Diabetic Retinopathy, lncRNAs, and Inflammation: A Dynamic, Interconnected Network
Abstract
1. Introduction
2. Inflammation and Diabetic Retinopathy (DR)
2.1. NF-κB
2.2. Cytokines and Chemokines
2.3. Complement System
3. LncRNAs: Novel, Emerging, Regulatory RNA Molecules
4. LncRNAs and DR
5. LncRNAs as Novel Regulators of Inflammation in DR
5.1. MALAT1
5.2. MIAT
5.3. ANRIL
5.4. H19
5.5. BDNF-AS
5.6. MEG3
5.7. RNCR3
5.8. HOTTIP
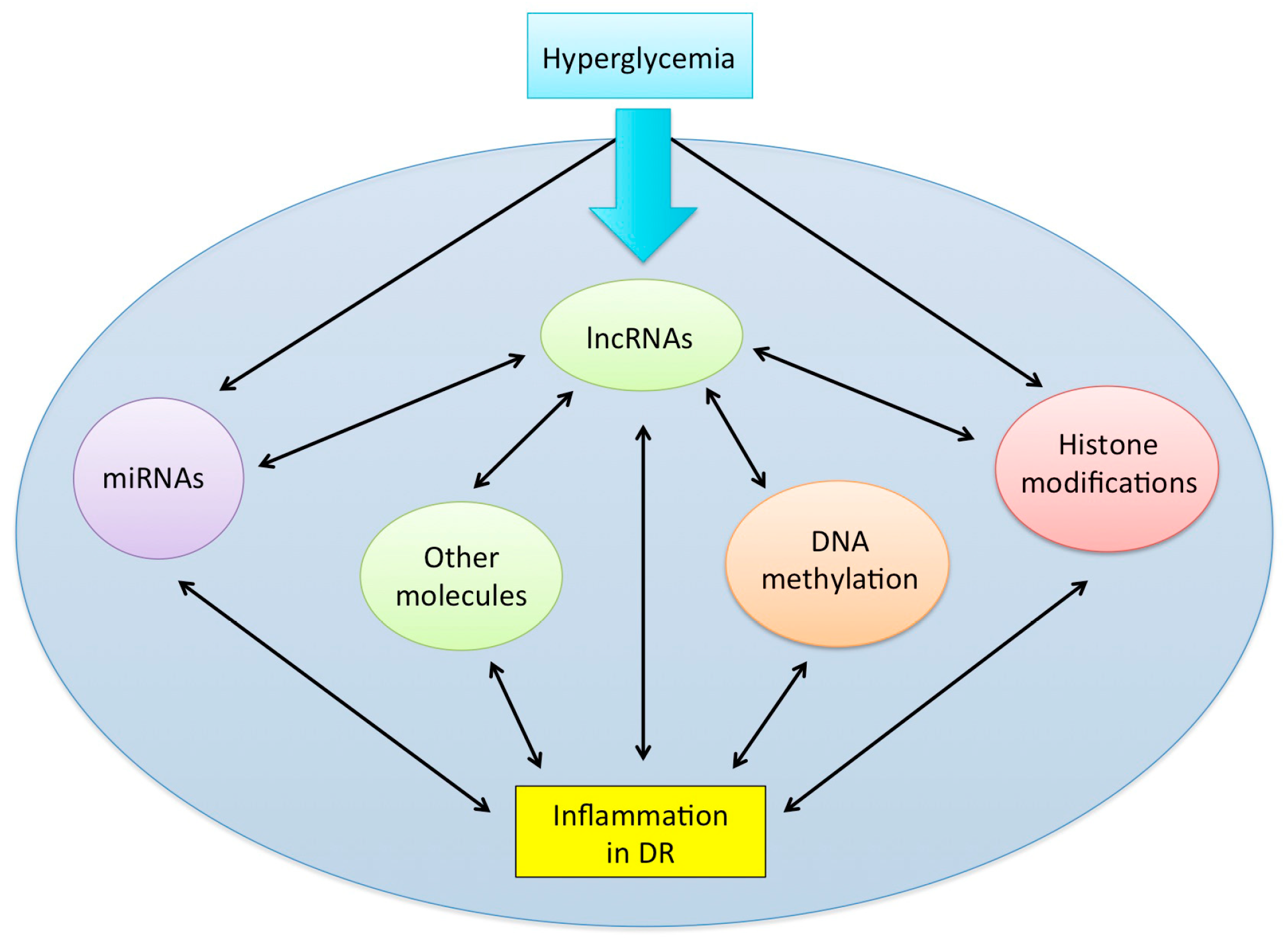
6. Other Epigenetic Players Involved in the Cross-Talk between lncRNAs and Inflammation: The Missing Puzzle Pieces?
6.1. DNA Methylation
6.2. Histone Modifications
6.3. miRNAs
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chan, J.C.N.; Malik, V.; Jia, W.; Kadowaki, T.; Yajnik, C.S.; Yoon, K.; Hu, F. Diabetes in Asia: Epidemiology, risk factors, and pathophysiology. JAMA 2009, 301, 2129–2140. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.E.; Sicree, R.A.; Zimmet, P.Z. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res. Clin. Pract. 2010, 87, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Werfalli, M.; Engel, M.E.; Musekiwa, A.; Kengne, A.P.; Levitt, N.S. The prevalence of type 2 diabetes among older people in Africa: A systematic review. Lancet Diabetes Endocrinol. 2016, 4, 72–84. [Google Scholar] [CrossRef]
- Yoon, K.H.; Lee, J.H.; Kim, J.W.; Cho, J.H.; Choi, Y.H.; Ko, S.H.; Zimmet, P.; Son, H.Y. Epidemic obesity and type 2 diabetes in Asia. Lancet 2006, 368, 1681–1688. [Google Scholar] [CrossRef]
- Sattar, N.; Gill, J.M. Type 2 diabetes in migrant south Asians: Mechanisms, mitigation, and management. Lancet Diabetes Endocrinol. 2015, 3, 1004–1016. [Google Scholar] [CrossRef]
- Leasher, J.L.; Bourne, R.A.A.; Flaxman, S.R.; Jonas, J.B.; Keefe, J.; Kovin, N.; Pesudovs, K.; Price, H.; White, R.A.; Wong, T.Y.; et al. Global estimates on the number of people blind or visually impaired by diabetic retinopathy: A meta-analysis from 1990 to 2010. Diabetes Care 2016, 39, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Yau, J.W.Y.; Rogers, S.L.; Kawasaki, R.; Lamoureux, E.L.; Kowalski, J.W.; Bek, T.; Chen, S.; Dekker, J.M.; Fletcher, A.; Grauslund, J.; et al. For the Meta-analysis for Eye Disease (META-EYE) Study Group. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care 2012, 35, 556–564. [Google Scholar] [CrossRef]
- Nentwich, M.M.; Ulbig, M.W. Diabetic retinopathy—Ocular complications of diabetes mellitus. World J. Diabetes 2015, 6, 489–499. [Google Scholar] [CrossRef]
- Prokofyeva, E.; Zrenner, E. Epidemiology of major eye diseases leading to blindness in Europe: A literature review. Ophthalmic Res. 2012, 7, 171–188. [Google Scholar] [CrossRef]
- Flaxman, S.R.; Bourne, R.R.A.; Resnikoff, S.; Ackland, P.; Braithwaite, T.; Cicinelli, M.V.; Das, A.; Jonas, J.B.; Keeffe, J.; Kempen, J.H.; et al. Vision Loss Expert Group of the Global Burden of Disease Study. Global causes of blindness and distance vision impairment 1990-2020: A systematic review and meta-analysis. Lancet Glob. Health 2017, 5, 1221–1234. [Google Scholar] [CrossRef]
- Wong, T.Y.; Mwamburi, M.; Klein, R.; Larsen, M.; Flynn, H.; Hernandez-Medina, M.; Ranganathan, G.; Wirostko, B.; Pleil, A.; Mitchell, P. Rates of progression in diabetic retinopathy during different time periods: A systematic review and meta-analysis. Diabetes Care 2009, 32, 2307–2313. [Google Scholar] [CrossRef] [PubMed]
- The Diabetes Control and Complications Trial Research Group. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 1993, 329, 977–986. [Google Scholar] [CrossRef] [PubMed]
- UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet 1998, 352, 837–853. [Google Scholar] [CrossRef]
- Tan, G.S.; Cheung, N.; Simo, R.; Cheung, G.C.; Wong, T.Y. Diabetic macular oedema. Lancet Diabetes Endocrinol. 2017, 5, 143–155. [Google Scholar] [CrossRef]
- Antonetti, D.A.; Klein, R.; Gardner, T.W. Diabetic retinopathy. N. Engl. J. Med. 2012, 366, 1227–1239. [Google Scholar] [CrossRef]
- Keenan, H.A.; Costacou, T.; Sun, J.K.; Doria, A.; Cavellerano, J.; Coney, J.; King, G.L. Clinical factors associated with resistance to microvascular complications in diabetic patients of extreme disease duration. Diabetes Care 2007, 30, 1995–1997. [Google Scholar] [CrossRef] [PubMed]
- Fong, D.S.; Aiello, L.; Gardner, T.W.; Kin, G.L.; Blankenship, G.; Cavallerano, J.D.; Klein, R. Retinopathy in diabetes. Diabetes Care 2004, 27, S84–S87. [Google Scholar] [CrossRef]
- Aiello, L.P.; Gardner, T.W.; King, G.L.; Blankenship, G.; Cavallerano, J.D.; Ferris, R.L.; Klein, R. Diabetic retinopathy. Diabetes Care 1998, 21, 143–156. [Google Scholar] [CrossRef]
- Kristinsson, J.K. Diabetic retinopathy. Screening and prevention of blindness. A doctoral thesis. Acta Ophthalmol. Scand. Suppl. 1997, 223, 1–76. [Google Scholar]
- Klein, R.; Klein, B.E.; Moss, S.E.; Davis, M.D.; DeMets, D.L. The Wisconsin epidemiologic study of diabetic retinopathy. III. Prevalence and risk of diabetic retinopathy when age at diagnosis is 30 or more years. Arch. Ophthalmol. 1984, 102, 527–532. [Google Scholar] [CrossRef]
- Stefánsson, E.; Bek, T.; Porta, M.; Larsen, N.; Kristinsson, J.K.; Agardh, E. Screening and prevention of diabetic blindness. Acta Ophthalmol. Scand. 2000, 78, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.J.; Maguire, J.I.; Regillo, C.D.; Spirn, M.J.; Tasman, W. The Wills Eye Manual: Office and Emergency Room Diagnosis and Treatment of Eye Disease, 7th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2017; pp. 610–621. [Google Scholar]
- Miller, J.W.; Adamis, A.P.; Shima, D.T.; D’Amore, P.A.; Moulton, R.S.; O’Reilly, M.S.; Folkman, J.; Dvorak, H.F.; Brown, L.F.; Berse, B.; et al. Vascular endothelial growth factor/vascular permeability factor is temporally and spatially correlated with ocular angiogenesis in a primate model. Am. J. Pathol. 1994, 145, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Davis-Smyth, T. The biology of vascular endothelial growth factor. Endocr. Rev. 1997, 18, 4–25. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.A.; Kermany, D.S.; Waters, J.; Jansen, M.E.; Tyler, L. Diabetic macular edema: It is more than just VEGF. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Adamis, A.P. Is diabetic retinopathy an inflammatory disease? Br. J. Ophthalmol. 2002, 86, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Kern, T.S. Contributions of Inflammatory Processes to the Development of the Early Stages of Diabetic Retinopathy. Exp. Diabetes Res. 2007, 2007, 95103. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Sarthy, V.; Kern, T. Interaction between NO and COX pathways in retinal cells exposed to elevated glucose and retina of diabetic rats. Am. J. Physiol. 2004, 287, R735–R741. [Google Scholar] [CrossRef] [PubMed]
- Ellis, E.A.; Guberski, D.L.; Hutson, B.; Grant, M.B. Time course of NADH oxidase, inducible nitric oxide synthase and peroxynitrite in diabetic retinopathy in the BBZ/WOR rat. Nitric Oxide 2002, 6, 295–304. [Google Scholar] [CrossRef]
- Du, Y.; Smith, M.A.; Miller, C.M.; Kern, T.S. Diabetes-induced nitrative stress in the retina, and correction by aminoguanidine. J. Neurochem. 2002, 80, 771–779. [Google Scholar] [CrossRef]
- Zheng, L.; Szabo, C.; Kern, T.S. Poly(ADP-ribose) polymerase is involved in the development of diabetic retinopathy via regulation of nuclear factor-B. Diabetes 2004, 53, 2960–2967. [Google Scholar] [CrossRef]
- Miyamoto, K.; Khosrof, S.; Bursell, S.E.; Rohan, R.; Murata, T.; Clermont, A.C.; Aiello, L.P.; Ogura, Y.; Adamis, A.P. Prevention of leukostasis and vascular leakage in streptozotocin-induced diabetic retinopathy via intercellular adhesion molecule-1 inhibition. Proc. Natl. Acad. Sci. USA 1999, 96, 10836–10841. [Google Scholar] [CrossRef] [PubMed]
- Joussen, A.M.; Poulaki, V.; Qin, W.; Kirchhof, B.; Mitsiades, N.; Wiegand, S.J.; Rudge, J.; Yancopoulos, G.D.; Adamis, A.P. Retinal vascular endothelial growth factor induces intercellular adhesion molecule-1 and endothelial nitric oxide synthase expression and initiates early diabetic retinal leukocyte adhesion in vivo. Am. J. Pathol. 2002, 160, 501–509. [Google Scholar] [CrossRef]
- Kowluru, R.A.; Odenbach, S. Role of interleukin-1beta in the development of retinopathy in rats: Effect of antioxidants. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4161–4166. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A.; Odenbach, S. Role of interleukin-1beta in the pathogenesis of diabetic retinopathy. Br. J. Ophthalmol. 2004, 88, 1343–1347. [Google Scholar] [CrossRef] [PubMed]
- Carmo, A.; Cunha-Vaz, J.G.; Carvalho, A.P.; Lopes, M.C. L-arginine transport in retinas from streptozotocin diabetic rats: Correlation with the level of IL-1 beta and NO synthase activity. Vis. Res. 1999, 39, 3817–3823. [Google Scholar] [CrossRef]
- Gerhardinger, C.; Costa, M.B.; Coulombe, M.C.; Toth, I.; Hoehn, T.; Grosu, P. Expression of acute-phase response proteins in retinal Muller cells in diabetes. Investig. Ophthalmol. Vis. Sci. 2005, 46, 349–357. [Google Scholar] [CrossRef]
- Joussen, A.M.; Huang, S.; Poulaki, V.; Camphausen, K.; Beecken, W.D.; Kirchhof, B.; Adamis, A.P. In vivo retinal gene expression in early diabetes. Investig. Ophthalmol. Vis. Sci. 2001, 42, 3047–3057. [Google Scholar]
- Du, X.; Stocklauser-Farber, K.; Rosen, P. Generation of reactive oxygen intermediates, activation of NF- kappaB, and induction of apoptosis in human endothelial cells by glucose: Role of nitric oxide synthase? Free Radic. Biol. Med. 1999, 27, 752–763. [Google Scholar] [CrossRef]
- Romeo, G.; Liu, W.; Asnaghi, V.; Kern, T.S.; Lorenzi, M. Activation of nuclear factor-kappa B induced by diabetes and high glucose regulates a proapoptotic program in retinal pericytes. Diabetes 2002, 51, 2241–2248. [Google Scholar] [CrossRef]
- Nagai, N.; Izumi-Nagai, K.; Oike, Y.; Koto, T.; Satofuka, S.; Ozawa, Y.; Yamashiro, K.; Inoue, M.; Tsubota, K.; Umezawa, K.; et al. Suppression of Diabetes-Induced Retinal Inflammation by Blocking the Angiotensin II Type 1 Receptor or Its Downstream Nuclear Factor- B Pathway. Retin. Cell Biol. 2007, 48, 4342–4350. [Google Scholar] [CrossRef]
- Zheng, L.; Howell, S.J.; Hatala, D.A.; Huang, K.; Kern, T.S. Salicylate-Based Anti-Inflammatory Drugs Inhibit the Early Lesion of Diabetic Retinopathy. Diabetes 2007, 56, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A.; Koppolu, P.; Chakrabarti, S.; Chen, S. Diabetes-induced Activation of Nuclear Transcriptional Factor in the Retina, and its Inhibition by Antioxidants. Free Radic. Res. 2003, 37, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Boss, J.D.; Singh, P.K.; Pandya, H.K.; Tosi, J.; Kim, C.; Tewari, A.; Juzych, M.S.; Abrams, G.W.; Kumar, A. Assessment of neurotrophins and inflammatory mediators in vitreous of patients with diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5594–5603. [Google Scholar] [CrossRef] [PubMed]
- Yuukia, T.; Kandab, T.; Kimuraa, Y.; Kotajimac, N.; Tamurac, J.; Kobayashic, I.; Kishia, S. Inflammatory cytokines in vitreous fluid and serum of patients with diabetic vitreoretinopathy. J. Diabetes Complicat. 2001, 15, 257–259. [Google Scholar] [CrossRef]
- Doganay, S.; Evereklioglu, C.; Er, H.; Türköz, Y.; Sevinç, A.; Mehmet, N.; Savli, H. Comparison of serum NO, TNF-alpha, IL-1beta, sIL-2R, IL-6 and IL-8 levels with grades of retinopathy in patients with diabetes mellitus. Eye (Lond) 2002, 16, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Hwang, D.; Song, X.; Tao, Y. Association between Aqueous Cytokines and Diabetic Retinopathy Stage. J. Ophthalmol. 2017, 2017, 9402198. [Google Scholar] [CrossRef]
- Mastropasqua, R.; D’Aloisio, R.; Di Nicola, M.; Di Martino, G.; Lamolinara, A.; Di Antonio, L.; Tognetto, D.; Toto, L. Relationship between aqueous humor cytokine level changes and retinal vascular changes after intravitreal aflibercept for diabetic macular edema. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Scuderi, S.; D’amico, A.G.; Federico, C.; Saccone, S.; Magro, G.; Bucolo, C.; Drago, F.; D’Agata, V. Different retinal expression patterns of IL-1α, IL-1β, and their receptors in a rat model of type 1 STZ-induced diabetes. J. Mol. Neurosci. 2015, 56, 431–439. [Google Scholar] [CrossRef]
- Endo, H.; Naito, T.; Asahara, T.; Kajima, M.; Shiota, H. Cytokines in the vitreous fluid of patients with proliferative diabetic retinopathy—Vascular endothelial growth factor and platelet-derived growth factor are elevated in proliferative diabetic retinopathy. Nippon Ganka Gakkai Zasshi 2000, 104, 711–716. [Google Scholar] [CrossRef]
- Voronov, E.; Shouval, D.S.; Krelin, Y.; Cagnano, E.; Benharroch, D.; Iwakura, Y.; Dinarello, C.A.; Apte, R.N. IL-1 is required for tumor invasiveness and angiogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 2645–2650. [Google Scholar] [CrossRef]
- Leali, D.; Dell’Era, P.; Stabile, H.; Sennino, B.; Chambers, A.F.; Naldini, A.; Sozzani, S.; Nico, B.; Ribatti, D.; Presta, M. Osteopontin (ETA-1) and fibroblast growth factor-2 cross-talk in angiogenesis. J. Immunol. 2003, 171, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Busik, J.V.; Mohr, S.; Grant, M.B. Hyperglycemia-induced reactive oxygen species toxicity to endothelial cells is dependent on paracrine mediators. Diabetes 2008, 57, 1952–1965. [Google Scholar] [CrossRef] [PubMed]
- Aplin, A.C.; Gelati, M.; Fogel, E.; Carnevale, E.; Nicosia, R.F. Angiopoietin-1 and vascular endothelial growth factor induce expression of inflammatory cytokines before angiogenesis. Physiol. Genom. 2006, 27, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Esselman, W.J.; Jump, D.B.; Busik, J.V. Anti-inflammatory effect of docosahexaenoic acid on cytokine-induced adhesion molecule expression in human retinal vascular endothelial cells. Investig. Ophthalmol. Vis. Sci. 2005, 46, 4342–4347. [Google Scholar] [CrossRef] [PubMed]
- Valle, A.; Giamporcaro, G.M.; Scavini, M.; Stabilini, A.; Grogan, P.; Bianconi, E.; Sebastiani, G.; Masini, M.; Maugeri, N.; Porretti, L.; et al. Reduction of circulating neutrophils precedes and accompanies type 1 diabetes. Diabetes 2013, 62, 2072–2077. [Google Scholar] [CrossRef] [PubMed]
- Stitt, A.W.; Curtis, T.M.; Chen, M.; Medina, R.J.; McKay, G.J.; Jenkins, A.; Gardiner, T.A.; Lyons, T.J.; Hammes, H.P.; Simo, R.; et al. The progress in understanding and treatment of diabetic retinopathy. Prog. Retin. Eye Res. 2016, 51, 156–186. [Google Scholar] [CrossRef] [PubMed]
- Engerman, R.L. Pathogenesis of diabetic retinopathy. Diabetes 1989, 38, 1203–1206. [Google Scholar] [CrossRef]
- Durham, J.T.; Herman, I.M. Microvascular modifications in diabetic retinopathy. Curr. Diab. Rep. 2001, 11, 25–264. [Google Scholar] [CrossRef]
- Imai, H.; Singh, R.S.; Fort, P.E.; Gardner, T.W. Neuroprotection for diabetic retinopathy. Dev. Ophthalmol. 2009, 44, 56–68. [Google Scholar] [CrossRef]
- Lange, C.; Storkebaum, E.; de Almodovar, C.R.; Dewerchin, M.; Carmeliet, P. Vascular endothelial growth factor: A neurovascular target in neurological diseases. Nat. Rev. Neurol. 2016, 12, 439–454. [Google Scholar] [CrossRef]
- Gilbert, R.E.; Vranes, D.; Berka, J.L.; Kelly, D.J.; Cox, A.; Wu, L.L.; Stacker, S.A.; Cooper, M.E. Vascular endothelial growth factor and its receptors in control and diabetic rat eyes. Lab. Investig. 1998, 78, 1017–1027. [Google Scholar] [PubMed]
- Gerl, V.; Bohl, J.; Pitz, S.; Stoffelns, B. Extensive deposits of complement C3d and C5b-9 in the choriocapillaris of eyes of patients with diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1104–1108. [Google Scholar]
- Zhang, J.; Gerhardinger, C.; Lorenzi, M. Early complement activation and decreased levels of glycosylphosphatidylinositol-anchored complement inhibitors in human and experimental diabetic retinopathy. Diabetes 2002, 51, 3499–3504. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Goldfine, A.; Krumrei, N.; Grubissich, L.; Acosta, J.; Chorey, M.; Hays, A.P.; Halperin, J.A. Glycation inactivation of the complement regulatory protein CD59: A possible role in the pathogenesis of the vascular complications of human diabetes. Diabetes 2004, 53, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrovskii, Y.A. Antithrombin III, C1 inhibitor, methylglyoxal, and poly- morphonuclear leukocytes in the development of vascular complications in diabetes mellitus. Thromb. Res. 1992, 67, 179–189. [Google Scholar] [CrossRef]
- Gao, B.B.; Chen, X.; Timothy, N.; Aiello, L.P.; Feener, E.P. Characterization of the Vitreous Proteome in Diabetes without Diabetic Retinopathy and Diabetes with Proliferative Diabetic Retinopathy. J. Proteome Res. 2008, 7, 2516–2525. [Google Scholar] [CrossRef] [PubMed]
- García-Ramírez, M.; Canals, F.; Hernández, C.; Colomé, N.; Ferrer, C.; Carrasco, E.; García-Arumí, J.; Simó, R. Proteomic analysis of human vitreous fluid by fluorescence- based difference gel electrophoresis (DIGE): A new strategy for identifying potential candidates in the pathogenesis. Diabetologia 2007, 50, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; McLeod, S.; Merges, C.A.; Lutty, G.A. Choriocapillaris degeneration and related pathologic changes in human diabetic eyes. Arch. Ophthalmol. 1998, 116, 589–597. [Google Scholar] [CrossRef]
- Cheng, L.; Bu, H.; Portillo, J.A.; Li, Y.; Subauste, C.S.; Huang, S.S.; Kern, T.S.; Lin, F. Modulation of retinal Müller cells by complement receptor C5aR. Investig. Ophthalmol. Vis. Sci. 2013, 54, 8191–8198. [Google Scholar] [CrossRef]
- Kowluru, R.A.; Zhong, Q.; Santos, J.M. Matrix metalloproteinases in diabetic retinopathy: Potential role of MMP-9. Expert Opin. Investig. Drugs. 2012, 21, 797–805. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Fang, J.; Zhou, H.; Liu, X.; Su, S.B. High glucose induces and activates Toll-like receptor 4 in endothelial cells of diabetic retinopathy. Diabetol. Metab. Syndr. 2015, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Losiewicz, M.K.; Fort, P.E. Diabetes Impairs the neuroprotective properties of retinal Alpha-crystallins. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5034–5042. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Gloss, B.S.; Dinger, M.E. Realizing the significance of noncoding functionality in clinical genomics. Exp. Mol. Med. 2018, 50, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigó, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; Thurman, R.E.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Perry, R.B.-T.; Ulitsky, I. The functions of long noncoding RNAs in development and stem cells. Development 2016, 143, 3882–3894. [Google Scholar] [CrossRef]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, 1–33. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Cech, T.R. RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, L.; Wang, L.; Liu, Z.; Li, Q.; Yao, B.; Wang, C.; Chen, T.; Tu, K.; Liu, Q. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/β-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Olgun, G.; Sahin, O.; Tastan, O. Discovering lncRNA mediated sponge interactions in breast cancer molecular subtypes. BMC Genom. 2018, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Orom, U.A.; Cesaroni, M.; Beringer, M.; Taatjes, D.J.; Blobel, G.A.; Shiekhattar, R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013, 494, 497–501. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef]
- Miao, H.; Wang, L.; Zhan, H.; Dai, J.; Chang, Y.; Wu, F.; Liu, T.; Liu, Z.; Gao, C.; Li, L.; et al. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019, 15, 1–24. [Google Scholar] [CrossRef]
- Wilk, R.; Hu, J.; Blotsky, D.; Krause, H.M. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016, 30, 594–609. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL: A regulator of VEGF in diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Tao, Z.F.; Li, X.M.; Zhang, H.; Yao, J.; Jiang, Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, X.; Liu, J.; Wang, J.; Zhang, Y.; Zhang, T.; Zhang, H. Effect of intravitreal conbercept treatment on the expression of Long Noncoding RNAs and mRNAs in Proliferative Diabetic Retinopathy Patients. Acta Ophthalmol. 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Awata, T.; Yamashita, H.; Kurihara, S.; Morita-Ohkubo, T.; Miyashita, Y.; Katayama, S.; Mori, K.; Yoneya, S.; Kohda, M.; Okazaki, Y.; et al. A genome-wide association study for diabetic retinopathy in a Japanese population: Potential association with a long intergenic non-coding RNA. PLoS ONE 2014, 9, 1–9. [Google Scholar] [CrossRef]
- Wang, W.J.; Tay, H.G.; Soni, R.; Perumal, G.S.; Goll, M.G.; Macaluso, F.P.; Asara, J.M.; Amack, J.D.; Tsou, M.F. CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat. Cell Biol. 2013, 15, 591–601. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Z.; Xie, T.; Zhang, X.; Dai, T. Inhibition of BDNF-AS provides neuroprotection for retinal ganglion cells against ischemic injury. PLoS ONE 2016, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, F.; Xiao, H.; Han, F. Long noncoding RNA BDNF-AS inversely regulated BDNF and modulated high-glucose induced apoptosis in human retinal pigment epithelial cells. J. Cell Biochem. 2018, 119, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Biswas, S.; Feng, B.; Chen, S.; Gonder, J.; Chakrabarti, S. lncRNA H19 prevents endothelial–mesenchymal transition in diabetic retinopathy. Diabetologia 2019, 62, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, Y.X. LncRNA HOTTIP improves diabetic retinopathy by regulating the p38-MAPK pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2941–2948. [Google Scholar] [PubMed]
- Liu, J.Y.; Yao, J.; Li, X.M.; Song, Y.C.; Wang, X.Q.; Li, Y.J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Thomas, A.A.; Chen, S.; Aref-Eshghi, E.; Feng, B.; Gonder, J.; Sadikovic, B.; Chakrabarti, S. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Qin, H.; Leng, Y.; Li, X.; Zhang, L.; Bai, D.; Meng, Y.; Wang, J. LncRNA MEG3 overexpression inhibits the development of diabetic retinopathy by regulating TGF-β1 and VEGF. Exp. Ther. Med. 2018, 16, 2337–2342. [Google Scholar] [CrossRef]
- Tong, P.; Peng, Q.H.; Gu, L.M.; Xie, W.W.; Li, W.J. LncRNA-MEG3 alleviates high glucose induced inflammation and apoptosis of retina epithelial cells via regulating miR-34a/SIRT1 axis. Exp. Mol. Pathol. 2019, 107, 102–109. [Google Scholar] [CrossRef]
- Qiu, G.Z.; Tian, W.; Fu, H.T.; Li, C.P.; Liu, B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016, 471, 135–141. [Google Scholar] [CrossRef]
- Yan, B.; Yao, J.; Liu, J.Y.; Li, X.M.; Wang, X.Q.; Li, Y.J.; Tao, Z.F.; Song, Y.C.; Chen, Q.; Jiang, Q. LncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Chen, J.; Lin, S.; Cai, D.; Chen, C.; Chen, Z. Long non-coding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing miR-29b and regulating cell apoptosis. Biosci. Rep. 2017, 37, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, C.P.; Wang, J.J.; Shan, K.; Liu, X.; Yan, B. RNCR3 knockdown inhibits diabetes mellitus-induced retinal reactive gliosis. Biochem. Biophys. Res. Commun. 2016, 479, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Li, C.P.; Liu, C.; Liu, X.; Yan, B. RNCR3: A regulator of diabetes mellitus-related retinal microvascular dysfunction. Biochem. Biophys. Res. Commun. 2017, 482, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, C.; Liu, Y.; Wu, C. 17β-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem. Biophys. Res. Commun. 2014, 445, 388–393. [Google Scholar] [CrossRef]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Liu, Y.; Xu, W.; Sun, Y.; Lu, J.; Wang, F.; Wei, M.; Shen, J.; Hou, J.; Gao, X.; et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 2013, 190, 2278–2287. [Google Scholar] [CrossRef]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef]
- Yao, J.; Wang, X.Q.; Li, Y.J.; Shan, K.; Yang, H.; Wang, Y.N.; Yao, M.D.; Liu, C.; Li, X.M.; Shen, Y.; et al. Long non-coding RNA MALAT1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol. Med. 2016, 8, 1–17. [Google Scholar] [CrossRef]
- Zhang, Q.S.; Wang, Z.H.; Zhang, J.L.; Duan, Y.L.; Li, G.F.; Zheng, D.L. Beta-asarone protects against MPTP-induced Parkinson’s disease via regulating long non-coding RNA MALAT1 and inhibiting α-synuclein protein expression. Biomed. Pharmacother. 2016, 83, 153–159. [Google Scholar] [CrossRef]
- Watts, R.; Johnsen, V.L.; Shearer, J.; Hittel, D.S. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am. J. Physiol. Physiol. 2013, 304, C995–C1001. [Google Scholar] [CrossRef] [PubMed]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [PubMed]
- Eißmann, M.; Gutschner, T.; Hämmerle, M.; Günther, S.; Caudron-Herger, M.; Groβ, M.; Schirmacher, P.; Rippe, K.; Zörnig, M.; Braun, T.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.D.; Biswas, S.; Feng, B.; Chakrabarti, S. MALAT1: A regulator of inflammatory cytokines in diabetic complications. Endocrinol. Diabetes Metab. 2018, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, R.; Li, X.; Fan, M.; Lin, J.; Zhen, J.; Chen, L.; Lv, Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J. Cell Mol. Med. 2017, 21, 2732–2747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gu, H.; Chen, J.; Zhou, X. Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int. J. Cardiol. 2016, 202, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Tang, J.; Li, G.; Berti-Mattera, L.; Lee, C.A.; Bartkowski, D.; Gale, D.; Monahan, J.; Niesman, M.R.; Alton, G.; et al. Effects of p38 MAPK inhibition on early stages of diabetic retinopathy and sensory nerve function. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2158–2164. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef]
- Liu, P.; Jia, S.B.; Shi, J.M.; Li, W.J.; Tang, L.S.; Zhu, X.H.; Tong, P. LncRNA-MALAT1 promotes neovascularization in diabetic retinopathy through regulating miR-125b/VE-cadherin axis. Biosci. Rep. 2019, 39, 1–26. [Google Scholar] [CrossRef]
- Capitão, M.; Soares, R. Angiogenesis and Inflammation Crosstalk in Diabetic Retinopathy. J. Cell Biochem. 2016, 117, 2443–2453. [Google Scholar] [CrossRef] [PubMed]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Zheng, F.S.; Yang, W.; Lu, J. Bin. The long non-coding RNA MIAT regulates zinc finger E-box binding homeobox 1 expression by sponging miR-150 and promoteing cell invasion in non-small-cell lung cancer. Gene 2017, 633, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Blackshaw, S.; Harpavat, S.; Trimarchi, J.; Cai, L.; Huang, H.; Kuo, W.P.; Weber, G.; Lee, K.; Fraioli, R.E.; Cho, S.H.; et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004, 2, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Aprea, J.; Prenninger, S.; Dori, M.; Ghosh, T.; Monasor, L.S.; Wessendorf, E.; Zocher, S.; Massalini, S.; Alexopoulou, D.; Lesche, M.; et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013, 32, 3145–3160. [Google Scholar] [CrossRef]
- Shen, Y.; Dong, L.F.; Zhou, R.M.; Yao, J.; Song, Y.C.; Yang, H.; Jiang, Q.; Yan, B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: A clinical and in vitro study. J. Cell Mol. Med. 2016, 20, 537–548. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, W.; Jin, M.; Chen, J.; Xu, W.; Kong, X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, D.Y.; Sha, W.G.; Shen, L.; Lu, G.Y.; Yin, X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem. Biophys. Res. Commun. 2015, 468, 726–732. [Google Scholar] [CrossRef]
- Pasmant, E.; Laurendeau, I.; Héron, D.; Vidaud, M.; Vidaud, D.; Bièche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef]
- Broadbent, H.M.; Peden, J.F.; Lorkowski, S.; Goel, A.; Ongen, H.; Green, F.; Clarke, R.; Collins, R.; Franzosi, M.G.; Tognoni, G.; et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008, 17, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, X.; Shen, J. ANRIL: A pivotal tumor suppressor long non-coding RNA in human cancers. Tumor Biol. 2016, 37, 5657–5661. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL regulates production of extracellular matrix proteins and vasoactive factors in diabetic complications. Am. J. Physiol. Metab. 2017, 314, E191–E200. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, L.R.; Loomis, S.J.; Kang, J.H.; Yaspan, B.L.; Abdrabou, W.; Budenz, D.L.; Chen, T.C.; Delbono, E.; Friedman, D.S.; Gaasterland, D.; et al. CDKN2B-AS1 genotype-glaucoma feature correlations in primary open-angle glaucoma patients from the United States. Am. J. Ophthalmol. 2013, 155, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Han, X.; Wittfeldt, A.; Sun, J.; Liu, C.; Wang, X.; Gan, L.M.; Cao, H.; Liang, Z. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 2016, 13, 98–108. [Google Scholar] [CrossRef]
- Pachnis, V.; Belayew, A.; Tilghman, S.M. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. USA 2006, 81, 5523–5527. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Yan, Y.; Hu, M.; Chen, X.; Wang, Y.; Dai, Y.; Wu, D.; Wang, Y.; Zhuang, Z.; Xia, H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2017, 2016, 129–136. [Google Scholar] [CrossRef]
- Luo, M.; Li, Z.; Wang, W.; Zeng, Y.; Liu, Z.; Qiu, J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013, 333, 213–221. [Google Scholar] [CrossRef]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 1–11. [Google Scholar] [CrossRef]
- Song, X.; Luo, X.; Gao, Q.; Wang, Y.; Gao, Q.; Long, W. Dysregulation of LncRNAs in Placenta and Pathogenesis of Preeclampsia. Curr. Drug Targets 2017, 18, 115–1170. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, H.; Fan, Z.; Li, G.; Ma, Q.; Tao, Z.; Wang, R.; Feng, J.; Luo, Y. Long Noncoding RNA H19 Promotes Neuroinflammation in Ischemic Stroke by Driving Histone Deacetylase 1-Dependent M1 Microglial Polarization. Stroke 2017, 48, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cao, B.; Han, D.; Sun, M.; Feng, J. Long Non-coding RNA H19 Induces Cerebral Ischemia Reperfusion Injury via Activation of Autophagy. Aging Dis. 2017, 8, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Han, C.L.; Ge, M.; Liu, Y.P.; Zhao, X.M.; Wang, K.L.; Chen, N.; Hu, W.; Zhang, J.G.; Li, L.; Meng, F.G. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Ding, Y.; Jin, X.; Xu, S.; Zhang, H. Long Non-coding RNA H19 Promotes Corneal Neovascularization by Targeting MicroRNA-29c. Biosci. Rep. 2019, 39, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.X. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 322–328. [Google Scholar] [PubMed]
- Biswas, S.; Chakrabarti, S. Increased Extracellular Matrix Protein Production in Chronic Diabetic Complications: Implications of Non-Coding RNAs. Non-Coding RNA 2019, 5, 30. [Google Scholar] [CrossRef]
- Roy, S.; Amin, S.; Roy, S. Retinal fibrosis in diabetic retinopathy. Exp. Eye Res. 2016, 142, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, C.; Cattaneo, E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat. Rev. Neurol. 2009, 5, 311–322. [Google Scholar] [CrossRef]
- Hackett, S.F.; Friedman, Z.; Freund, J.; Schoenfeld, C.; Curtis, R.; DiStefano, P.S.; Campochiaro, P.A. A splice variant of trkB and brain-derived neurotrophic factor are co- expressed in retinal pigmented epithelial cells and promote differentiated characteristics. Brain Res. 1998, 789, 201–212. [Google Scholar] [CrossRef]
- Kaviarasan, K.; Jithu, M.; Arif Mulla, M.; Sharma, T.; Sivasankar, S.; Das, U.N.; Angayarkanni, N. Low blood and vitreal BDNF, LXA4 and altered Th1/Th2 cytokine balance are potential risk factors for diabetic retinopathy. Metabolism 2015, 64, 958–966. [Google Scholar] [CrossRef]
- Ola, M.S.; Nawaz, M.I.; El-Asrar, A.A.; Abouammoh, M.; Alhomida, A.S. Reduced levels of brain derived neurotrophic factor (BDNF) in the serum of diabetic retinopathy patients and in the retina of diabetic rats. Cell Mol. Neurobiol. 2013, 33, 359–367. [Google Scholar] [CrossRef] [PubMed]
- da Rocha, S.T.; Edwards, C.A.; Ito, M.; Ogata, T.; Ferguson-Smith, A.C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008, 24, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Jiang, Q.; Wang, X.Q.; Wang, Y.N.; Yang, H.; Yao, M.D.; Liu, C.; Li, X.M.; Yao, J.; Liu, B.; et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–126. [Google Scholar] [CrossRef]
- Lian, Y.; Cai, Z.; Gong, H.; Xue, S.; Wu, D.; Wang, K. HOTTIP: A critical oncogenic long non-coding RNA in human cancers. Mol. Biosyst. 2016, 12, 3247–3253. [Google Scholar] [CrossRef]
- Giacco, F.; Brownlee, M. Oxidative stress and diabetic complications. Circ. Res. 2010, 107, 1058–1070. [Google Scholar] [CrossRef]
- Ceriello, A.; Esposito, K.; Ihnat, M.; Thorpe, J.; Giugliano, D. Long-term glycemic control influences the long-lasting effect of hyperglycemia on endothelial function in type 1 diabetes. J. Clin. Endocrinol. Metab. 2009, 94, 2751–2756. [Google Scholar] [CrossRef]
- Aiello, L.P. Diabetic retinopathy and other ocular findings in the diabetes control and complications trial/epidemiology of diabetes interventions and complications study. Diabetes Care 2014, 37, 17–23. [Google Scholar] [CrossRef]
- Villeneuve, L.M.; Natarajan, R. The role of epigenetics in the pathology of diabetic complications. Am. J. Physiol. Physiol. 2010, 299, F14–F25. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Chen, Z.; Genuth, S.; Paterson, A.; Zhang, L.; Wu, X.; Li, S.M.; Cleary, P.; Riggs, A.; Harlan, D.M.; et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 2014, 63, 1748–1762. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A.; Santos, J.M.; Mishra, M. Epigenetic Modifications and Diabetic Retinopathy. Biomed. Res. Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Felsenfeld, G. A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 2014, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Miranda, T.B.; Jones, P.A. DNA methylation: The nuts and bolts of repression. J. Cell Physiol. 2007, 213, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, N.M.; Silva, A.S.; Wanderley, D.Q.E.I.; Modesto, F.J.; Alves, P.G.C.; Ferreira, D.N.R.; Pordeus, L.R.; de Carvalho, C.M.; Paulo, D.O.N.; Camati, P.D. Analysis of the DNA methylation profiles of miR-9-3, miR-34a, and miR-137 promoters in patients with diabetic retinopathy and nephropathy. J. Diabetes Complicat. 2018, 32, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A.; Shan, Y.; Mishra, M. Dynamic DNA methylation of matrix metalloproteinase-9 in the development of diabetic retinopathy. Lab. Investig. 2016, 96, 1040–1049. [Google Scholar] [CrossRef]
- Mishra, M.; Kowluru, R.A. The role of DNA methylation in the metabolic memory phenomenon associated with the continued progression of diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5748–5757. [Google Scholar] [CrossRef]
- Agardh, E.; Lundstig, A.; Perfilyev, A.; Volkov, P.; Freiburghaus, T.; Lindholm, E.; Rönn, T.; Agardh, C.D.; Ling, C. Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 2015, 13, 1–9. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Eberharter, A.; Becker, P. Histone acetylation: A switch between repressive and permissive chromatin. EMBO Rep. 2002, 3, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Sawan, C.; Herceg, Z. Histone Modifications and Cancer. Adv. Genet. 2010, 70, 57–85. [Google Scholar] [CrossRef] [PubMed]
- El-Osta, A.; Brasacchio, D.; Yao, D.; Pocai, A.; Jones, P.L.; Roeder, R.G.; Cooper, M.E.; Brownlee, M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 2008, 205, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Reddy, M.A.; Yuan, H.; Lanting, L.; Kato, M.; Natarajan, R. Epigenetic Histone Methylation Modulates Fibrotic Gene Expression. J. Am. So. Nephrol. 2010, 21, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, L.M.; Reddy, M.A.; Lanting, L.L.; Wang, M.; Meng, L.; Natarajan, R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc. Natl. Acad. Sci. USA 2008, 105, 9047–9052. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Wu, X.; Zhang, L.; Yuan, Y.C.; Riggs, A.D.; Natarajan, R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J. Biol. Chem. 2007, 282, 13854–13863. [Google Scholar] [CrossRef]
- Zhong, Q.; Kowluru, R.A. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes 2011, 60, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Zhong, Q.; Kowluru, R.A. Epigenetic modifications of Keap1 regulate its interaction with the protective factor Nrf2 in the development of diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 7256–7265. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sidoli, S.; Zhang, W.; Wang, Q.; Wang, L.; Jensen, O.N.; Guo, L.; Zhao, X.; Zheng, L. Abnormal levels of histone methylation in the retinas of diabetic rats are reversed by minocycline treatment. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Margueron, R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 2017, 42, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.A.; Feng, B.; Chakrabarti, S. Polycomb repressive complex 2 regulates MiR-200b in retinal endothelial cells: Potential relevance in diabetic retinopathy. PLoS ONE 2015, 10, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Feng, B.; George, B.; Chakrabarti, R.; Chen, M.; Chakrabarti, S. Transcriptional coactivator p300 regulates glucose-induced gene expression in endothelial cells. Am. J. Physiol. Metab. 2009, 298, E127–E137. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Feng, B.; Chen, S.; Chu, Y.; Chakrabarti, S. Mechanisms of endothelial to mesenchymal transition in the retina in diabetes. Investig. Ophthalmol. Vis. Sci. 2014, 55, 7321–7331. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Cao, Y.; Chen, S.; Chu, X.; Chu, Y.; Chakrabarti, S. MiR-200b mediates endothelial-to-mesenchymal transition in diabetic cardiomyopathy. Diabetes 2016, 65, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Li, Z.; Wu, Z.; Aau, M.; Guan, P.; Karuturi, R.K.; Liou, Y.C.; Yu, Q. Context-Specific Regulation of NF-κB Target Gene Expression by EZH2 in Breast Cancers. Mol. Cell 2011, 43, 798–810. [Google Scholar] [CrossRef]
- Shivdasani, R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood 2006, 108, 3646–3653. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.N.; Srivastava, D. microRNAs as developmental regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A Brief Review on the Mechanisms of miRNA Regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Romaine, S.P.R.; Tomaszewski, M.; Condorelli, G.; Samani, N.J. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart 2015, 101, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Wang, W.X.; Rajeev, B.W. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008, 18, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, B.; Lumayag, S.; Cowan, C.; Xu, S. microRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4402–4409. [Google Scholar] [CrossRef]
- McArthur, K.; Feng, B.; Wu, Y.; Chen, S.; Chakrabarti, S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes 2011, 60, 1314–1323. [Google Scholar] [CrossRef]
- Chen, S.; Feng, B.; Thomas, A.A.; Chakrabarti, S. MiR-146a regulates glucose induced upregulation of inflammatory cytokines extracellular matrix proteins in the retina and kidney in diabetes. PLoS ONE 2017, 12, 1–17. [Google Scholar] [CrossRef]
- Mortuza, R.; Feng, B.; Chakrabarti, S. MiR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia 2014, 57, 1037–1046. [Google Scholar] [CrossRef]
| lncRNA | Reported Functions in DR | Implications in Other Epigenetic Mechanisms |
|---|---|---|
| ANRIL |
|
|
| BDNF-AS |
|
|
| H19 |
| |
| HOTTIP |
|
|
| MALAT1 |
|
|
| MEG3 |
|
|
| MIAT |
|
|
| RNCR3 |
|
|
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, S.; Sarabusky, M.; Chakrabarti, S. Diabetic Retinopathy, lncRNAs, and Inflammation: A Dynamic, Interconnected Network. J. Clin. Med. 2019, 8, 1033. https://doi.org/10.3390/jcm8071033
Biswas S, Sarabusky M, Chakrabarti S. Diabetic Retinopathy, lncRNAs, and Inflammation: A Dynamic, Interconnected Network. Journal of Clinical Medicine. 2019; 8(7):1033. https://doi.org/10.3390/jcm8071033
Chicago/Turabian StyleBiswas, Saumik, Marie Sarabusky, and Subrata Chakrabarti. 2019. "Diabetic Retinopathy, lncRNAs, and Inflammation: A Dynamic, Interconnected Network" Journal of Clinical Medicine 8, no. 7: 1033. https://doi.org/10.3390/jcm8071033
APA StyleBiswas, S., Sarabusky, M., & Chakrabarti, S. (2019). Diabetic Retinopathy, lncRNAs, and Inflammation: A Dynamic, Interconnected Network. Journal of Clinical Medicine, 8(7), 1033. https://doi.org/10.3390/jcm8071033





