MicroRNAs as Signaling Mediators and Biomarkers of Drug- and Chemical-Induced Liver Injury
Abstract
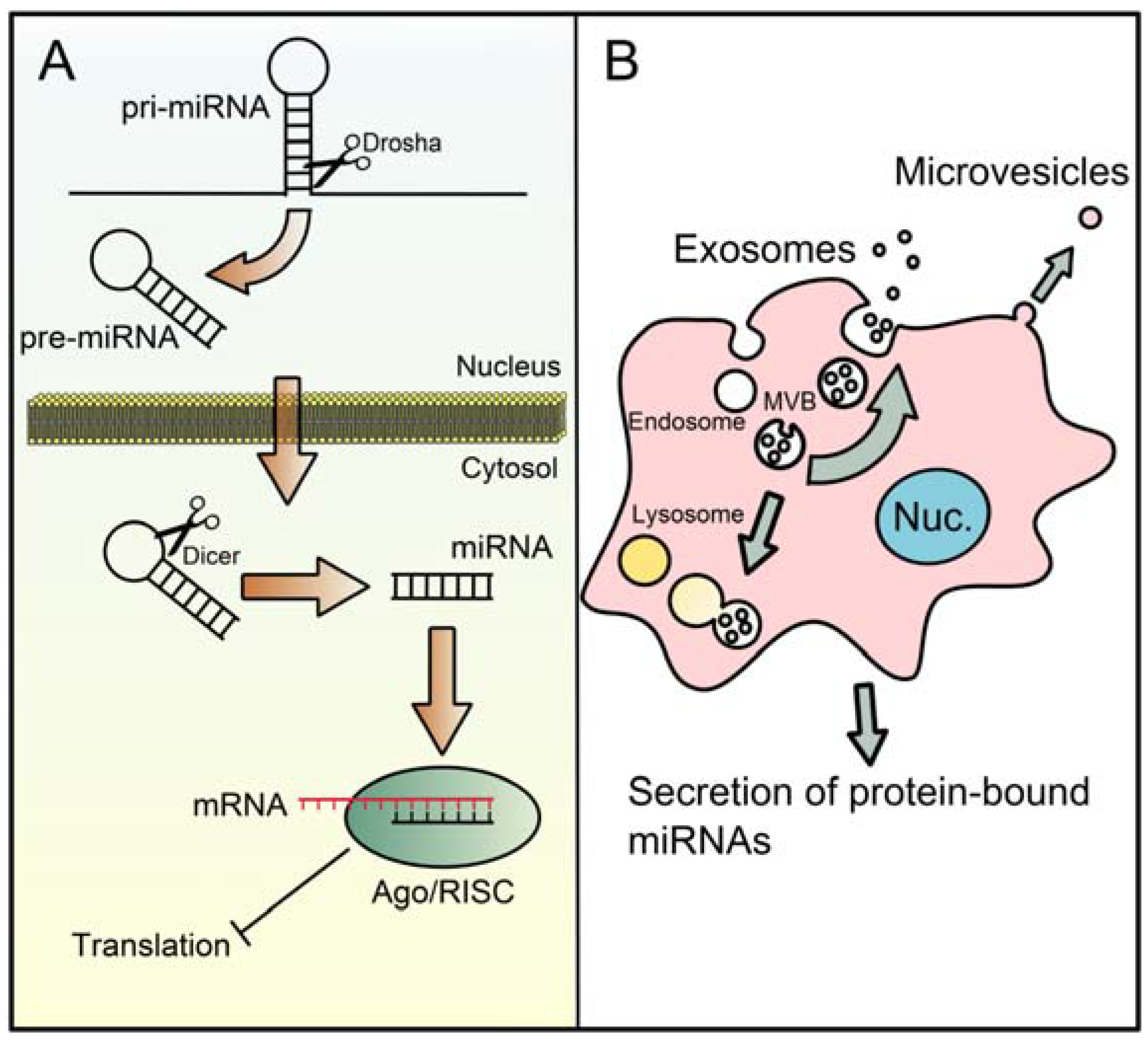
:1. Introduction
2. The microRNA Machinery
3. MicroRNAs in the Mechanisms of DILI
| Study Type | Effect | Putative miRNA(s) Involved | References |
|---|---|---|---|
| In vivo | Liver injury | Decrease: miR-101a, -106b, -298, -370, -491-5p miR-122? | [21,32,33,39,45] |
| Fibrosis | Decrease: miR-29 Increase: miR-21 | [43] [44] | |
| Liver regeneration | Pro-regenerative: miR-21 | [35,36,38] | |
| In vitro | Cell injury | Decrease: Let-7b, miR-124 | [40] |
| Cell proliferation | Pro-proliferative: miR-21 | [44] |
4. MicroRNAs as Biomarkers of DILI
| Human Studies (hsa-) | Human (hsa-) and Mouse (mur-) d | ||
|---|---|---|---|
| Exact, All Studies a | Exact, Adult Studies b | Similar, Adult and Child Studies c | |
| miR-122 | miR-122 | miR-193a/b-5p | miR-122 |
| miR-125b-5p | miR-378a/b/c/e/g/h/i | miR-192 | |
| miR-483-5p | miR-30a/d-5p | miR-125b | |
| miR-202-3p | |||
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chalasani, N.; Björnsson, E. Risk factors for idiosyncratic drug-induced liver injury. Gastroenterology 2010, 138, 2246–2259. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M. Etiologies of acute liver failure. Semin. Liver Dis. 2008, 28, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Senior, J.R. Evolution of the Food and Drug Administration approach to liver safety assessment for new drugs: Current status and challenges. Drug Saf. 2014, 37 (Suppl. 1), S9–S17. [Google Scholar] [PubMed]
- United State Food and Drug Administration (US FDA). Guidance for Industry: Drug-Induced Liver Injury: Premarketing Clinical Evaluation; United States Food and Drug Administration: Silver Spring, MD, USA, 2009.
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.E.; Cheng, X. Structural and biochemical advances in mammalian RNAi. J. Cell. Biochem. 2006, 99, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Borralho, P.M.; Rodrigues, C.M.; Steer, C.J. Mitochondrial microRNAs and their potential role in cell function. Curr. Pathobiol. Rep. 2014, 2, 123–132. [Google Scholar] [CrossRef]
- Wheeler, B.M.; Heimberg, A.M.; Moy, V.N.; Sperling, E.A.; Holstein, T.W.; Heber, S.; Peterson, K.J. The deep evolution of metazoan microRNAs. Evol. Dev. 2009, 11, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Kren, B.T.; Wong, P.Y.; Sarver, A.; Zhang, X.; Zeng, Y.; Steer, C.J. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009, 6, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Li, L.M.; Tang, R.; Hou, D.X.; Chen, X.; Zhang, C.Y.; Zen, K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010, 20, 1076–1078. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, X.; Zhang, H.; Liang, X.; Xiang, Y.; Yu, C.; Zen, K.; Li, Y.; Zhang, C.Y. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J. Lipid Res. 2009, 50, 1756–1765. [Google Scholar] [CrossRef] [PubMed]
- Vincent, R.; Sanyal, A. Recent advances in understanding of NASH: microRNAs as both biochemical markers and players. Curr. Pathobiol. Rep. 2014, 2, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Soblewski, C.; Calo, N.; Portius, D.; Foti, M. MicroRNAs in fatty liver disease. Semin. Liver Dis. 2015, 35, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Aly, H.H.; Tajima, A.; Inoue, I.; Shimotohno, K. Regulation of the hepatitis C virus genome replication by miR-199a. J. Hepatol. 2009, 50, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Thirion, M.; Ochiya, T. Roles of microRNAs in the hepatitis B virus infection and related diseases. Viruses 2013, 5, 2690–2703. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Ishibashi, O.; Kawahigashi, Y.; Mishima, T.; Kosuge, T.; Mizuguchi, Y.; Shimizu, T.; Arima, Y.; Yokomuro, S.; Yoshida, H.; et al. Identification of obstructive jaundice-related microRNAs in mouse liver. Hepatogastroenterology 2010, 57, 1013–1023. [Google Scholar] [PubMed]
- Dai, B.H.; Geng, L.; Wang, Y.; Sui, C.J.; Cie, F.; Shen, W.F.; Yang, J.M. microRNA-199a-5p protects hepatocytes from bile acid-induced sustained endoplasmic reticulum stress. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef]
- Fukushima, T.; Hamada, Y.; Yamada, H.; Horii, I. Changes of micro-RNA expression in rat liver treated by acetaminophen or carbon tetrachloride—Regulating role of micro-RNA for RNA expression. J. Toxicol. Sci. 2007, 32, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Yamaura, Y.; Nakajima, M.; Tastumi, N.; Takagi, S.; Fukami, T.; Tsuneyama, K.; Yokoi, T. Changes in the expression of miRNAs at the pericentral and periportal regions of the rat liver in response to hepatocellular injury: Comparison with the changes in the expression of plasma miRNAs. Toxicology 2014, 322, 89–98. [Google Scholar] [CrossRef] [PubMed]
- McGill, M.R.; Williams, C.D.; Xie, Y.; Ramachandran, A.; Jaeschke, H. Acetaminophen-induced liver injury in rats and mice: Comparison of protein adducts, mitochondrial dysfunction, and oxidative stress in the mechanism of toxicity. Toxicol. Appl. Pharmacol. 2012, 264, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef] [PubMed]
- McGill, M.R.; Yan, H.M.; Ramachandran, A.; Murray, G.J.; Rollins, D.E.; Jaeschke, H. HepaRG cells: A human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology 2011, 53, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; McGill, M.R.; Dorko, K.; Kumer, S.C.; Schmitt, T.M.; Forster, J.; Jaeschke, H. Mechanisms of acetaminophen-induced cell death in primary human hepatocytes. Toxicol. Appl. Pharmacol. 2014, 279, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Lind, R.C.; Gandolfi, A.; Hall, P.D. Age and gender influence halothane-associated hepatotoxicity in strain 13 guinea pigs. Anesthesiology 1989, 71, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Farrell, G.C.; Frost, L.; Tapner, M.; Field, J.; Weltman, M.; Mahoney, J. Halothane-induced liver injury in guinea-pigs: Importance of cytochrome P450 enzyme activity and hepatic blood flow. J. Gastroenterol. Hepatol. 1996, 11, 594–601. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Cheng, L.; Reilly, T.P.; Wegmann, D.; Ju, C. Role of neutrophils in a mouse model of halothane-induced liver injury. Hepatology 2006, 44, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; You, Q.; Yin, H.; Holt, M.P.; Ju, C. Involvement of natural killer T cells in halothane-induced liver injury in mice. Biochem. Pharmacol. 2010, 80, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Uetrecht, J.; Naisbitt, D.J. Idiosyncratic adverse drug reactions: Current concepts. Pharmacol. Rev. 2013, 65, 779–808. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Yano, A.; Fukami, T.; Nakajima, M.; Yokoi, T. Involvement of miRNAs in the early phase of halothane-induced liver injury. Toxicology 2014, 319, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, W.; Higashiyama, W.; Tohyama, C. Involvement of microRNAs in dioxin-induced liver damage in the mouse. Toxicol. Sci. 2011, 122, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lian, J.; Feng, Y.; Srinivas, S.; Guo, Z.; Zhong, H.; Zhuang, Z.; Wang, S. Genome-Wide miRNA-profiling of aflatoxin B1-induced hepatic injury using deep sequencing. Toxicol. Lett. 2014, 226, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Dippold, R.P.; Vadigepalli, R.; Gonye, G.E.; Hoek, J.B. Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am. J. Gastrointest. Liver Physiol. 2012, 303, G733–G743. [Google Scholar] [CrossRef]
- Dippold, R.P.; Vadigepalli, R.; Gonye, G.E.; Patra, B.; Hoek, J.B. Chronic ethanol feeding alters miRNA expression dynamics during liver regeneration. Alcohol. Clin. Exp. Res. 2013, 37 (Suppl. 1), E59–E69. [Google Scholar] [PubMed]
- Meng, F.; Henson, R.; Wehbe-Jank, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.E.; Ferreira, D.M.; Zhang, X.; Borralho, P.M.; Sarver, A.L.; Zeng, Y.; Steer, C.J.; Kren, B.T.; Rodrigues, C.M. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G887–G897. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Kim, T.H.; Natarajan, A.; Wang, S.S.; Choi, J.; Wu, J.; Zern, M.A.; Venugopal, S.K. Acute liver injury upregulates microRNA-491-5p in mice, and its overexpression sensitizes Hep G2 cells for tumour necrosis factor-alpha-induced apoptosis. Liver Int. 2010, 30, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Tian, Q.; Steuerwald, N.M.; Schrum, L.W.; Bonkovsky, H.L. The let-7 microRNA enhances heme oxygenase-1 by suppressing Bach1 and attenuates oxidant injury in human hepatocytes. Biochim. Biophys. Acta 2012, 1819, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liu, H.; Chen, F.; Li, D.; Zhao, Y. MiR-214 promotes the alcohol-induced oxidative stress via down-regulation of glutathione reductase and cytochrome P450 oxidoreductase in liver cells. Alcohol. Clin. Exp. Res. 2014, 38, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yi, S.; Deng, Y.; Cheng, J.; Wu, X.; Liu, W.; Tai, Y.; Chen, G.; Zhang, Q.; Yang, Y.; et al. MiR-124 protects human hepatic L02 cells from H2O2-induced apoptosis by targeting Rab38 gene. Biochem. Biophys. Res. Commun. 2014, 450, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, L.; Wang, Y.; Zhang, M.; Li, L.; Zhu, D.; Li, X.; Gu, H.; Zhang, C.Y.; Zen, K.; et al. Protective role of estrogen-induced miRNA-29 expression in carbon tetrachloride-induced mouse liver injury. J. Biol. Chem. 2012, 287, 14851–14862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zha, Y.; Hu, W.; Huang, Z.; Gao, Z.; Zang, Y.; Chen, J.; Dong, L.; Zhang, J. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J. Biol. Chem. 2013, 288, 37082–37093. [Google Scholar] [CrossRef] [PubMed]
- Lardizábal, M.N.; Rodríguez, R.E.; Nocito, A.L.; Daniele, S.M.; Palatnik, J.F.; Veggi, L.M. Alteration of the microRNA-122 regulatory network in rat models of hepatotoxicity. Environ. Toxicol. Pharmacol. 2014, 37, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Bolleyn, J.; Fraczek, J.; Vinken, M.; Lizarraga, D.; Gaj, S.; van Delft, J.H.; Rogiers, V.; Vanhaecke, T. Effect of Trichostatin A on miRNA expression in cultures of primary rat hepatocytes. Toxicol. In Vitro 2011, 25, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jia, Y.; Zheng, R.; Guo, Y.; Wang, Y.; Guo, H.; Fei, M.; Sun, S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic disorders. Clin. Chem. 2010, 56, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Starkey Lewis, P.J.; Dear, J.; Platt, V.; Simpson, K.J.; Craig, D.G.; Antoine, D.J.; French, N.S.; Dhaun, N.; Webb, D.J.; Costello, E.M.; et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011, 54, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Woolbright, B.L.; Antoine, D.J.; Jenkins, R.E.; Bajt, M.L.; Park, B.K.; Jaeschke, H. Plasma biomarkers of liver injury and inflammation demonstrate a lack of apoptosis during obstructive cholestasis in mice. Toxicol. Appl. Pharmacol. 2013, 273, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Antoine, D.J.; Weemhoff, J.L.; Jenkins, R.E.; Farhood, A.; Park, B.K.; Jaeschke, H. Biomarkers distinguish apoptotic and necrotic cell death during hepatic ischemia/reperfusion injury in mice. Liver Transpl. 2014, 20, 1372–1382. [Google Scholar] [CrossRef] [PubMed]
- Laterza, O.F.; Scott, M.G.; Garrett-Engele, P.W.; Korenblat, K.M.; Lockwood, C.M. Circulating miR-122 as a potential biomarker of liver disease. Biomark. Med. 2013, 7, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.; Kanchagar, C.; Veksler-Lublinsky, I.; Lee, R.C.; McGill, M.R.; Jaeschke, H.; Curry, S.C.; Ambros, V.R. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc. Natl. Acad. Sci. USA 2014, 111, 12169–12174. [Google Scholar] [CrossRef] [PubMed]
- McGill, M.R.; Jaeschke, H. Mechanistic biomarkers in acetaminophen-induced hepatotoxicity and acute liver failure: From preclinical models to patients. Expert Opin. Drug Metab. Toxicol. 2014, 10, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Arrese, M.; Eguchi, A.; Feldstein, A.E. Circulating microRNAs: Emerging biomarkers of liver disease. Semin. Liver Dis. 2015, 35, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Harrill, A.H.; Roach, J.; Fier, I.; Eaddy, J.S.; Kurtz, C.L.; Antoine, D.J.; Spencer, D.M.; Kishimoto, T.K.; Pisetsky, D.S.; Park, B.K.; et al. The effects of heparins on the liver: Application of mechanistic serum biomarkers in a randomized study in healthy volunteers. Clin. Pharmacol. Ther. 2012, 92, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.W.; Chen, X.; Jiang, Z.Z.; Wang, T.; Wang, C.; Zhang, Y.; Wen, J.; Xue, M.; Zhu, D.; Zhang, Y.; et al. A panel of serum microRNAs as specific biomarkers for diagnosis of compound- and herb-induced liver injury in rats. PLoS ONE 2012, 7, e37395. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Petrasek, J.; Mundkur, S.; Catalano, D.; Levin, I.; Ward, J.; Alao, H.; Kodys, K.; Szabo, G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012, 56, 1946–1957. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.; Bala, S.; Petrasek, J.; Szabo, G. Plasma microRNA profiles distinguish lethal injury in acetaminophen toxicity: A research study. World J. Gastroenterol. 2012, 18, 2798–27804. [Google Scholar] [CrossRef] [PubMed]
- Krauskopf, J.; Caiment, F.; Claessen, S.M.; Johnson, K.J.; Warner, R.L.; Schomaker, S.J.; Burt, D.A.; Aubrecht, J.; Kleinjans, J.C. Application of high-throughput sequencing to circulating microRNAs reveals novel biomarkers for drug-induced liver injury. Toxicol. Sci. 2015, 143, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Salminen, W.F.; Shi, Q.; Greenhaw, J.; Gill, P.S.; Bhattacharyya, S.; Beger, R.D.; Mendrick, D.L.; Mattes, W.B.; James, L.P.; et al. Potential of extracellular microRNAs as biomarkers of acetaminophen toxicity in childen. Toxicol. Appl. Pharmacol. 2015. [CrossRef]
- Dear, J.W.; Antoine, D.J.; Starkey-Lewis, P.; Goldring, C.E.; Park, B.K. Early detection of paracetamol toxicity using circulating liver microRNAs and markers of cell necrosis. Br. J. Clin. Pharmacol. 2014, 77, 904–905. [Google Scholar] [CrossRef] [PubMed]
- Antoine, D.J.; Dear, J.W.; Lewis, P.S.; Platt, V.; Coyle, J.; Masson, M.; Thanacoody, R.H.; Gray, A.J.; Webb, D.J.; Moggs, J.G.; et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology 2013, 58, 777–787. [Google Scholar] [CrossRef] [PubMed]
- McGill, M.R.; Staggs, V.S.; Sharpe, M.R.; Lee, W.M.; Jaeschke, H.; Acute Liver Failure Study Group. Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology 2014, 60, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Antoine, D.J.; Jenkins, R.E.; Dear, J.W.; Williams, D.P.; McGill, M.R.; Sharpe, M.R.; Craig, D.G.; Simpson, K.J.; Jaeaschke, H.; Park, B.K.; et al. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J. Hepatol. 2012, 56, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Björnsson, E.; Nordlinder, H.; Olsson, R. Clinical characteristics and prognostic markers in disulfiram-induced liver injury. J. Hepatol. 2006, 44, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Falcon-Perez, J.M. Liver extracellular vesicles in health and disease. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef]
- Wang, K.; Li, H.; Yuan, Y.; Etheridge, A.; Zhou, Y.; Huang, D.; Wilmes, P.; Galas, D. The complex exogenous RNA spectra in human plasma: An interface with human gut biota? PLoS ONE 2012, 7, e51009. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Wang, W.X.; Wilfred, B.R.; Tang, G. Technical variables in high-throughput miRNA expression profiling: Much work remains to be done. Biochim. Biophys. Acta 2008, 1779, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Chugh, P.; Dittmer, D.P. Potential pitfalls in microRNA profiling. Wiley Interdiscip. Rev. RNA 2012, 3, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Qi, R.; Weiland, M.; Gao, X.H.; Zhou, L.; Mi, Q.S. Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology 2012, 55, 1640–1642. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Edelman, J.J.; Kao, S.C.; Vallely, M.P.; van Zandwijk, N.; Reid, G. The impact of hemolysis on cell-fre microRNA biomarkers. Front. Genet. 2013, 4. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McGill, M.R.; Jaeschke, H. MicroRNAs as Signaling Mediators and Biomarkers of Drug- and Chemical-Induced Liver Injury. J. Clin. Med. 2015, 4, 1063-1078. https://doi.org/10.3390/jcm4051063
McGill MR, Jaeschke H. MicroRNAs as Signaling Mediators and Biomarkers of Drug- and Chemical-Induced Liver Injury. Journal of Clinical Medicine. 2015; 4(5):1063-1078. https://doi.org/10.3390/jcm4051063
Chicago/Turabian StyleMcGill, Mitchell R., and Hartmut Jaeschke. 2015. "MicroRNAs as Signaling Mediators and Biomarkers of Drug- and Chemical-Induced Liver Injury" Journal of Clinical Medicine 4, no. 5: 1063-1078. https://doi.org/10.3390/jcm4051063
APA StyleMcGill, M. R., & Jaeschke, H. (2015). MicroRNAs as Signaling Mediators and Biomarkers of Drug- and Chemical-Induced Liver Injury. Journal of Clinical Medicine, 4(5), 1063-1078. https://doi.org/10.3390/jcm4051063







