Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness
Abstract
1. Introduction
2. Materials and Methods
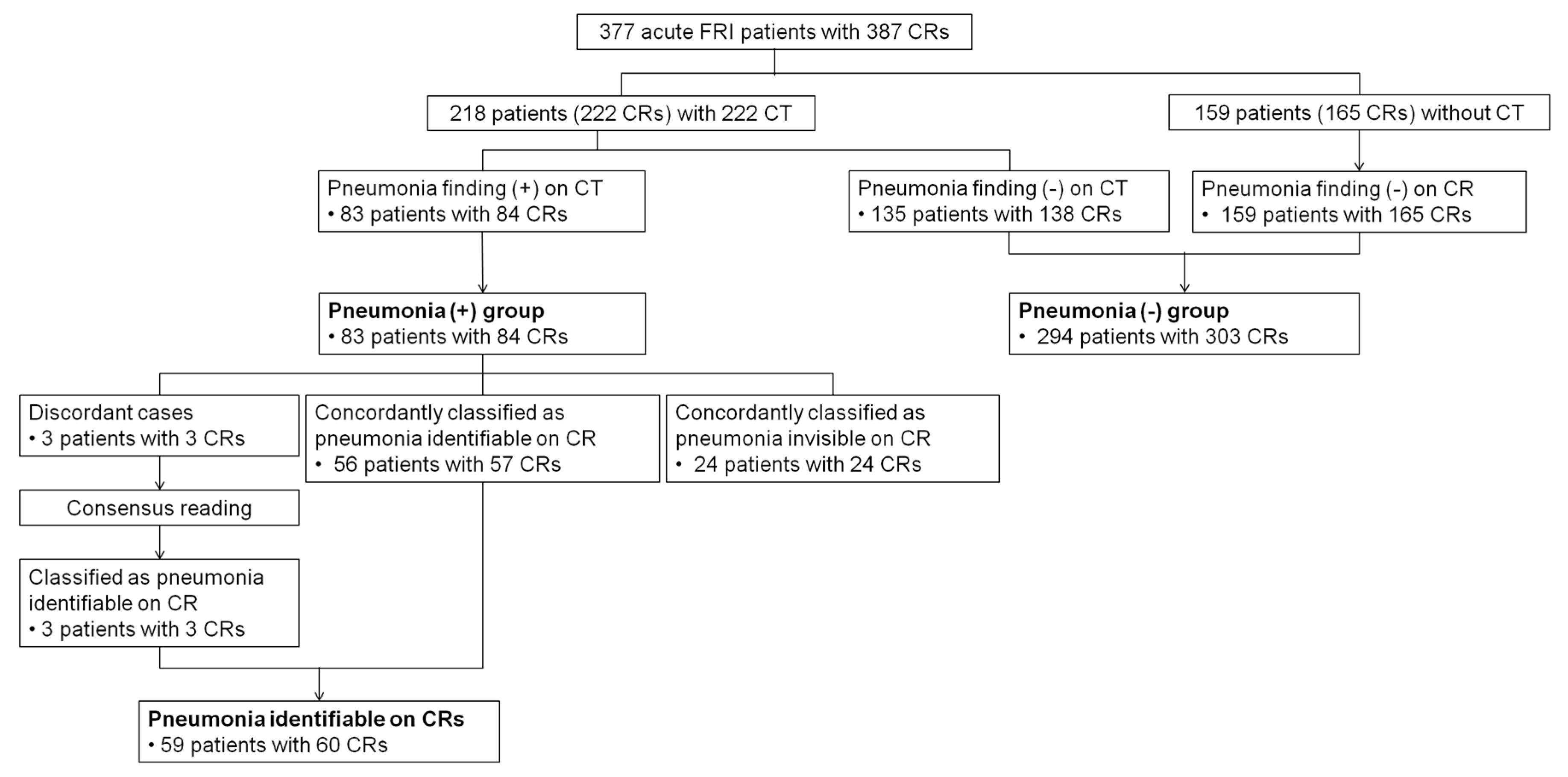
2.1. Patients and CR Collection
2.2. Laboratory Testing and Pathogen Detection
2.3. DL Algorithm
2.4. Reference Standards
2.5. Observer Performance Assessment
2.6. Statistical Analysis
3. Results
3.1. Pneumonia Detection Performance of the Deep-Learning Algorithm on CRs
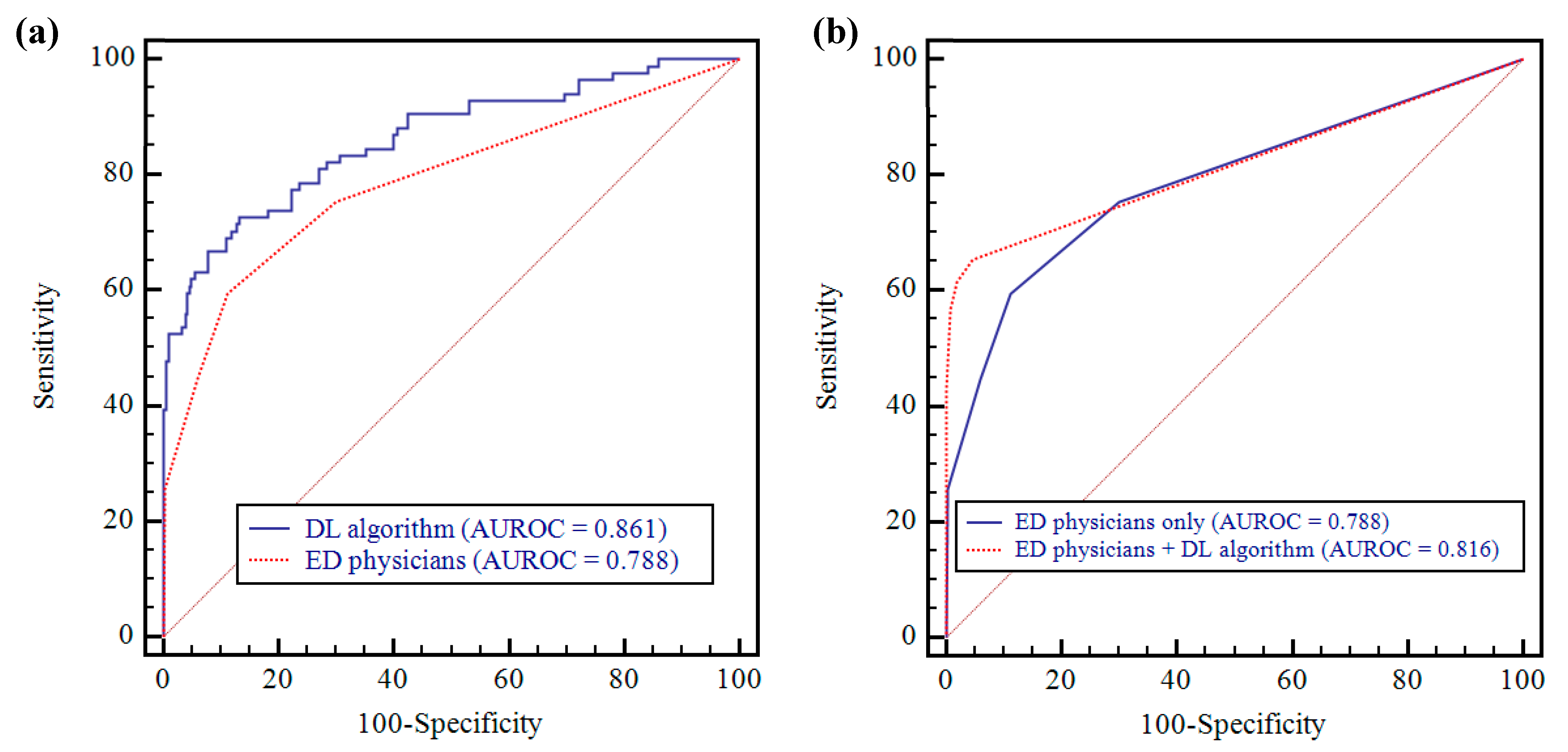
3.2. Performance Comparison between Deep-Learning Algorithm and Physicians
3.3. Performance Comparison between Physicians-only and Physicians Aided by the Algorithm
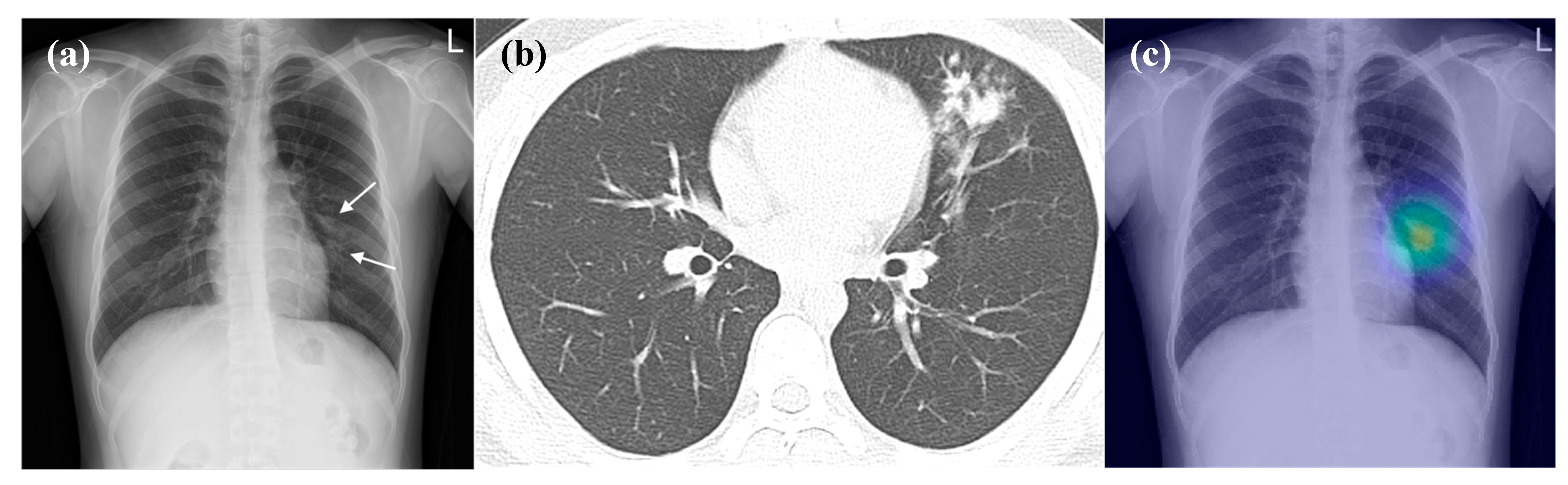
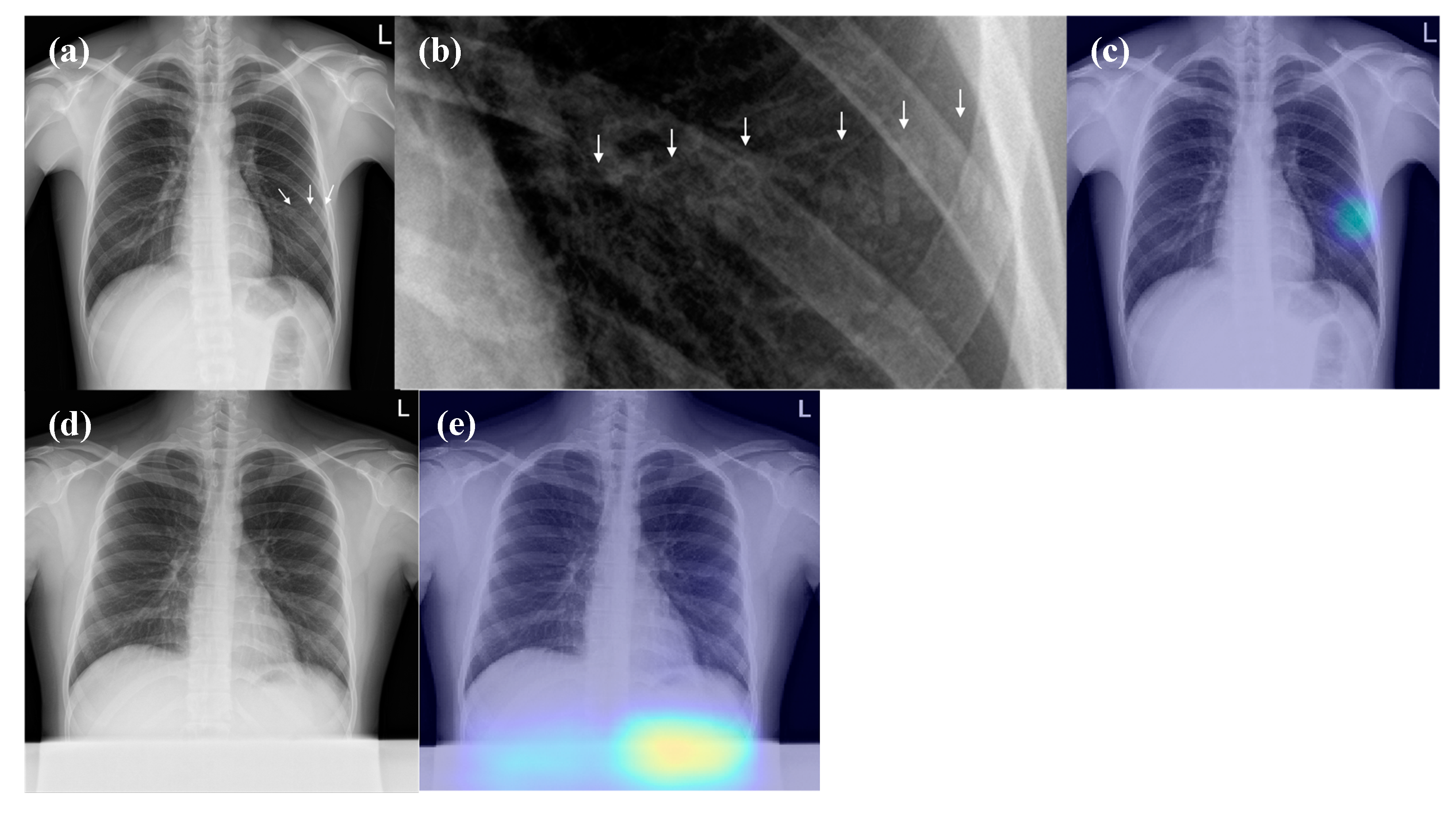
3.4. False-Positive Interpretations of DL Algorithm (Detection of Pneumonia on CRs)
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ARI | acute respiratory illness |
| FRI | febrile respiratory illness |
| ED | emergency department |
| CR | chest radiograph |
| DL | deep-learning |
References
- Ferkol, T.; Schraufnagel, D. The global burden of respiratory disease. Ann. Am. Thorac. Soc. 2014, 11, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Hooker, E.A.; Mallow, P.J.; Oglesby, M.M. Characteristics and Trends of Emergency Department Visits in the United States (2010–2014). J. Emerg. Med. 2019, 56, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Jokerst, C.; Chung, J.H.; Ackman, J.B.; Carter, B.; Colletti, P.M.; Crabtree, T.D.; de Groot, P.M.; Iannettoni, M.D.; Maldonado, F.; McComb, B.L.; et al. ACR Appropriateness Criteria((R)) Acute Respiratory Illness in Immunocompetent Patients. J. Am. Coll. Radiol. 2018, 15, S240–S251. [Google Scholar] [CrossRef] [PubMed]
- Sandrock, C.; Stollenwerk, N. Acute febrile respiratory illness in the ICU: Reducing disease transmission. Chest 2008, 133, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.; Mysko, W.K.; Weller, G.E.; Renard, R.; Gitlin, J.N.; Bluemke, D.A.; Magid, D.; Kelen, G.D.; Scott, W.W., Jr. Interpretation of Emergency Department radiographs: A comparison of emergency medicine physicians with radiologists, residents with faculty, and film with digital display. AJR Am. J. Roentgenol. 2000, 175, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Gatt, M.E.; Spectre, G.; Paltiel, O.; Hiller, N.; Stalnikowicz, R. Chest radiographs in the emergency department: Is the radiologist really necessary? Postgrad. Med. J. 2003, 79, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Al aseri, Z. Accuracy of chest radiograph interpretation by emergency physicians. Emerg. Radiol. 2009, 16, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Petinaux, B.; Bhat, R.; Boniface, K.; Aristizabal, J. Accuracy of radiographic readings in the emergency department. Am. J. Emerg. Med. 2011, 29, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.J.; Nam, J.G.; Lim, W.H.; Park, S.J.; Jeong, Y.S.; Kang, J.H.; Hong, E.K.; Kim, T.M.; Goo, J.M.; Park, S.; et al. Deep Learning for Chest Radiograph Diagnosis in the Emergency Department. Radiology 2019, 293, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.W.; Cheung, C.Y.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; San Yeo, I.Y.; Lee, S.Y.; et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA 2017, 318, 2211–2223. [Google Scholar] [CrossRef] [PubMed]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.G.; Park, S.; Hwang, E.J.; Lee, J.H.; Jin, K.N.; Lim, K.Y.; Vu, T.H.; Sohn, J.H.; Hwang, S.; Goo, J.M.; et al. Development and Validation of Deep Learning-based Automatic Detection Algorithm for Malignant Pulmonary Nodules on Chest Radiographs. Radiology 2019, 290, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.J.; Jang, J.K.; Lee, S.S.; Sung, Y.S.; Shim, W.H.; Kim, H.S.; Yun, J.; Choi, J.Y.; Lee, Y.; Kang, B.K.; et al. Development and Validation of a Deep Learning System for Staging Liver Fibrosis by Using Contrast Agent-enhanced CT Images in the Liver. Radiology 2018, 289, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.J.; Park, S.; Jin, K.N.; Kim, J.I.; Choi, S.Y.; Lee, J.H.; Goo, J.M.; Aum, J.; Yim, J.J.; Cohen, J.G.; et al. Development and Validation of a Deep Learning-Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs. JAMA Netw. Open 2019, 2, e191095. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Han, X.; Jiang, N.; Cao, Y.; Alwalid, O.; Gu, J.; Fan, Y.; Zheng, C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]

| All Patients (n = 377) | Pneumonia (n = 83) | Non-Pneumonia (n = 294) | p Value * | ||
|---|---|---|---|---|---|
| Characteristics | |||||
| Age, years | 20.0 (20.0–21.0) | 20.0 (20.0–21.0) | 20·0 (20.0–21.0) | 0.737 | |
| >50 | 1 (0.3%) | 0 (0.0%) | 1 (0.3%) | 1.000 | |
| ≤50 | 376 (99.7%) | 83 (100.0%) | 293 (99.7%) | ||
| Sex | 1.000 | ||||
| Male | 375 (99.5%) | 83 (100.0%) | 291 (99.3%) | ||
| Female | 2 (0.5%) | 0 (0.0%) | 2 (0.7%) | ||
| Symptoms | |||||
| Fever | 377 (100.0%) | 83 (100.0%) | 294 (100.0%) | NA | |
| Maximum temperature, °C | 38.6 (38.3–39.1) | 38.6 (38.4–39.1) | 38.6 (38.3–39.0) | 0.669 | |
| 38–39 | 282 (74.8%) | 61 (73.5%) | 221 (75.2%) | 0.775 | |
| >39 | 95 (25.2%) | 22 (26.5%) | 73 (24.8%) | ||
| Dyspnea | 6 (1.6%) | 4 (4.8%) | 2 (0.7%) | 0.023 | |
| Cough | 377 (100.0%) | 83 (100.0%) | 294 (100.0%) | NA | |
| Sputum | 287 (76.1%) | 63 (75.9%) | 224 (76.2%) | 1.000 | |
| Rhinorrhea | 200 (53.1%) | 39 (47.0%) | 161 (54.8%) | 0.216 | |
| Sore throat | 275 (73.0%) | 50 (60.2%) | 225 (76.5%) | 0.005 | |
| Headache | 202 (53.6%) | 43 (51.8%) | 159 (54.1%) | 0.803 | |
| Nausea | 69 (18.3%) | 18 (21.7%) | 51 (17.3%) | 0.421 | |
| Vomiting | 23 (6.1%) | 9 (10.8%) | 14 (4.8%) | 0.064 | |
| Diarrhea | 22 (5.8%) | 5 (6.0%) | 17 (5.8%) | 1.000 | |
| AUROC (95% CI) | p Value | Sensitivity (95% CI) | p Value | Specificity (95% CI) | p Value | Reading Time (min) | |
|---|---|---|---|---|---|---|---|
| DL algorithm | 0.861 (0.823–0.894) | NA | 0.583 * (0.471–0.690) | NA | 0.944 * (0.912–0.967) | NA | 13 |
| Session 1 (ED physicians only) | |||||||
| Observer 1 | 0.788 (0.743–0.827) | 0.019 a | 0.595 (0.483–0.701) | 1.000 a | 0.690 (0.634–0.741) | <0.0001 a | 156 |
| Observer 2 | 0.814 (0.771–0.851) | 0.132 a | 0.500 (0.389–0.611) | 0.119 a | 0.974 (0.949–0.989) | 0.093 a | 160 |
| Observer 3 | 0.808 (0.766–0.846) | 0.043 a | 0.500 (0.389–0.611) | 0.065 a | 0.997 (0.982–1.000) | 0.0001 a | 179 |
| Group | 0.788 (0.763–0.811) | 0.017 a | 0.532 (0.468–0.595) | 0.053 a | 0.887 (0.864–0.907) | <0.0001 a | 165 |
| Session 2 (ED physicians with DL algorithm assistance) | |||||||
| Observer 1 | 0.838 (0.798–0.874) | 0.111 b | 0.655 (0.543–0.755) | 0.302 b | 0.954 (0.924–0.975) | <0.0001 b | 97 |
| Observer 2 | 0.807 (0.765–0.846) | 0.801 b | 0.560 (0.447–0.668) | 0.227 b | 1.000 (0.988–1.000) | 0.008 b | 87 |
| Observer 3 | 0.806 (0.763–0.844) | 0.913 b | 0.583 (0.471–0.690) | 0.065 b | 0.990 (0.971–0.998) | 0.625 b | 119 |
| Group | 0.816 (0.793–0.838) | 0.068 b | 0.599 (0.536–0.660) | 0.008 b | 0.981 (0.970–0.989) | <0.0001 b | 101 |
| AUROC (95% CI) | p Value | Sensitivity (95% CI) | p Value | Specificity (95% CI) | p Value | |
|---|---|---|---|---|---|---|
| DL algorithm | 0.940 (0.910–0.962) | NA | 0.817 * (0.696–0.905) | NA | 0·944 * (0·912–0·967) | NA |
| Session 1 (ED physicians only) | ||||||
| Observer 1 | 0.856 (0.816–0.891) | 0.003 a | 0.833 (0.715–0.917) | 1.000 a | 0.690 (0.634–0.741) | <0.0001 a |
| Observer 2 | 0.887 (0.850–0.918) | 0.053 a | 0.700 (0.568–0·812) | 0.119 a | 0.974 (0.949–0.989) | 0.093 a |
| Observer 3 | 0.920 (0.887–0.946) | 0.455 a | 0.683 (0.550–0.797) | 0.022 a | 0.997 (0.982–1.000) | 0.0001 a |
| Group | 0.871 (0.849–0.890) | 0.007 a | 0.739 (0.668–0.801) | 0.034 a | 0.887 (0.864–0.907) | <0.0001 a |
| Session 2 (ED physicians with DL algorithm assistance) | ||||||
| Observer 1 | 0.936 (0.905–0.958) | 0.007 b | 0.867 (0.754–0.941) | 0.774 b | 0.954 (0.924–0.975) | <0.0001 b |
| Observer 2 | 0.907 (0.873–0.935) | 0.412 b | 0.783 (0.658–0.879) | 0.227 b | 1.000 (0.988–1.000) | 0.008 b |
| Observer 3 | 0.907 (0.872–0.934) | 0.609 b | 0.817 (0.696–0.905) | 0.022 b | 0.990 (0.971–0.998) | 0.625 b |
| Group | 0.916 (0.898–0.931) | 0.002 b | 0.822 (0.758–0.875) | 0.014 b | 0.981 (0.970–0.989) | <0.0001 b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Kim, J.Y.; Kim, G.H.; Kang, D.; Kim, I.J.; Seo, J.; Andrews, J.R.; Park, C.M. Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness. J. Clin. Med. 2020, 9, 1981. https://doi.org/10.3390/jcm9061981
Kim JH, Kim JY, Kim GH, Kang D, Kim IJ, Seo J, Andrews JR, Park CM. Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness. Journal of Clinical Medicine. 2020; 9(6):1981. https://doi.org/10.3390/jcm9061981
Chicago/Turabian StyleKim, Jae Hyun, Jin Young Kim, Gun Ha Kim, Donghoon Kang, In Jung Kim, Jeongkuk Seo, Jason R. Andrews, and Chang Min Park. 2020. "Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness" Journal of Clinical Medicine 9, no. 6: 1981. https://doi.org/10.3390/jcm9061981
APA StyleKim, J. H., Kim, J. Y., Kim, G. H., Kang, D., Kim, I. J., Seo, J., Andrews, J. R., & Park, C. M. (2020). Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness. Journal of Clinical Medicine, 9(6), 1981. https://doi.org/10.3390/jcm9061981





