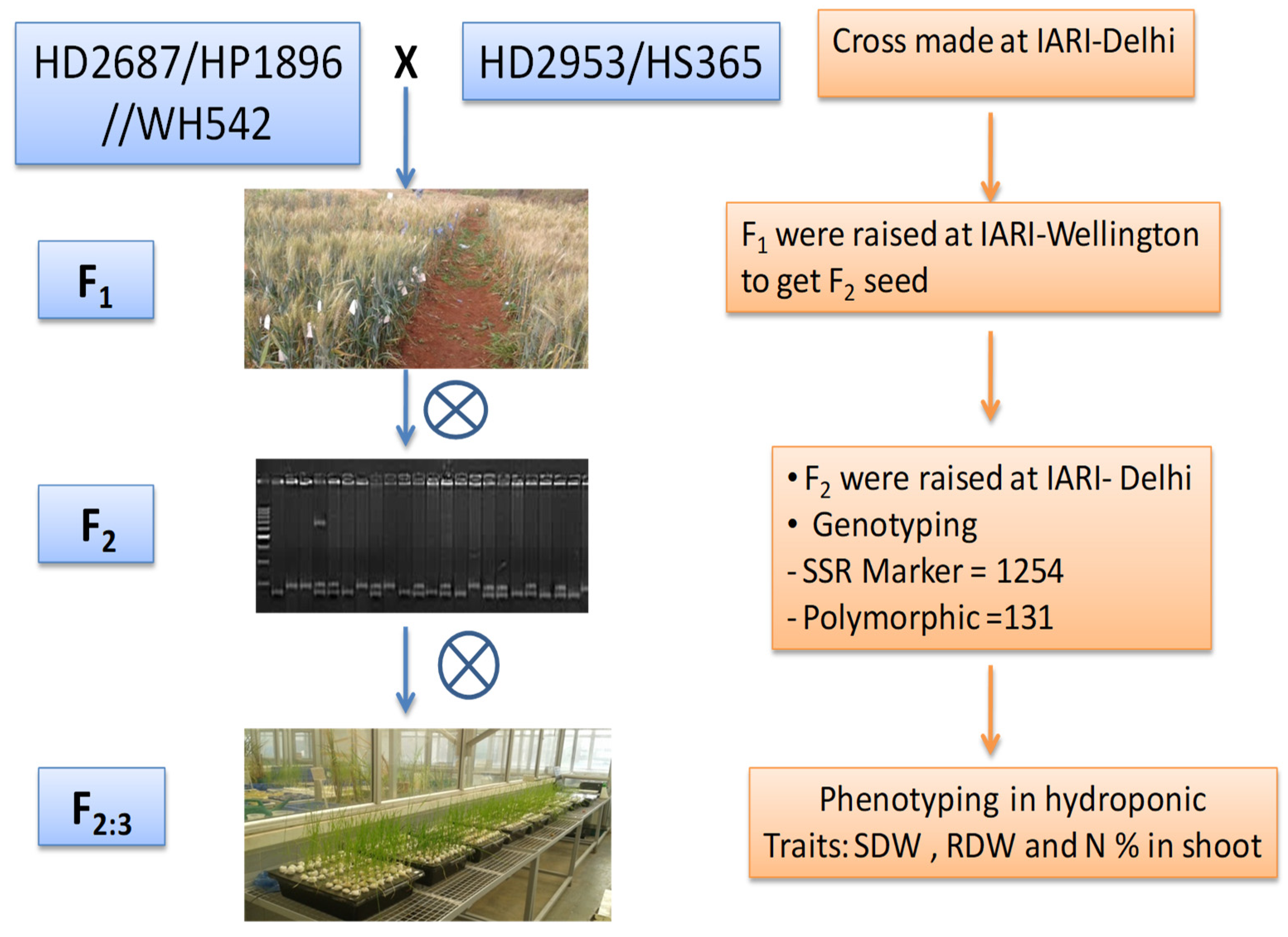
2.1. Plant Material
2.2. Genotyping
2.3. Phenotyping of F2:3 Populations under Hydroponic Condition
2.4. Statistical Analysis
2.5. Detection of Loci Linked with the Quantitative Traits
3. Results
3.1. Phenotypic Characterization of NUE and Related Traits in Two Parents and F2:3 Population
3.2. Construction of Genetic Map and Mapping of QTLs for NUE Related Traits
3.3. Detection of Small-Effect QTLs
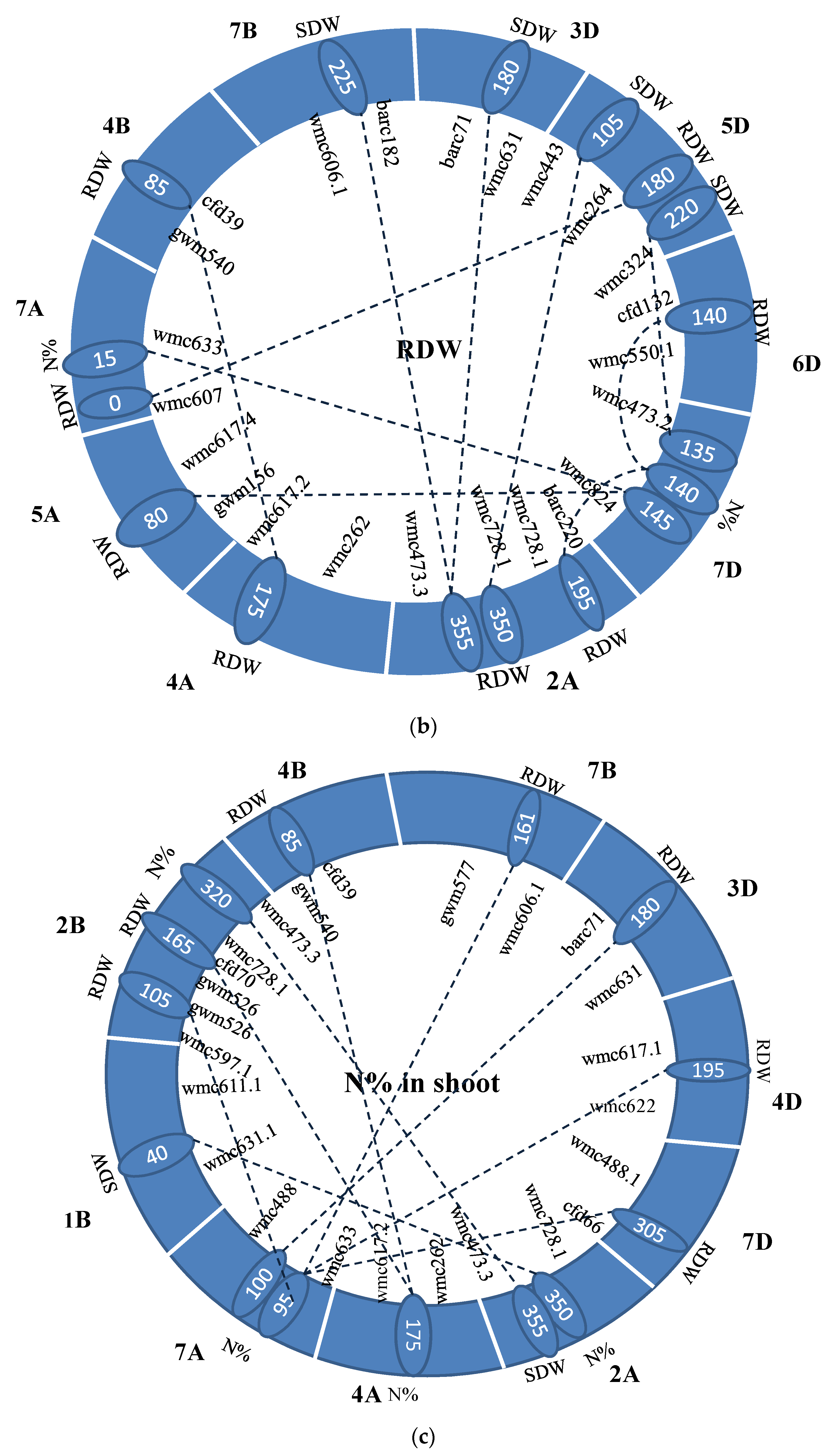
3.4. Identification of Epistasis QTLs
4. Discussion
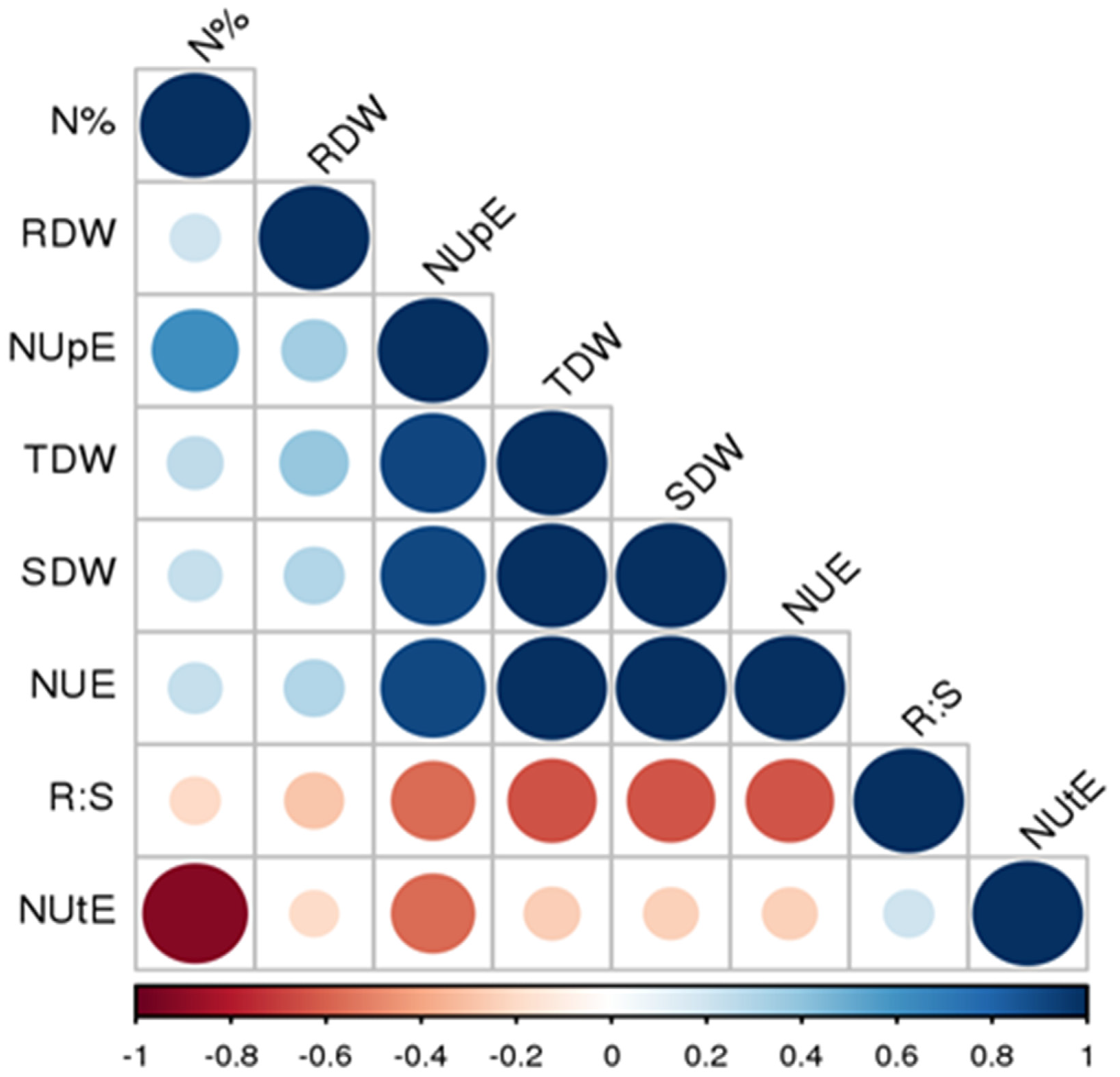
4.1. Phenotypic Performance and Association Analysis among NUE Related Traits
4.2. Identification of QTLs for NUE Related Traits
4.3. Epistasis QTL Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Available online: http://www.fao.org/faostat/en/#data (accessed on 20 July 2020).
- Yadav, R.; Singh, S.S.; Jain, N.; Singh, G.P.; Prabhu, K.V. Wheat production in India: Technologies to face future challenges. J. Agric. Sci. 2010, 2, 164. [Google Scholar] [CrossRef] [Green Version]
- Marschner, P. Rhizosphere Biology. Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Academic Press: Cambridge, MA, USA, 2012; pp. 369–388. [Google Scholar]
- Robertson, G.P.; Vitousek, P.M. Nitrogen in agriculture: Balancing the cost of an essential resource. Annu. Rev. Environ. Resour. 2009, 34, 97–125. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.A.; Zentner, R.P.; Selles, F.; McConkey, B.G.; Dyck, F.B. Nitrogen management for spring wheat grown annually on zero-tillage: Yields and nitrogen use efficiency. Agron. J. 1993, 85, 107–114. [Google Scholar] [CrossRef]
- Matson, P.A.; Naylor, R.; Ortiz-Monasterio, I. Integration of environmental, agronomic, and economic aspects of fertilizer management. Science 1998, 280, 112–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjan, R.; Yadav, R. Targeting nitrogen use efficiency for sustained production of cereal crops. J. Plant Nutr. 2019, 42, 1086–1113. [Google Scholar] [CrossRef]
- Malyan, S.K.; Bhatia, A.; Kumar, A.; Gupta, D.K.; Singh, R.; Kumar, S.S.; Tomer, R.; Kumar, O.; Jain, N. Methane production, oxidation and mitigation: A mechanistic understanding and comprehensive evaluation of influencing factors. Sci. Total Environ. 2016, 572, 874–896. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Gaikwad, K.B.; Bhattacharyya, R. Breeding wheat for yield maximization under conservation agriculture. Indian J. Genet. Plant Breed. 2017, 77, 185–198. [Google Scholar] [CrossRef]
- Cassman, K.G.; Dobermann, A.; Walters, D.T.; Yang, H. Meeting cereal demand while protecting natural resources and improving environmental quality. Annu. Rev. Environ. Resour. 2003, 28, 315–358. [Google Scholar] [CrossRef] [Green Version]
- Ministry of Agriculture and Farmers Welfare, Government of India. 2019. Available online: https://eands.dacnet.nic.in/Advance_Estimate/4th%20Adv%20Estimates%202018-19%20Eng.pdf (accessed on 20 July 2020).
- Tewatia, R.K.; Chanda, T.K. Trends in fertilizer nitrogen production and consumption in India. In The Indian Nitrogen Assessment: Sources of Reactive Nitrogen, Environmental and Climate Effects, and Management Options and Policies; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Sylvester-Bradley, R.; Stokes, D.T.; Scott, R.K.; Willington, V.B.A. A physiological analysis of the diminishing responses of winter wheat to applied nitrogen. Asp. Appl. Biol. 1990, 25, 289–300. [Google Scholar]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and interpretation of factors which contribute to the efficiency of nitrogen utilization. J. Agron. 1982, 74, 562–564. [Google Scholar] [CrossRef]
- Ranjan, R.; Yadav, R.; Kumar, A.; Mandal, S.N. Contributing traits for nitrogen use efficiency in selected wheat genotypes and corollary between screening methodologies. Acta Agric. Scand. Sect. B 2019, 69, 588–595. [Google Scholar] [CrossRef]
- Kant, S.; Bim, Y.M.; Rothstein, S.J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2010, 62, 1499–1509. [Google Scholar] [CrossRef] [Green Version]
- Quarrie, S.A.; Steed, A.; Calestani, C.; Semikhodskii, A.; Lebreton, C.; Chinoy, C.; Steele, D.; Pljevljakusić, D.; Waterman, E.; Weyen, J.; et al. A high-density genetic map of hexaploid wheat (Triticumaestivum L.) from the cross Chinese Spring× SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet. 2005, 110, 865–880. [Google Scholar] [CrossRef]
- An, D.; Su, J.; Liu, Q.; Zhu, Y.; Tong, Y.; Li, J.; Jing, R.; Li, B.; Li, Z. Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.). Plant Soil 2006, 284, 73–84. [Google Scholar] [CrossRef]
- Laperche, A.; Brancourt-Hulmel, M.; Heumez, E.; Gardet, O.; Le Gouis, J. Estimation of genetic parameters of a DH wheat population grown at different N stress levels characterized by probe genotypes. Theor. Appl. Genet. 2006, 112, 797–807. [Google Scholar] [CrossRef]
- Laperche, A.; Brancourt-Hulmel, M.; Heumez, E.; Gardet, O.; Hanocq, E.; Devienne-Barret, F.; Le Gouis, J. Using genotype× nitrogen interaction variables to evaluate the QTL involved in wheat tolerance to nitrogen constraints. Theor. Appl. Genet. 2007, 115, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Habash, D.Z.; Bernard, S.; Schondelmaier, J.; Weyen, J.; Quarrie, S.A. The genetics of nitrogen use in hexaploid wheat: N utilization, development and yield. Theor. Appl. Genet. 2007, 114, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Le Gouis, J.; Fontaine, J.X.; Laperche, A.; Heumez, E.; Devienne-Barret, F.; Brancourt-Hulmel, M.; Dubois, F.; Hirel, B. Genetic analysis of wheat nitrogen use efficiency: Coincidence between QTL for agronomical and physiological traits. In Proceedings of the 11th International Wheat Genetics Symposium, Brisbane, Australia, 24–29 August 2008; pp. 916–918. [Google Scholar]
- Fontaine, J.X.; Ravel, C.; Pageau, K.; Heumez, E.; Dubois, F.; Hirel, B.; Le Gouis, J. A quantitative genetic study for elucidating the contribution of glutamine synthetase, glutamate dehydrogenase and other nitrogen-related physiological traits to the agronomic performance of common wheat. Theor. Appl. Genet. 2009, 119, 645–662. [Google Scholar] [CrossRef] [PubMed]
- García-Suárez, J.; Röder, M.; Díaz de León, J. Identification of QTLs and associated molecular markers of agronomic traits in wheat (Triticum aestivum L.) under two conditions of nitrogen fertilization. Cereal Res. Commun. 2010, 38, 459–470. [Google Scholar] [CrossRef]
- Guo, Y.; Kong, F.M.; Xu, Y.F.; Zhao, Y.; Liang, X.; Wang, Y.Y.; An, D.G.; Li, S.S. QTL mapping for seedling traits in wheat grown under varying concentrations of N., P and K nutrients. Theor. Appl. Genet. 2012, 124, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, R.; Tong, Y.; Zhao, H.; Xie, Q.; Liu, D.; Zhang, A.; Li, B.; Xu, H.; An, D. Mapping QTLs for yield and N-related traits in wheat: Influence of nitrogen and phosphorus fertilization on QTL expression. Theor. Appl. Genet. 2014, 127, 59–72. [Google Scholar] [CrossRef]
- Ren, Y.; Qian, Y.; Xu, Y.; Zou, C.; Liu, D.; Zhao, X.; Zhang, A.; Tong, Y. Characterization of QTLs for root traits of wheat grown under different nitrogen and phosphorus supply levels. Front. Plant Sci. 2017, 8, 2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Cui, Y.; Han, Q.; Fang, W.; Li, J.; Tian, J. Discovery of consistent QTLs of wheat spike-related traits under nitrogen treatment at different development stages. Front. Plant Sci. 2017, 8, 2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monostori, I.; Szira, F.; Tondelli, A.; Arendas, T.; Gierczik, K.; Cattivelli, L.; Galiba, G.; Vagujfalvi, A. Genome-wide association study and genetic diversity analysis on nitrogen use efficiency in a Central European winter wheat (Triticum aestivum L.) collection. PLoS ONE 2017, 12, e0189265. [Google Scholar] [CrossRef]
- Zhang, M.; Gao, M.; Zheng, H.; Yuan, Y.; Zhou, X.; Guo, Y.; Zhang, G.; Zhao, Y.; Kong, F.; An, Y.; et al. QTL mapping for nitrogen use efficiency and agronomic traits at the seedling and maturity stages in wheat. Mol. Breed. 2019, 39, 71. [Google Scholar] [CrossRef]
- Brasier, K.; Ward, B.; Smith, J.; Seago, J.; Oakes, J.; Balota, M.; Davis, P.; Fountain, M.; Brown-Guedira, G.; Sneller, C.; et al. Identification of quantitative trait loci associated with nitrogen use efficiency in winter wheat. PLoS ONE 2020, 15, e0228775. [Google Scholar] [CrossRef] [PubMed]
- Waines, J.G.; Ehdaie, B. Domestication and crop physiology: Roots of green-revolution wheat. Ann. Bot. 2007, 100, 991–998. [Google Scholar] [CrossRef] [Green Version]
- Sanguineti, M.C.; Li, S.; Maccaferri, M.; Corneti, S.; Rotondo, F.; Chiari, T.; Tuberosa, R. Genetic dissection of seminal root architecture in elite durum wheat germplasm. Ann. Appl. Biol. 2007, 151, 291–305. [Google Scholar] [CrossRef]
- Canè, M.A.; Maccaferri, M.; Nazemi, G.; Salvi, S.; Francia, R.; Colalongo, C.; Tuberosa, R. Association mapping for root architectural traits in durum wheat seedlings as related to agronomic performance. Mol. Breed. 2014, 34, 1629–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, J.A.; Wingen, L.U.; Griffiths, M.; Pound, M.P.; Gaju, O.; Foulkes, M.J.; Le Gouis, J.; Griffiths, S.; Bennett, M.J.; King, J.; et al. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Exp. Bot. 2015, 66, 2283–2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccaferri, M.; El-Feki, W.; Nazemi, G.; Salvi, S.; Canè, M.A.; Colalongo, M.C.; Stefanelli, S.; Tuberosa, R. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J. Exp. Bot. 2016, 67, 1161–1178. [Google Scholar] [CrossRef] [PubMed]
- Iannucci, A.; Marone, D.; Russo, M.A.; De Vita, P.; Miullo, V.; Ferragonio, P.; Blanco, A.; Gadaleta, A.; Mastrangelo, A.M. Mapping QTL for Root and shoot morphological traits in a durum wheat × T. dicoccum segregating population at seedling stage. Int. J. Genom. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, R.; Wingen, L.U.; Snape, J.W.; Dolan, L. Mapping of quantitative trait loci for root hair length in wheat identifies loci that co-locate with loci for yield components. J. Exp. Bot. 2016, 67, 4535–4543. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, R.; Yadav, R.; Gaikwad, K.; Kumar, M.; Kumar, N.; Babu, P.; Pandey, R.; Joshi, A.K. Genetic variability for root traits and its role in adaptation under conservation agriculture in spring wheat. Indian J. Genet. Plant Breed. 2021, 81, 24–33. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [Green Version]
- Somers, D.J.; Isaac, P.; Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 1105–1114. [Google Scholar] [CrossRef]
- Song, Q.J.; Shi, J.R.; Singh, S.; Fickus, E.W.; Costa, J.M.; Lewis, J.; Gill, B.S.; Ward, R.; Cregan, P.B. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 2005, 110, 550–560. [Google Scholar] [CrossRef]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.H.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [CrossRef]
- Pestsova, E.; Ganal, M.W.; Röder, M.S. Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 2000, 43, 689–697. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.; Edwards, K.; Isaac, P.; Korzun, V.; Gautier, M.F.; Joudrier, P.; Schlatter, A.; Dubcovsky, J.; De la Pena, R.; et al. Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat. Theor. Appl. Genet. 2002, 105, 413–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayalew, H.; Ma, X.; Yan, G. Screening wheat (Triticum spp.) genotypes for root length under contrasting water regimes: Potential sources of variability for drought resistance breeding. J. Agron. Crop Sci. 2015, 201, 189–194. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Sheoran, O.P.; Tonk, D.S.; Kaushik, L.S.; Hasija, R.C.; Pannu, R.S. Statistical Software Package for Agricultural Research Workers: Recent Advances in Information Theory; Hooda, D.S., Hasija, R.C., Eds.; Department of Mathematics Statistics, CCS HAU: Hisar, India, 1998; pp. 139–143. [Google Scholar]
- Zhang, X.; Sun, C.; Zhang, Z.; Dai, Z.; Chen, Y.; Yuan, X.; Yuan, Z.; Tang, W.; Li, L.; Hu, Z. Genetic dissection of main and epistatic effects of QTL based on augmented triple test cross design. PLoS ONE 2017, 12, e018905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Y.-J.; Zhang, Y.-W.; Zhang, J.; Feng, J.-Y.; Dunwell, J.M.; Zhang, Y.-M. An efficient multi-locus mixed model framework for the detection of small and linked QTLs in F2. Brief. Bioinform. 2019, 20, 1913–1924. [Google Scholar] [CrossRef]
- Zhang, Y.-W.; Wen, Y.-J.; Dunwell, J.M.; Zhang, Y.-M. QTL.gCIMapping.GUI v2.0: An R software for detecting small-effect and linked QTLs for quantitative traits in bi-parental segregation populations. Comput. Struct. Biotechnol. J. 2020, 18, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017, 5, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.T.; Dang, D.T.; Van Pham, C.; Bertin, P. QTL mapping for nitrogen use efficiency and related physiological and agronomical traits during the vegetative phase in rice under hydroponics. Euphytica 2016, 212, 473–500. [Google Scholar] [CrossRef]
- Hodge, A.; Berta, G.; Doussan, C.; Merchan, F.; Crespi, M. Plant rootgrowth, architecture and function. Plant Soil 2009, 321, 153–187. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Hawkesford, M.; Barraclough, P.B.; Holdsworth, M.J. Identifying traits to improve the nitrogen economy of wheat: Recent advances and future prospects. Field Crops Res. 2009, 114, 329–342. [Google Scholar] [CrossRef]
- McPhee, K. Variation for seedling root architecture in the core collection of pea germplasm. Crop Sci. 2005, 45, 1758–1763. [Google Scholar] [CrossRef]
- Hurd, E.A. Root study of three wheat varieties and their resistance to drought and damage by soil cracking. Can. J. Plant Sci. 1964, 44, 240–248. [Google Scholar] [CrossRef]
- Price, A.H.; Steele, K.A.; Moore, B.J.; Jones, R.G.W. Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes: II. Mapping quantitative trait loci for root morphology and distribution. Field Crops Res. 2002, 76, 25–43. [Google Scholar] [CrossRef]
- Lynch, J.P. Roots of the second green revolution. Aust. J. Bot. 2007, 55, 493–512. [Google Scholar] [CrossRef]
- Srividya, A.; Ramanarao, P.V.; Sridhar, S.; Jayaprada, M.; Anuradha, G.; Srilakshmi, B.; Reddy, H.K.; Hariprasad, A.S.; Siddiq, E.A.; Vemireddy, L.R. Molecular mapping of QTLs for drought related traits at seedling stage under PEG induced stress conditions in rice. Am. J. Plant Sci. 2011, 2, 190. [Google Scholar] [CrossRef] [Green Version]
- Gallais, A.; Hirel, B. An approach to the genetics of nitrogen use efficiency in maize. J. Exp. Bot. 2004, 55, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hitz, K. Breeding for Nitrogen Use Efficiency in Soft Red Winter Wheat. Master’s Thesis, University of Kentucky, Lexington, KY, USA, 2015. [Google Scholar]
- DoVale, J.C.; DeLima, R.O.; Fritsche-Neto, R. Breeding for nitrogen use efficiency. In Plant Breeding for Abiotic Stress Tolerance; Springer: Berlin/Heidelberg, Germany, 2012; pp. 53–65. [Google Scholar] [CrossRef]
- Kearsey, M.J.; Pooni, H.S. Genotype by environment interaction. In The Genetical Analysis of Quantitative Traits; Springer: New York, NY, USA, 1996; pp. 241–265. [Google Scholar]
- Sourdille, P.; Singh, S.; Cadalen, T.; Brown-Guedira, G.L.; Gay, G.; Qi, L.; Gill, B.S.; Dufour, P.; Murigneux, A.; Bernard, M. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genom. 2004, 4, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Fan, X.; Zhao, C.; Zhang, W.; Chen, M.; Ji, J.; Li, J. A novel genetic map of wheat: Utility for mapping QTL for yield under different nitrogen treatments. BMC Genet. 2014, 15, 57. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Xu, S. Mapping Quantitative Trait Loci in F2 Incorporating Phenotypes of F3 Progeny. Genetics 2004, 166, 1981–1993. [Google Scholar] [CrossRef]
- Sun, J.J.; Guo, Y.; Zhang, G.Z.; Gao, M.G.; Zhang, G.H.; Kong, F.M.; Zhao, Y.; Li, S.S. QTL mapping for seedling traits under different nitrogen forms in wheat. Euphytica 2013, 191, 319–331. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates, Inc.: Sunderland, MA, USA, 1998; pp. 535–557. [Google Scholar]
- Lei, L.; Li, G.; Zhang, H.; Powers, C.; Fang, T.; Chen, Y.; Wang, S.; Zhu, X.; Carver, B.F.; Yan, L. Nitrogen use efficiency is regulated by interacting proteins relevant to development in wheat. Plant Biotechnol. J. 2018, 16, 1214–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quraishi, U.M.; Abrouk, M.; Murat, F.; Pont, C.; Foucrier, S.; Desmaizieres, G.; Confolent, C.; Riviere, N.; Charmet, G.; Paux, E.; et al. Cross-genome map based dissection of a nitrogen use efficiency orthometa QTL in bread wheat unravels concerted cereal genome evolution. Plant J. 2011, 65, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Tao, Q.N.; Wu, P. QTLs underlying rice root to uptake NH4-N and NO3-N and rice N use efficiency at seedling stage. Plant Nutr. Fert. Sci. 2001, 7, 159–165. [Google Scholar]
- Ranjan, R.; Yadav, R. Genetics analysis of nitrogen use efficiency component traits under nitrogen-limiting environment. Cereal Res. Commun. 2020, 48, 431–439. [Google Scholar] [CrossRef]
- Piao, Z.; Han, L.; Koh, H.; Zhang, J.; Lu, J.; Li, P. Analysis of combining the ability of dry weight and nitrogen use efficiency in rice. Zhong Guo Shui Dao Ke Xue 2005, 19, 527–532. [Google Scholar]
- Hao, X.; Dong, Z.D.; Li, Y.; Cui, D.Q. Genetic model of nitrogen concentrations and nitrogen utilization efficiency of wheat in different water and nitrogen environment. J. Triticeae Crops 2006, 4, 103–106. [Google Scholar]
- Xiong, H.; Guo, H.; Zhou, C.; Guo, X.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Ding, Y.; Liu, L. A combined association mapping and t-test analysis of SNP loci and candidate genes involving in resistance to low nitrogen traits by a wheat mutant population. PLoS ONE 2009, 14, e0211492. [Google Scholar] [CrossRef]
- He, X.; Qu, B.; Li, W.; Zhao, X.; Teng, W.; Ma, W.; Ren, Y.; Li, B.; Li, Z.; Tong, Y. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 2015, 169, 1991–2005. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Sharma, M.; Kumar, S.; Tyagi, P.; Wani, S.H.; Gajula, M.N.V.P.; Singh, K.P. Functional and structural insightsinto candidate genes associated with nitrogen and phosphorus nutrition in wheat (Triticum aestivum L.). Int. J. Biolmacromol. 2018, 118, 76–91. [Google Scholar] [CrossRef]
- Xu, Y.; Crouch, J.H. Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.K.; Jiang, X.L.; Peng, T.; Shi, C.L.; Han, S.X.; Tian, B.; Zhu, Z.L.; Tian, J.C. Mapping quantitative trait loci with additive effects and additive × additive epistatic interactions for biomass yield, grain yield, and straw yield using a doubled haploid population of wheat (Triticumaestivum L.). Genet. Mol. Res. 2014, 13, 1412–1424. [Google Scholar] [CrossRef] [PubMed]
- Carlborg, Ö.; Haley, C.S. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 2004, 5, 618. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhu, J.; Lou, X.; Lu, Y. A method for marker-assisted selection based on QTLs with epistatic effects. Genetica 2003, 119, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.Q.; Ma, J.L.; Zhang, C.R.; Dong, H.F.; Xi, Z.Y.; Xia, Z.L.; Wu, J.Y. QTL mapping for test weight by using F2:3 population in maize. J. Genet. 2011, 90, 75–80. [Google Scholar] [CrossRef] [PubMed]

| Traits | Parent | F2:3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | Mean | Variance | SE | Minimum | Maximum | Skewness | W-Test | p-Value | |
| SDW | 0.970 | 0.575 | 0.654 | 0.048 | 0.016 | 0.298 | 1.788 | 1.071 | 0.965 | 0.0001 |
| RDW | 0.144 | 0.070 | 0.076 | 0.001 | 0.002 | 0.033 | 0.176 | 0.789 | 0.972 | 0.0008 |
| TDW | 1.115 | 0.645 | 0.729 | 0.05 | 0.016 | 0.344 | 1.852 | 0.969 | 0.969 | 0.0004 |
| R:S | 0.148 | 0.121 | 0.128 | 0.004 | 0.005 | 0.036 | 0.384 | 1.292 | 0.947 | 0.0000 |
| N% | 2.493 | 1.933 | 2.57 | 0.251 | 0.037 | 1.065 | 4.692 | 0.475 | 0.990 | 0.2677 |
| gN | 0.024 | 0.011 | 0.017 | 0 | 0.001 | 0.004 | 0.049 | 0.878 | 0.966 | 0.0002 |
| NUpE | 1.052 | 0.483 | 0.742 | 0.097 | 0.023 | 0.192 | 2.123 | 0.868 | 0.967 | 0.0002 |
| NUtE | 40.10 | 51.73 | 40.60 | 92.5 | 0.703 | 21.311 | 93.938 | 0.41 | 0.986 | 0.0783 |
| NUE | 42.18 | 25 | 28.41 | 91.1 | 0.698 | 12.957 | 77.725 | 1.07 | 0.965 | 0.0001 |
| Scheme | QTL | Position | Lt Marker | Rt Marker | LOD | PVE (%) | A (a) | D (d) | d/a | Lt CI | Rt CI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SDW | |||||||||||
| 1 | QSdw.iari-2A | 353 | wmc728.1 | wmc473.3 | 3.65 | 10.47 | 0.007 | 0.161 | 21.5 | 334.5 | 358 |
| 2 | QSdw.iari-7A | 0 | wmc607 | wmc633 | 3.15 | 5.01 | 0.051 | 0.068 | 1.3 | 0 | 14.5 |
| RDW | |||||||||||
| 1 | QRdw.iari-4A | 180 | wmc262 | wmc617.2 | 4.54 | 5.82 | −0.0008 | 0.024 | 31.1 | 166.5 | 190 |
| 2 | QRdw.iari-7A | 94 | wmc633 | wmc488 | 3.52 | 8.23 | −0.016 | −0.019 | 1.15 | 81.5 | 107.5 |
| TDW | |||||||||||
| 1 | QTdw.iari-2A | 358 | wmc728.1 | wmc473.3 | 4.04 | 10.64 | −0.0004 | 0.138 | 347 | 334.5 | 358 |
| 2 | QTdw.iari-4D | 254 | wmc617.1 | wmc622 | 3.25 | 8.6 | −0.011 | 0.133 | 12 | 232.5 | 254 |
| R:S | |||||||||||
| 1 | QR:S.iari-2A.1 | 41 | wmc779 | gwm249.1 | 4.6 | 2.92 | −0.058 | −0.055 | 0.945 | 26.5 | 57.5 |
| 2 | QR:S.iari-2A.2 | 292 | wmc728.1 | wmc473.3 | 4.39 | 3.52 | 0.058 | −0.059 | 1.02 | 269.5 | 319.5 |
| 3 | QR: S.iari-4A | 36 | wmc597.3 | cfa2026 | 3.66 | 2.9 | 0.051 | −0.052 | 1.01 | 25.5 | 45.5 |
| 4 | QR: S.iari-1B | 50 | wmc631.1 | wmc611.1 | 3.26 | 2.95 | 0.052 | −0.062 | 1.2 | 30.5 | 67.5 |
| 5 | QR: S.iari-3B | 234 | wmc527.1 | wmc754.1 | 3.16 | 2.98 | −0.049 | −0.066 | 1.3 | 222.5 | 245.5 |
| 6 | QR:S.iari-6B | 70 | gwm219 | wmc597.4 | 3.7 | 3.15 | 0.05 | −0.064 | 1.2 | 44.5 | 89.5 |
| 7 | QR:S.iari-2D | 322 | gwm261 | wmc503 | 3.1 | 3.06 | −0.051 | −0.058 | 1.12 | 300.5 | 343.5 |
| N% | |||||||||||
| 1 | QN%.iari-4B | 4 | wmc617 | gwm540 | 4.64 | 9.87 | −0.068 | 0.383 | 5.6 | 0 | 13.5 |
| 2 | QN%.iari-7D | 139 | wmc473.2 | wmc824 | 4.05 | 10.81 | −0.014 | 0.405 | 28 | 132.5 | 150.5 |
| NUpE | |||||||||||
| 1 | QNUpE.iari-2A | 358 | wmc728.1 | wmc473.3 | 5.52 | 10.96 | −0.007 | 0.22 | 28.5 | 343.5 | 358 |
| 2 | QNUpE.iari-4A | 185 | wmc262 | wmc617.2 | 3.44 | 11.18 | −0.001 | 0.219 | 219.9 | 171.5 | 190 |
| NUtE | |||||||||||
| 1 | QNUtE.iari-2A.1 | 36 | wmc779 | gwm249.1 | 7.94 | 3.68 | −14.714 | −14.362 | 0.9 | 25.5 | 50.5 |
| 2 | QNUtE.iari-2A.2 | 311 | wmc728.1 | wmc473.3 | 5.82 | 3.82 | −13.129 | −13.676 | 1.03 | 292.5 | 323.5 |
| 3 | QNUtE.iari-3A | 381 | wmc527 | wmc264.1 | 6.29 | 3.8 | 13.336 | −14.032 | 1.05 | 368.5 | 389.5 |
| 4 | QNUtE.iari-5A | 138 | wmc617.4 | gwm186 | 5.87 | 3.76 | 13.95 | −13.196 | 0.94 | 130.5 | 145.5 |
| 5 | QNUtE.iari-5B | 31 | wmc75 | wmc616 | 6.42 | 3.8 | 13.268 | −13.349 | 1.06 | 21.5 | 41.5 |
| 6 | QNUtE.iari-1D | 40 | cfa2129.2 | wmc609 | 3.03 | 1.21 | −4.359 | −4.597 | 1.05 | 23.5 | 59.5 |
| 7 | QNUtE.iari-2D | 322 | gwm261 | wmc503 | 6.63 | 3.76 | 14.163 | −14.587 | 1.02 | 301.5 | 333.5 |
| 8 | QNUtE.iari-4D | 144 | wmc473 | wmc617.1 | 6.32 | 3.68 | 14.02 | −13.692 | 0.97 | 135.5 | 152.5 |
| 9 | QNUtE.iari-7D.1 | 138 | wmc473.2 | wmc824 | 3.4 | 1.09 | −0.512 | −7.256 | 14.21 | 132.5 | 150.5 |
| 10 | QNUtE.iari-7D.2 | 251 | wmc824 | wmc488.1 | 7.7 | 3.79 | 14.062 | −13.991 | 0.99 | 239.5 | 262.5 |
| NUE | |||||||||||
| 1 | QNUE.iari-2A | 353 | wmc728.1 | wmc473.3 | 3.65 | 10.47 | 0.324 | 7.031 | 21.6 | 334.5 | 358 |
| 2 | QNUE.iari-7A | 0 | wmc607 | wmc633 | 3.15 | 5.01 | 2.238 | 2.963 | 1.3 | 0 | 14.5 |
| Trait | Chr | Position (cM) | LOD | Left_Marker | Right_Marker | PVE (%) |
|---|---|---|---|---|---|---|
| SDW | 18 | 254.75 | 3.3 | wmc617 | wmc622 | 3.71 |
| SDW | 21 | 145 | 6.7 | wmc473-2 | wmc824 | 6.64 |
| RDW | 7 | 196.92 | 5.2 | wmc606 | wmc606 | 2.40 |
| RDW | 17 | 85 | 5.0 | cfd152 | barc71 | 5.41 |
| TDW | 18 | 120 | 8.0 | wmc473 | wmc617-1 | 7.40 |
| TDW | 18 | 250 | 4.1 | wmc617-1 | wmc622 | 5.81 |
| R:S | 21 | 150 | 6.2 | wmc473-2 | wmc824 | 7.82 |
| N% | 6 | 15 | 5.4 | wmc617-3 | wmc256 | 2.65 |
| N% | 8 | 215 | 4.6 | wmc597 | cfa2147 | 2.24 |
| N% | 10 | 100 | 7.7 | barc77 | gwm247-1 | 5.76 |
| N% | 13 | 60 | 5.0 | gwm219 | wmc597-4 | 3.20 |
| N% | 16 | 20 | 8.5 | barc59 | wmc11 | 4.59 |
| NUpE | 11 | 20 | 4.0 | wmc617 | gwm540 | 3.83 |
| NUpE | 18 | 254.75 | 3.0 | wmc622 | wmc622 | 2.66 |
| NUtE | 6 | 15 | 8.3 | wmc617-3 | wmc256 | 7.82 |
| NUtE | 8 | 220 | 4.0 | wmc597 | cfa2147 | 3.29 |
| NUtE | 12 | 155 | 6.0 | barc59-1 | wmc728-2 | 6.06 |
| NUE | 18 | 254.75 | 3.2 | wmc622 | wmc622 | 3.57 |
| NUE | 21 | 145 | 6.3 | wmc473-2 | wmc824 | 6.19 |
| Trait | Chr 1 | Pos1 | Flanking Marker | Chr2 | Pos2 | Flanking Marker | LOD | R2(%) | A1 | D1 | A2 | D2 | AA | AD | DA | DD | Dd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SDW | 1A | 125 | wmc611-wmc550.2 | 2A | 250 | wmc728.1-wmc473.3 | 5.68 | 1.44 | 0.20 | −0.31 | 0.08 | −0.36 | 0.24 | −0.17 | −0.11 | 0.64 | OD |
| SDW | 2A | 10 | wmc779-gwm249.1 | 2A | 265 | wmc728.1-wmc473.3 | 7.20 | 1.56 | −0.37 | −0.23 | 0.13 | −0.29 | −0.25 | 0.54 | −0.28 | 0.10 | D |
| SDW | 2A | 330 | wmc728.1-wmc473.3 | 3A | 185 | gwm247-wmc550 | 5.55 | 1.77 | −0.31 | 0.09 | −0.30 | −0.31 | 0.31 | 0.34 | 0.38 | 0.14 | D |
| SDW | 6A | 125 | wmc754-gwm169 | 7A | 10 | wmc607-wmc633 | 6.18 | 1.06 | −0.17 | −0.19 | 0.15 | −0.13 | −0.08 | 0.30 | −0.11 | 0.34 | OD |
| SDW | 7A | 80 | wmc633-wmc488 | 7A | 250 | wmc606-wmc646.1 | 5.52 | 1.38 | 0.11 | −0.24 | −0.05 | −0.12 | −0.29 | −0.11 | −0.01 | 0.38 | OD |
| SDW | 2A | 235 | wmc728.1-wmc473.3 | 1B | 320 | wmc728-cfa2129.1 | 8.23 | 1.51 | 0.24 | −0.19 | −0.12 | −0.38 | −0.30 | −0.38 | 0.02 | 0.03 | A |
| SDW | 7A | 10 | wmc607-wmc633 | 4B | 295 | wmc754.1-wmc597.2 | 5.05 | 1.07 | 0.15 | −0.19 | −0.18 | −0.19 | −0.08 | −0.11 | 0.24 | 0.42 | OD |
| SDW | 2A | 325 | wmc728.1-wmc473.3 | 7D | 325 | wmc488.1-cfd66 | 5.58 | 1.63 | 0.31 | −0.07 | 0.30 | −0.27 | 0.31 | −0.30 | −0.35 | 0.45 | OD |
| SDW | 7A | 70 | wmc633-wmc488 | 7D | 500 | wmc646-cfd31 | 5.31 | 1.29 | 0.10 | −0.24 | −0.03 | −0.10 | −0.26 | −0.12 | 0.00 | 0.36 | OD |
| Trait | Chr1 | Pos1 | Flanking Marker | Trai * | Chr2 | Pos2 | Flanking Marker | LOD | R2 (%) | AA | AD | DA | DD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SDW | 2A | 220 | barc220-wmc728.1 | N% | 4B | 0 | wmc617-gwm540 | 6.24 | 0.65 | 0.21 | −0.13 | −0.12 | 0.21 |
| SDW | 2B | 190 | gwm526-cfd70 | N% | 4B | 5 | wmc617-gwm540 | 5.21 | 0.67 | −0.20 | 0.02 | −0.09 | 0.24 |
| SDW | 3A | 10 | wmc11.1-wmc559 | N% | 4B | 10 | wmc 617-gwm540 | 6.24 | 0.82 | −0.23 | 0.08 | −0.04 | 0.38 |
| SDW | 4B | 0 | wmc 617-gwm540 | N% | 6B | 20 | gwm219-wmc597.4 | 5.10 | 0.62 | 0.15 | −0.20 | 0.00 | 0.02 |
| SDW | 4B | 0 | wmc 617-gwm540 | N% | 2D | 50 | barc59-wmc11 | 5.64 | 0.71 | −0.20 | −0.07 | 0.02 | 0.20 |
| SDW | 2A | 0 | wmc779-gwm249.1 | RDW | 4A | 175 | wmc262-wmc617.2 | 5.37 | 0.53 | −0.23 | 0.22 | −0.24 | 0.11 |
| RDW | 2A | 355 | wmc728.1-wmc473.3 | SDW | 7B | 225 | wmc606.1-barc182 | 7.78 | 0.43 | 0.01 | −0.04 | 0.00 | −0.04 |
| RDW | 2A | 355 | wmc728.1-wmc473.3 | SDW | 3D | 180 | barc71-wmc631 | 8.21 | 0.49 | −0.03 | 0.00 | −0.01 | 0.04 |
| RDW | 2A | 350 | wmc728.1-wmc473.3 | SDW | 5D | 105 | wmc443-wmc264 | 7.96 | 0.56 | −0.03 | 0.01 | −0.02 | −0.01 |
| RDW | 7A | 0 | wmc607-wmc633 | SDW | 5D | 180 | wmc264-wmc324 | 6.00 | 0.32 | 0.00 | 0.01 | 0.02 | 0.03 |
| RDW | 2A | 195 | barc220-wmc728.1 | SDW | 2A | 350 | wmc728.1-wmc473.3 | 10.11 | 0.52 | −0.03 | −0.01 | 0.01 | 0.04 |
| RDW | 5D | 220 | wmc264-wmc324 | N% | 7D | 135 | wmc473.2-wmc824 | 6.40 | 0.28 | 0.02 | 0.01 | 0.02 | 0.02 |
| RDW | 2A | 195 | barc220-wmc728.1 | N% | 7D | 140 | wmc473.2-wmc824 | 9.86 | 0.51 | −0.03 | −0.01 | 0.01 | 0.04 |
| RDW | 6D | 140 | cfd132-wmc550.1 | N% | 7D | 140 | wmc473.2-wmc824 | 7.88 | 0.53 | 0.03 | 0.01 | 0.00 | 0.05 |
| RDW | 5A | 80 | gwm156-wmc617.4 | N% | 7D | 145 | wmc473.2-wmc824 | 9.31 | 0.61 | −0.01 | −0.01 | 0.04 | 0.05 |
| RDW | 7A | 15 | wmc607-wmc633 | N% | 7D | 145 | wmc473.2-wmc824 | 8.21 | 0.48 | 0.02 | 0.02 | 0.02 | 0.03 |
| RDW | 4A | 175 | wmc262-wmc617.2 | RDW | 4B | 85 | gwm540-cfd39 | 5.14 | 0.64 | 0.01 | 0.01 | 0.00 | -0.02 |
| N% | 2B | 320 | wmc728.1-wmc473.3 | SDW | 2A | 355 | wmc728.1-wmc473.3 | 10.93 | 0.39 | −0.20 | −0.86 | 0.51 | −0.01 |
| N% | 2A | 350 | wmc728.1-wmc473.3 | SDW | 1B | 40 | wmc631.1-wmc611.1 | 11.43 | 0.43 | −0.78 | 0.48 | −0.20 | −0.13 |
| N% | 7A | 100 | wmc633-wmc488 | RDW | 2B | 105 | wmc597.1-gwm526 | 8.96 | 0.42 | 0.65 | −0.64 | −0.08 | 0.22 |
| N% | 4A | 175 | wmc262-wmc617.2 | RDW | 2B | 165 | gwm526-cfd70 | 9.57 | 0.36 | −0.68 | -0.15 | 0.55 | 0.13 |
| N% | 4A | 175 | wmc262-wmc617.2 | RDW | 4B | 85 | gwm540-cfd39 | 9.72 | 0.40 | −0.67 | −0.23 | 0.49 | −0.20 |
| N% | 7A | 95 | wmc633-wmc488 | RDW | 7B | 165 | gwm577-wmc606.1 | 10.04 | 0.41 | 0.05 | −0.72 | −0.64 | −0.19 |
| N% | 7A | 100 | wmc633-wmc488 | RDW | 3D | 180 | barc71-wmc631 | 11.97 | 0.41 | 0.09 | −0.72 | −0.60 | 0.21 |
| N% | 7A | 95 | wmc633-wmc488 | RDW | 4D | 195 | wmc617.1-wmc622 | 12.90 | 0.42 | 0.64 | −0.70 | −0.06 | −0.04 |
| N% | 7A | 95 | wmc633-wmc488 | RDW | 7D | 305 | wmc488.1-cfd66 | 13.35 | 0.47 | 0.56 | −1.03 | −0.02 | 0.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranjan, R.; Yadav, R.; Jain, N.; Sinha, N.; Bainsla, N.K.; Gaikwad, K.B.; Kumar, M. Epistatic QTLsPlay a Major Role in Nitrogen Use Efficiency and Its Component Traits in Indian Spring Wheat. Agriculture 2021, 11, 1149. https://doi.org/10.3390/agriculture11111149
Ranjan R, Yadav R, Jain N, Sinha N, Bainsla NK, Gaikwad KB, Kumar M. Epistatic QTLsPlay a Major Role in Nitrogen Use Efficiency and Its Component Traits in Indian Spring Wheat. Agriculture. 2021; 11(11):1149. https://doi.org/10.3390/agriculture11111149
Chicago/Turabian StyleRanjan, Rumesh, Rajbir Yadav, Neelu Jain, Nivedita Sinha, Naresh Kumar Bainsla, Kiran B. Gaikwad, and Manjeet Kumar. 2021. "Epistatic QTLsPlay a Major Role in Nitrogen Use Efficiency and Its Component Traits in Indian Spring Wheat" Agriculture 11, no. 11: 1149. https://doi.org/10.3390/agriculture11111149
APA StyleRanjan, R., Yadav, R., Jain, N., Sinha, N., Bainsla, N. K., Gaikwad, K. B., & Kumar, M. (2021). Epistatic QTLsPlay a Major Role in Nitrogen Use Efficiency and Its Component Traits in Indian Spring Wheat. Agriculture, 11(11), 1149. https://doi.org/10.3390/agriculture11111149






