Negative Effect of Reduced NME1 Expression on Recurrence-Free Survival in Early Stage Non-Small Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Method
2.1. Study Population
2.2. Immunohistochemistry
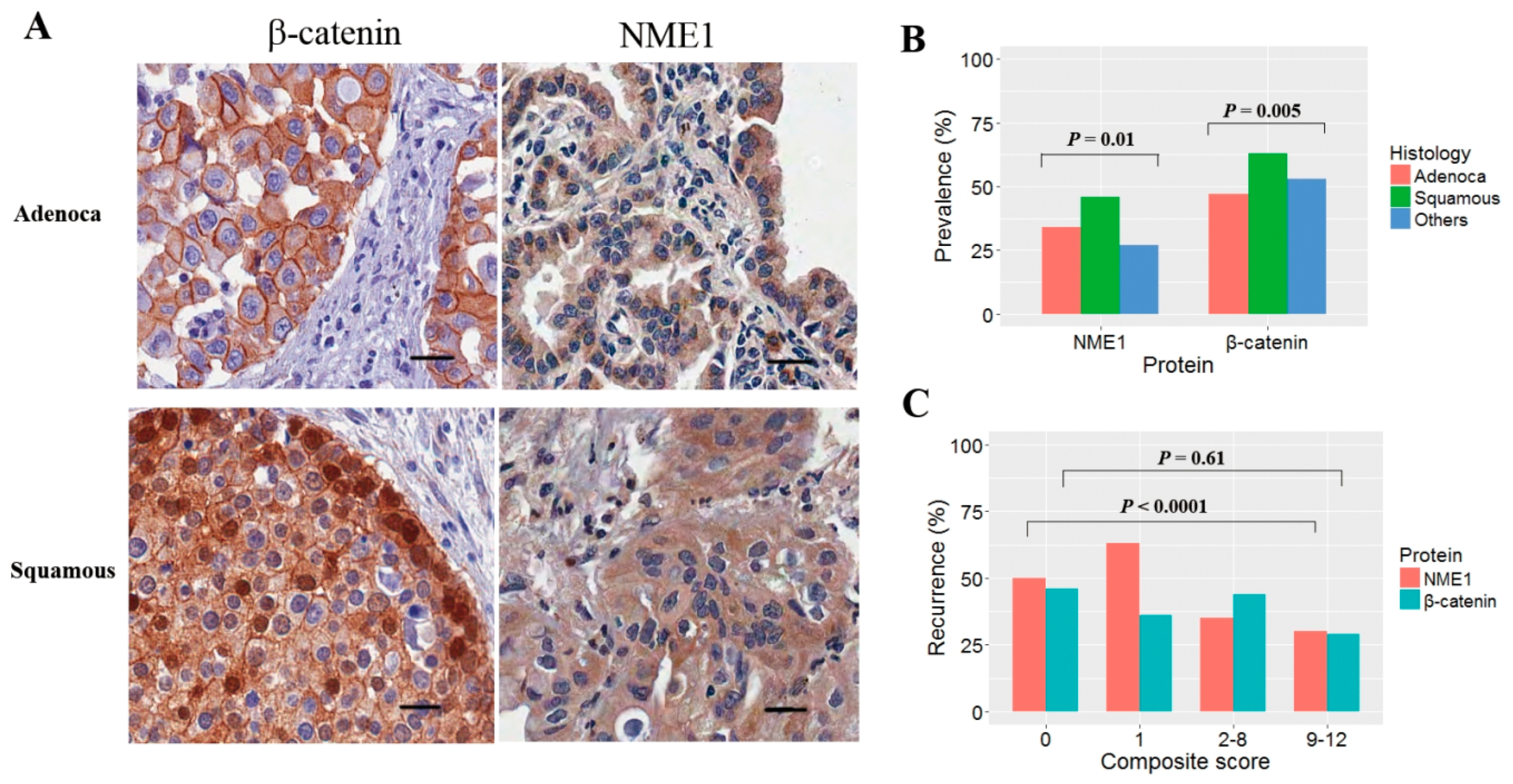
2.3. Interpretation of Immunohistochemical Staining
2.4. Study Design
2.5. Statistical Analyses
3. Results
3.1. Clinicopathological Characteristics
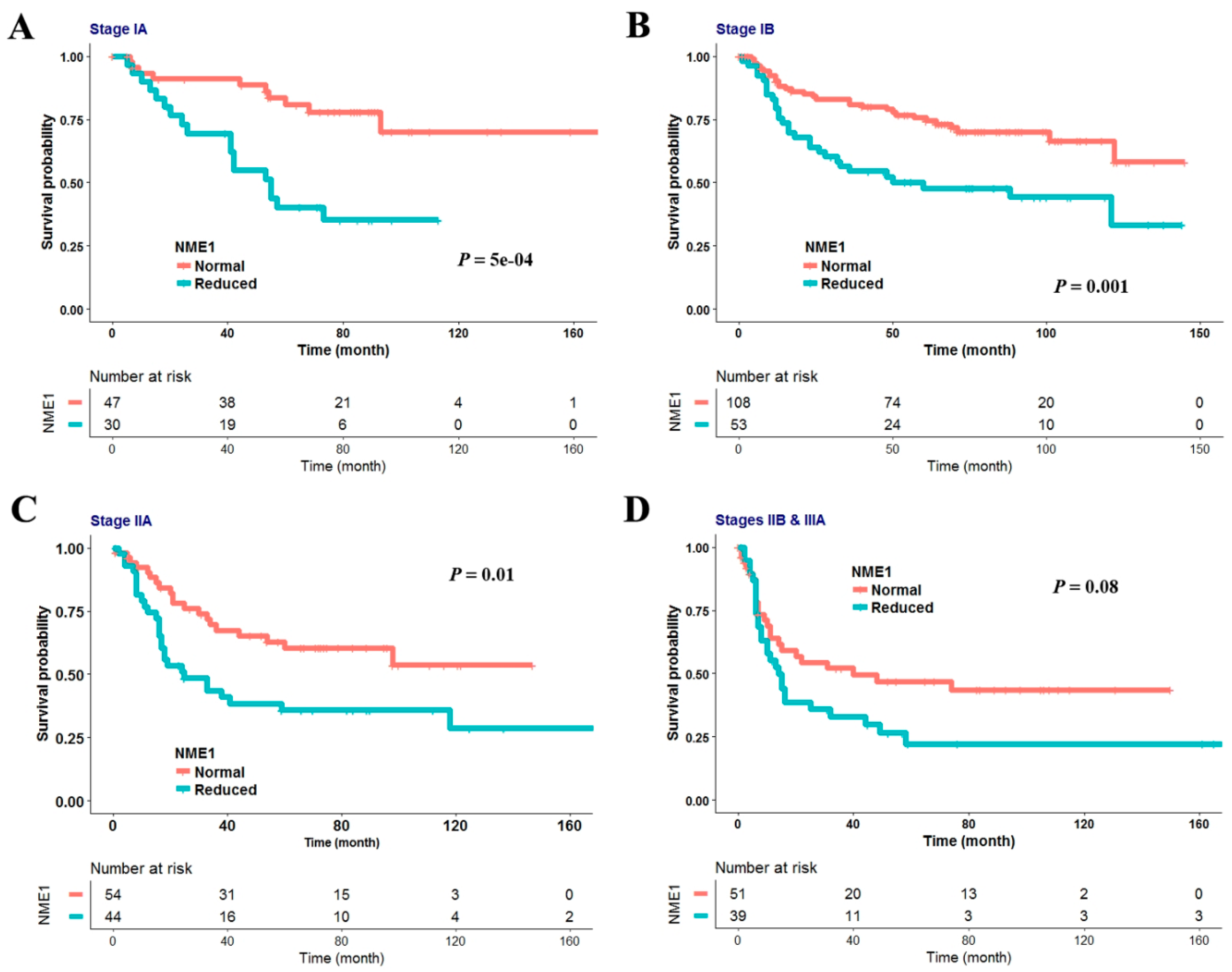
3.2. Reduced NME1 Expression Is Significantly Associated with Poor RFS Irrespective of Histology or Pathologic Stage
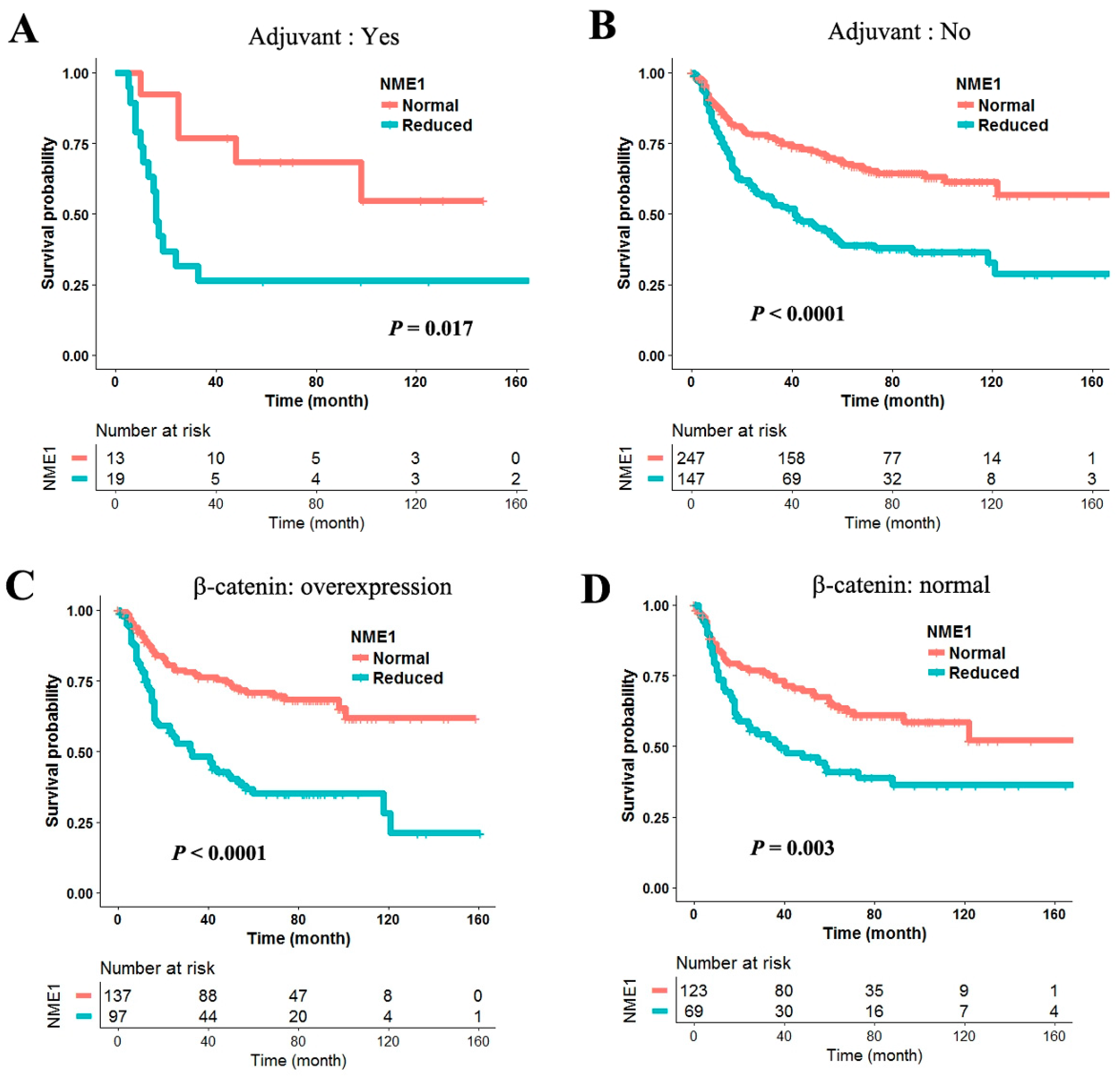
3.3. Negative Effect of Reduced NME1 Expression on RFS in Patients with Cisplatin-Based Adjuvant Chemotherapy and β-Catenin Overexpression
3.4. Multivariate Cox Proportional Hazards Analysis
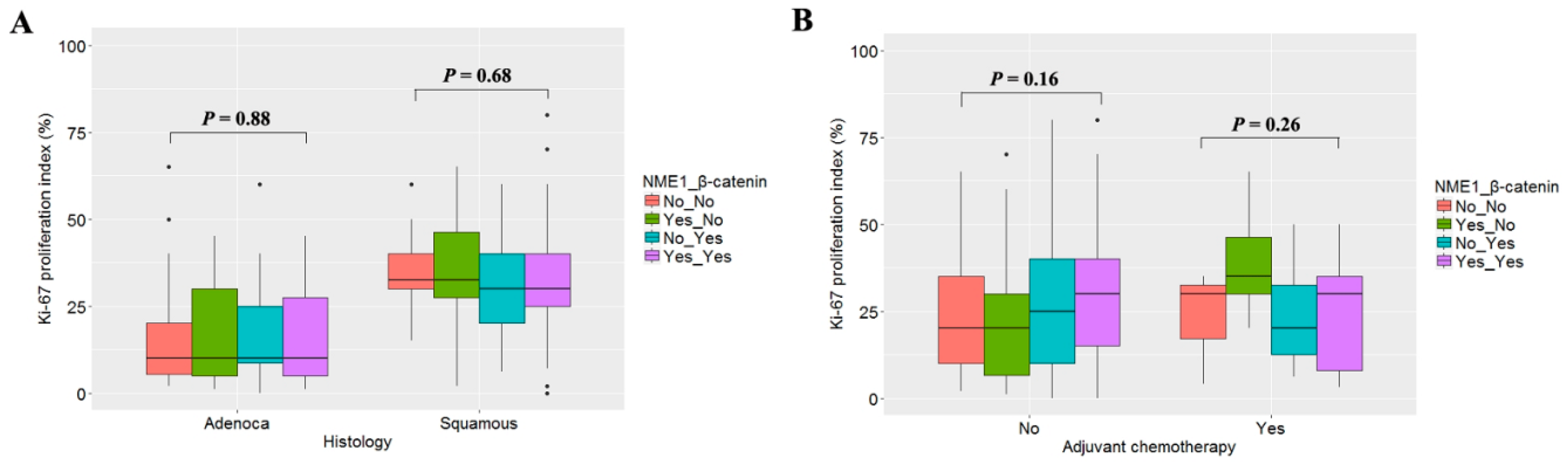
3.5. Relationship between Ki-67 Labeling Index and Expression of NME1 and β-Catenin
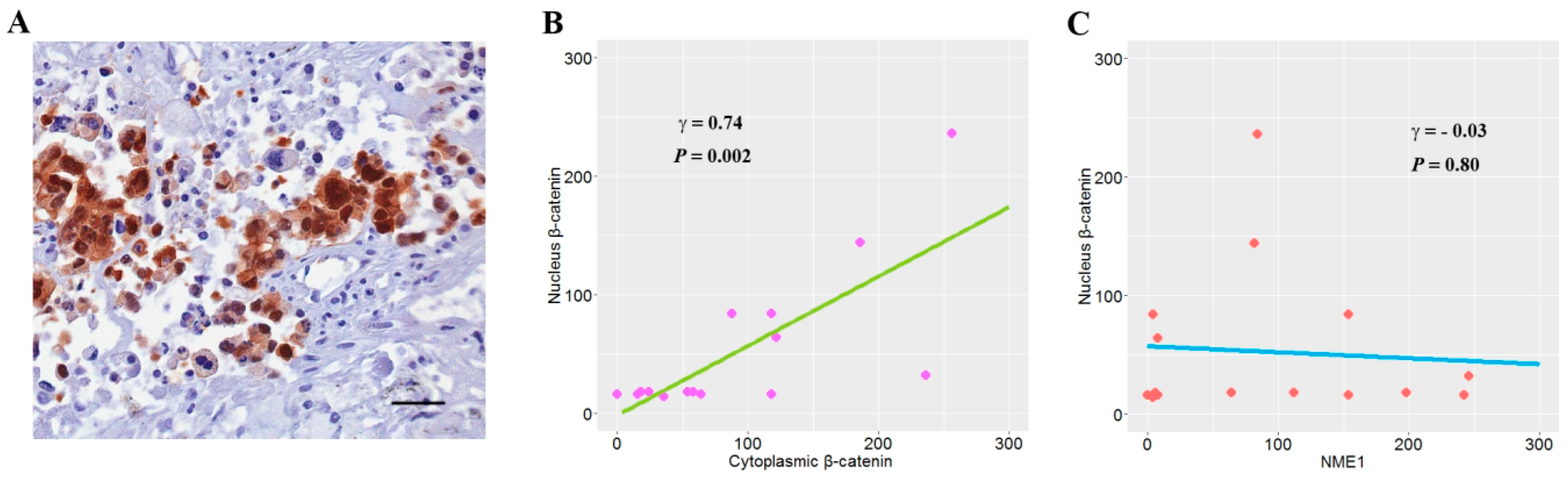
3.6. The Relationship between NME1 and Nuclear β-Catenin Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pignon, J.-P.; Tribodet, H.; Scagliotti, G.V.; Douillard, J.-Y.; Shepherd, F.A.; Stephens, R.J.; Dunant, A.; Torri, V.; Rosell, R.; Seymour, L.; et al. Lung Adjuvant Cisplatin Evaluation: A Pooled Analysis by the LACE Collaborative Group. J. Clin. Oncol. 2008, 26, 3552–3559. [Google Scholar] [CrossRef] [PubMed]
- Arriagada, R.; Dunant, A.; Pignon, J.-P.; Bergman, B.; Chabowski, M.; Grunenwald, D.; Kozlowski, M.; Le Péchoux, C.; Pirker, R.; Pinel, M.-I.S.; et al. Long-Term Results of the International Adjuvant Lung Cancer Trial Evaluating Adjuvant Cisplatin-Based Chemotherapy in Resected Lung Cancer. J. Clin. Oncol. 2010, 28, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S.; Bevilacqua, G.; Kopper, L.; Thorgeirsson, U.P.; Talmadge, J.E.; Liotta, L.A.; Sobel, M.E. Evidence for a Novel Gene Associated with Low Tumor Metastatic Potential. J. Natl. Cancer Inst. 1988, 80, 200–204. [Google Scholar] [CrossRef]
- Boissan, M.; Dabernat, S.; Peuchant, E.; Schlattner, U.; Lascu, I.; Lacombe, M.-L. The mammalian Nm23/NDPK family: From metastasis control to cilia movement. Mol. Cell. Biochem. 2009, 329, 51–62. [Google Scholar] [CrossRef]
- Goncharuk, V.N.; Del-Rosario, A.; Kren, L.; Anwar, S.; E Sheehan, C.; Carlson, J.A.; Ross, J.S. Co-downregulation of PTEN, KAI-1, and nm23-H1 tumor/metastasis suppressor proteins in non-small cell lung cancer. Ann. Diagn. Pathol. 2004, 8, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Ayabe, T.; Tomita, M.; Matsuzaki, Y.; Ninomiya, H.; Hara, M.; Shimizu, T.; Edagawa, M.; Onitsuka, T.; Hamada, M. Micrometastasis and expression of nm23 messenger RNA of lymph nodes from lung cancer and the postoperative clinical outcome. Ann. Thorac. Cardiovasc. Surg. 2004, 10, 152–159. [Google Scholar]
- Sarris, M.; Scolyer, R.A.; Konopka, M.; Thompson, J.F.; Harper, C.G.; Lee, C.S. Cytoplasmic expression of nm23 predicts the potential for cerebral metastasis in patients with primary cutaneous melanoma. Melanoma Res. 2004, 14, 23–27. [Google Scholar] [CrossRef]
- Khan, I.; Gril, B.; Steeg, P.S. Metastasis Suppressors NME1 and NME2 Promote Dynamin 2 Oligomerization and Regulate Tumor Cell Endocytosis, Motility, and Metastasis. Cancer Res. 2019, 79, 4689–4702. [Google Scholar] [CrossRef]
- Khera, L.; Paul, C.; Kaul, R. Hepatitis C Virus E1 protein promotes cell migration and invasion by modulating cellular metastasis suppressor Nm23-H1. Virology 2017, 506, 110–120. [Google Scholar] [CrossRef]
- Guan-Zhen, Y.; Ying, C.; Can-Rong, N.; Guo-Dong, W.; Jian-Xin, Q.; Jie-Jun, W. Reduced protein expression of metastasis-related genes (nm23, KISS1, KAI1 and p53) in lymph node and liver metastases of gastric cancer. Int. J. Exp. Pathol. 2007, 88, 175–183. [Google Scholar] [CrossRef]
- Yang, T.; Chen, B.Z.; Li, D.F.; Wang, H.M.; Lin, X.S.; Wei, H.F.; Zeng, Y.M. Reduced NM23 Protein Level Correlates with Worse Clinicopathologic Features in Colorectal Cancers: A Meta-Analysis of Pooled Data. Medicine (Baltimore) 2016, 95, e2589. [Google Scholar] [PubMed]
- Rasool, R.U.; Nayak, D.; Chakraborty, S.; Jamwal, V.L.; Mahajan, V.; Katoch, A.; Faheem, M.M.; Iqra, Z.; Amin, H.; Gandhi, S.G.; et al. Differential regulation of NM23-H1 under hypoxic and serum starvation conditions in metastatic cancer cells and its implication in EMT. Eur. J. Cell Boil. 2017, 96, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Horak, C.; Mendoza, A.; Vega-Valle, E.; Albaugh, M.; Graff-Cherry, C.; McDermott, W.G.; Hua, E.; Merino, M.J.; Steinberg, S.M.; Khanna, C.; et al. Nm23-H1 Suppresses Metastasis by Inhibiting Expression of the Lysophosphatidic Acid Receptor EDG2. Cancer Res. 2007, 67, 11751–11759. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Steeg, P.S. The relationship of NM23 (NME) metastasis suppressor histidine phosphorylation to its nucleotide diphosphate kinase, histidine protein kinase and motility suppression activities. Oncotarget 2017, 9, 10185–10202. [Google Scholar] [CrossRef]
- Che, G.; Chen, J.; Liu, L.; Wang, Y.; Li, L.; Qin, Y.; Zhou, Q. Transfection of nm23-H1 increased expression of beta-Catenin, E-Cadherin and TIMP-1 and decreased the expression of MMP-2, CD44v6 and VEGF and inhibited the metastatic potential of human non-small cell lung cancer cell line L9981. Neoplasma 2006, 53, 530–537. [Google Scholar]
- You, D.-J.; Park, C.R.; Lee, H.B.; Moon, M.J.; Kang, J.-H.; Lee, C.; Oh, S.-H.; Ahn, C.; Seong, J.Y.; Hwang, J.-I. A Splicing Variant of NME1 Negatively Regulates NF-κB Signaling and Inhibits Cancer Metastasis by Interacting with IKKβ. J. Boil. Chem. 2014, 289, 17709–17720. [Google Scholar] [CrossRef]
- Boissan, M.; De Wever, O.; Lizarraga, F.; Wendum, D.; Poincloux, R.; Chignard, N.; Desbois-Mouthon, C.; Dufour, S.; Nawrocki-Raby, B.; Birembaut, P.; et al. Implication of Metastasis Suppressor NM23-H1 in Maintaining Adherens Junctions and Limiting the Invasive Potential of Human Cancer Cells. Cancer Res. 2010, 70, 7710–7722. [Google Scholar] [CrossRef]
- Hansen, M.T.; Forst, B.; Cremers, N.; Quagliata, L.; Ambartsumian, N.; Grum-Schwensen, B.; Klingelhöfer, J.; Abdul-Al, A.; Herrmann, P.; Osterland, M.; et al. A link between inflammation and metastasis: Serum amyloid A1 and A3 induce metastasis, and are targets of metastasis-inducing S100A4. Oncogene 2014, 34, 424–435. [Google Scholar] [CrossRef]
- He, W.; He, S.; Wang, Z.; Shen, H.; Fang, W.; Zhang, Y.; Qian, W.; Lin, M.; Yuan, J.; Wang, J.; et al. Astrocyte elevated gene-1(AEG-1) induces epithelial-mesenchymal transition in lung cancer through activating Wnt/β-catenin signaling. BMC Cancer 2015, 15, 107. [Google Scholar] [CrossRef]
- Shukla, S.; Sinha, S.; Khan, S.; Kumar, S.; Singh, K.; Mitra, K.; Maurya, R.; Meeran, S.M. Cucurbitacin B inhibits the stemness and metastatic abilities of NSCLC via downregulation of canonical Wnt/β-catenin signaling axis. Sci. Rep. 2016, 6, 21860. [Google Scholar] [CrossRef]
- Lu, J.; Du, C.; Yao, J.; Wu, B.; Duan, Y.; Zhou, L.; Xu, N.; Zhou, F.; Gu, L.; Zhou, H.; et al. C/EBPα Suppresses Lung Adenocarcinoma Cell Invasion and Migration by Inhibiting β-Catenin. Cell. Physiol. Biochem. 2017, 42, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Y.; Li, M.-Y.; Ng, C.S.H.; Yang, S.-L.; Wang, S.S.; Zou, C.; Dong, Y.; Du, J.; Long, X.; et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol. Cancer 2017, 16, 124. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, Z.; Zhang, X.; He, J.; Pan, Y.; Hao, F.; Xie, L.; Li, Q.; Qiu, X.; Wang, E. Inhibition of cytoplasmic GSK 3β increases cisplatin resistance through activation of Wnt/β catenin signaling in A549/DDP cells. Cancer Lett. 2013, 336, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, J.; Li, H.; Wang, J. β-Catenin signaling pathway regulates cisplatin resistance in lung adenocarcinoma cells by upregulating Bcl-xl. Mol. Med. Rep. 2016, 13, 2543–2551. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, J.-W.; Han, J.; Shim, Y.M.; Park, J.; Kim, D.-H. Cohypermethylation of p16 and FHIT Promoters as a Prognostic Factor of Recurrence in Surgically Resected Stage I Non–Small Cell Lung Cancer. Cancer Res. 2006, 66, 4049–4054. [Google Scholar] [CrossRef][Green Version]
- Edge, S.B.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.L.; Troth, A. American Joint Committee on Cancer. In AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010; pp. 253–270. [Google Scholar]
- Kim, J.S.; Han, J.; Shim, Y.M.; Park, J.; Kim, D.H. Aberrant methylation of H-cadherin (CDH13) promoter is associated with tumor progression in primary nonsmall cell lung cancer. Cancer 2005, 104, 1825–1833. [Google Scholar]
- MacKinnon, M. p53, c-erbB-2 and nm23 expression have no prognostic significance in primary pulmonary adenocarcinoma. Eur. J. Cardio-Thoracic Surg. 1997, 11, 838–842. [Google Scholar] [CrossRef]
- Tomita, M.; Ayabe, T.; Matsuzaki, Y.; Onitsuka, T. Immunohistochemical analysis of nm23-H1 gene product in node-positive lung cancer and lymph nodes. Lung Cancer 1999, 24, 11–16. [Google Scholar] [CrossRef]
- Yang, M.; Sun, Y.; Sun, J.; Wang, Z.; Zhou, Y.; Yao, G.; Gu, Y.; Zhang, H.; Zhao, H. Differentially expressed and survival-related proteins of lung adenocarcinoma with bone metastasis. Cancer Med. 2018, 7, 1081–1092. [Google Scholar] [CrossRef]
- Ohta, Y.; Nozawa, H.; Tanaka, Y.; Oda, M.; Watanabe, Y. Increased vascular endothelial growth factor and vascular endothelial growth factor–c and decreased nm23 expression associated with microdissemination in the lymph nodes in stage i non–small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2000, 119, 804–813. [Google Scholar] [CrossRef]
- Katakura, H.; Tanaka, F.; Oyanagi, H.; Miyahara, R.; Yanagihara, K.; Otake, Y.; Wada, H. Clinical significance of nm23 expression in resected pathologic-stage I, non-small cell lung cancer. Ann. Thorac. Surg. 2002, 73, 1060–1064. [Google Scholar] [CrossRef]
- Liu, C.; Liu, J.; Wang, X.; Mao, W.; Jiang, L.; Ni, H.; Mo, M.; Wang, W. Prognostic impact of nm23-H1 and PCNA expression in pathologic stage I non-small cell lung cancer. J. Surg. Oncol. 2011, 104, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Otsuki, Y.; Tanaka, M.; Yoshii, S.; Kawazoe, N.; Nakaya, K.; Sugimura, H. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc. Natl. Acad. Sci. USA 2001, 98, 4385–4390. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, E.; Ota, T.; Tsukuda, K.; Okita, A.; Matsuoka, K.; Murakami, M.; Doihara, H.; Shimizu, N. nm23-H1 reduces in vitro cell migration and the liver metastatic potential of colon cancer cells by regulating myosin light chain phosphorylation. Int. J. Cancer 2004, 108, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Meneses, P.I.; Lan, K.; Robertson, E.S. The suppressor of metastasis Nm23-H1 interacts with the Cdc42 Rho family member and the pleckstrin homology domain of oncoprotein Dbl-1 to suppress cell migration. Cancer Boil. Ther. 2008, 7, 677–688. [Google Scholar] [CrossRef]
- You, J.; Chang, R.; Liu, B.; Zu, L.; Zhou, Q. Nm23-H1 was involved in regulation of KAI1 expression in high-metastatic lung cancer cells L9981. J. Thorac. Dis. 2016, 8, 1217–1226. [Google Scholar] [CrossRef]
- Leonard, M.K.; Novak, M.; Snyder, D.; Snow, G.; Pamidimukkala, N.; McCorkle, J.R.; Yang, X.H.; Kaetzel, D. The metastasis suppressor NME1 inhibits melanoma cell motility via direct transcriptional induction of the integrin beta-3 gene. Exp. Cell Res. 2019, 374, 85–93. [Google Scholar] [CrossRef]
- Subramanian, C.; Robertson, E.S. The Metastatic Suppressor Nm23-H1 Interacts with EBNA3C at Sequences Located between the Glutamine- and Proline-Rich Domains and Can Cooperate in Activation of Transcription. J. Virol. 2002, 76, 8702–8709. [Google Scholar] [CrossRef][Green Version]
- Curtis, C.D.; Likhite, V.S.; McLeod, I.X.; Yates, J.R.; Nardulli, A.M. Interaction of the Tumor Metastasis Suppressor Nonmetastatic Protein 23 Homologue H1 and Estrogen Receptor Alters Estrogen-Responsive Gene Expression. Cancer Res. 2007, 67, 10600–10607. [Google Scholar] [CrossRef]
- Horak, C.; Lee, J.H.; Elkahloun, A.G.; Boissan, M.; Dumont, S.; Maga, T.K.; Arnaud-Dabernat, S.; Palmieri, D.; Stetler-Stevenson, W.G.; Lacombe, M.-L.; et al. Nm23-H1 Suppresses Tumor Cell Motility by Down-regulating the Lysophosphatidic Acid Receptor EDG2. Cancer Res. 2007, 67, 7238–7246. [Google Scholar] [CrossRef]
- Choudhuri, T.; Murakami, M.; Kaul, R.; Sahu, S.K.; Mohanty, S.; Verma, S.; Kumar, P.; Robertson, E.S. Nm23-H1 can induce cell cycle arrest and apoptosis in B cells. Cancer Boil. Ther. 2010, 9, 1065–1078. [Google Scholar] [CrossRef]
- Ferguson, A.W.; Flatow, U.; Macdonald, N.J.; Larminat, F.; A Bohr, V.; Steeg, P.S. Increased sensitivity to cisplatin by nm23-transfected tumor cell lines. Cancer Res. 1996, 56, 2931–2935. [Google Scholar]
- Ma, D.; McCorkle, J.R.; Kaetzel, D. The Metastasis Suppressor NM23-H1 Possesses 3′-5′ Exonuclease Activity. J. Boil. Chem. 2004, 279, 18073–18084. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-F.; Chang, C.-J.; Chiu, J.-H.; Lin, C.-P.; Li, W.-Y.; Chang, S.-Y.; Chu, P.-Y.; Tai, S.-K.; Chen, Y.-J. NM23-H1 expression of head and neck squamous cell carcinoma in association with the response to cisplatin treatment. Oncotarget 2014, 5, 7392–7405. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Jin, H.; Peng, M.; Li, B.; She, M.; Zhu, T.; Wen, S.; Qin, D.-C. Association between NME1 polymorphisms and cancer susceptibility: A meta-analysis based on 1644 cases and 2038 controls. Pathol. Res. Pract. 2018, 214, 467–474. [Google Scholar] [CrossRef] [PubMed]
| Adjuvant Chemotherapy | β-Catenin | Reduced NME1 | HR | 95% CI | p-Value |
|---|---|---|---|---|---|
| Overexpression | Expression | ||||
| No | No | No | 1 | ||
| (N = 176) | Yes | 1.89 | 1.21–2.95 | 0.005 | |
| Yes | No | 1 | |||
| (N = 217) | Yes | 2.54 | 1.66–3.89 | <0.0001 | |
| Yes | No | No | 1 | ||
| (N = 16) | Yes | 5.59 | 0.62–50.91 | 0.13 | |
| Yes | No | 1 | |||
| (N = 16) | Yes | 15.52 | 2.94–82.38 | 0.001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Kim, Y.; Lee, B.B.; Kim, D.; Lee, O.-J.; Jeong, P.; Kim, W.-J.; Cho, E.Y.; Han, J.; Shim, Y.M.; et al. Negative Effect of Reduced NME1 Expression on Recurrence-Free Survival in Early Stage Non-Small Cell Lung Cancer. J. Clin. Med. 2020, 9, 3067. https://doi.org/10.3390/jcm9103067
Kim D, Kim Y, Lee BB, Kim D, Lee O-J, Jeong P, Kim W-J, Cho EY, Han J, Shim YM, et al. Negative Effect of Reduced NME1 Expression on Recurrence-Free Survival in Early Stage Non-Small Cell Lung Cancer. Journal of Clinical Medicine. 2020; 9(10):3067. https://doi.org/10.3390/jcm9103067
Chicago/Turabian StyleKim, Dohun, Yujin Kim, Bo Bin Lee, Dongho Kim, Ok-Jun Lee, Pildu Jeong, Wun-Jae Kim, Eun Yoon Cho, Joungho Han, Young Mog Shim, and et al. 2020. "Negative Effect of Reduced NME1 Expression on Recurrence-Free Survival in Early Stage Non-Small Cell Lung Cancer" Journal of Clinical Medicine 9, no. 10: 3067. https://doi.org/10.3390/jcm9103067
APA StyleKim, D., Kim, Y., Lee, B. B., Kim, D., Lee, O.-J., Jeong, P., Kim, W.-J., Cho, E. Y., Han, J., Shim, Y. M., & Kim, D.-H. (2020). Negative Effect of Reduced NME1 Expression on Recurrence-Free Survival in Early Stage Non-Small Cell Lung Cancer. Journal of Clinical Medicine, 9(10), 3067. https://doi.org/10.3390/jcm9103067





