Unraveling the Skin Microbiome in Hidradenitis Suppurativa: Implications for Treatment and Disease Progression
Abstract
1. Introduction
2. Results
- A.
- Samples collected from superficial HS lesions
- Samples collected from superficial lesions located in the axillary region
- 2.
- Samples collected from superficial lesions located in the inguinal and gluteal regions
- 3.
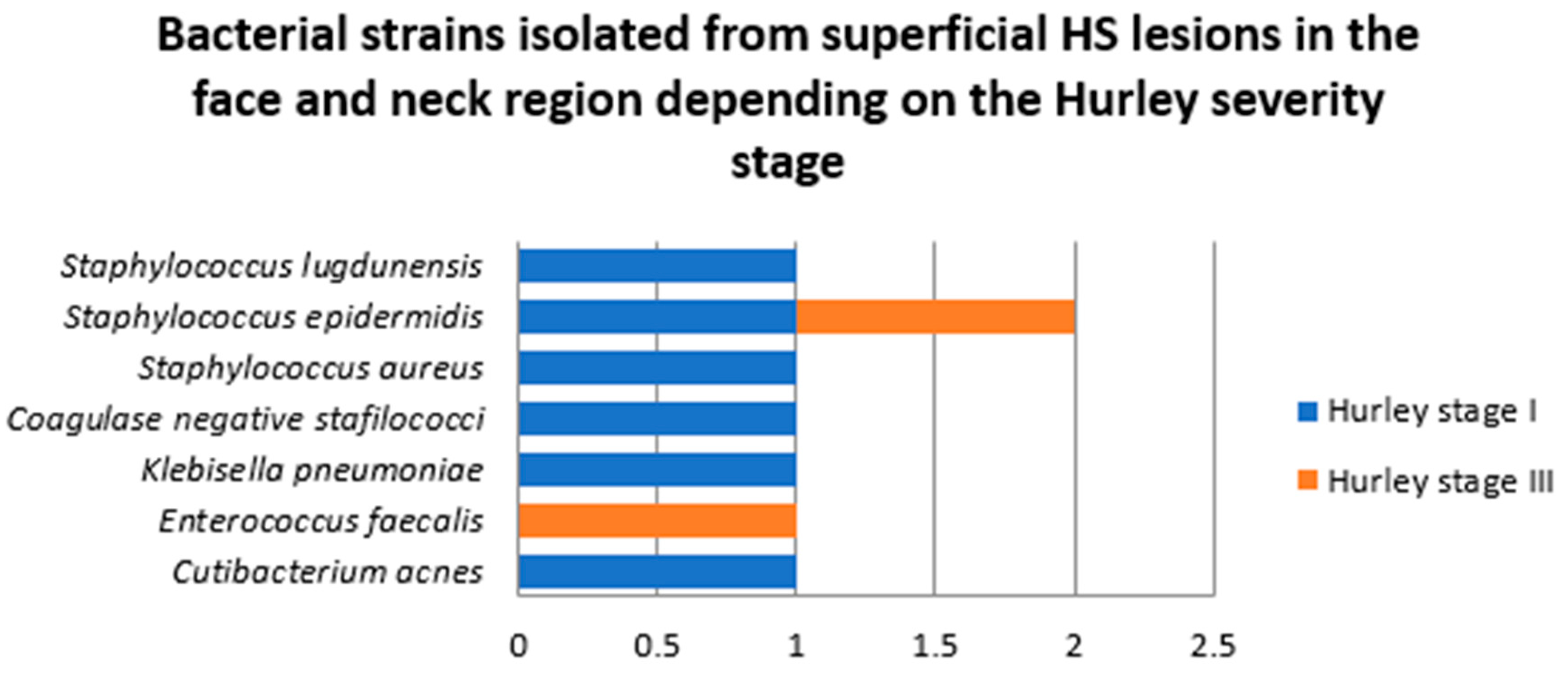
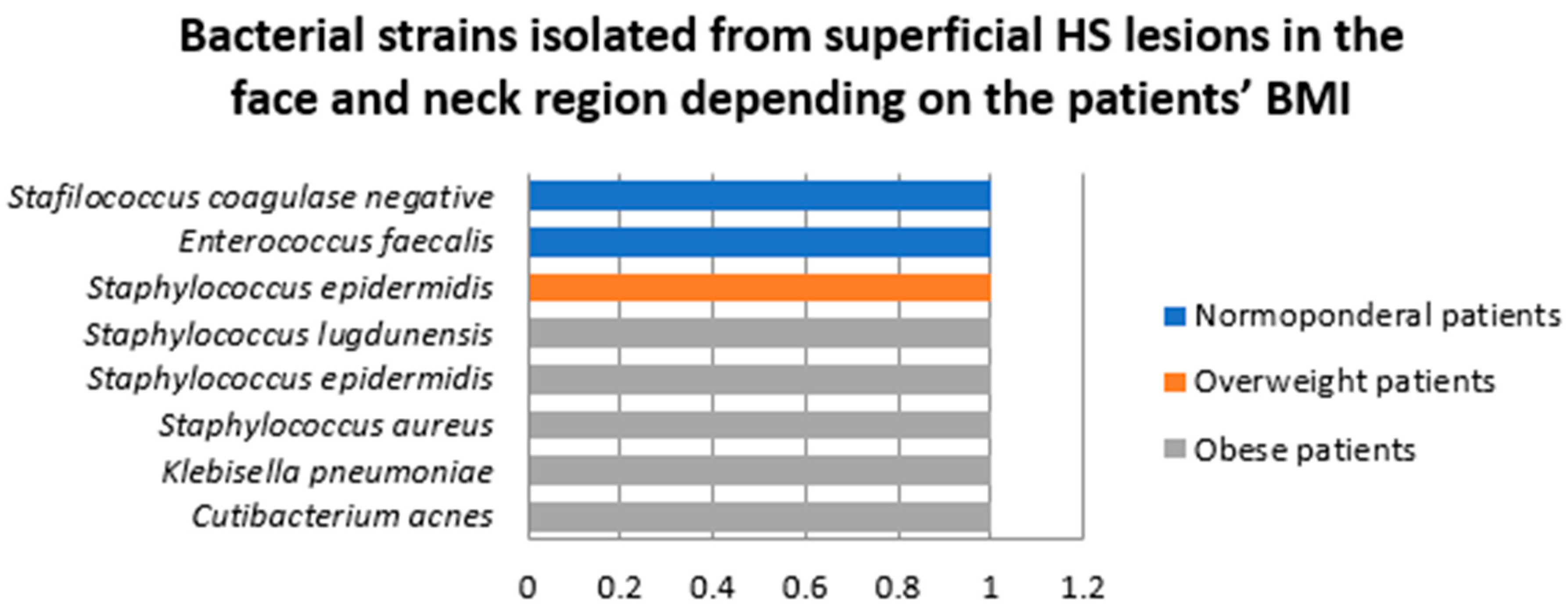
- Samples collected from superficial lesions located in face and neck region
- B.
- Samples collected from deep HS lesions
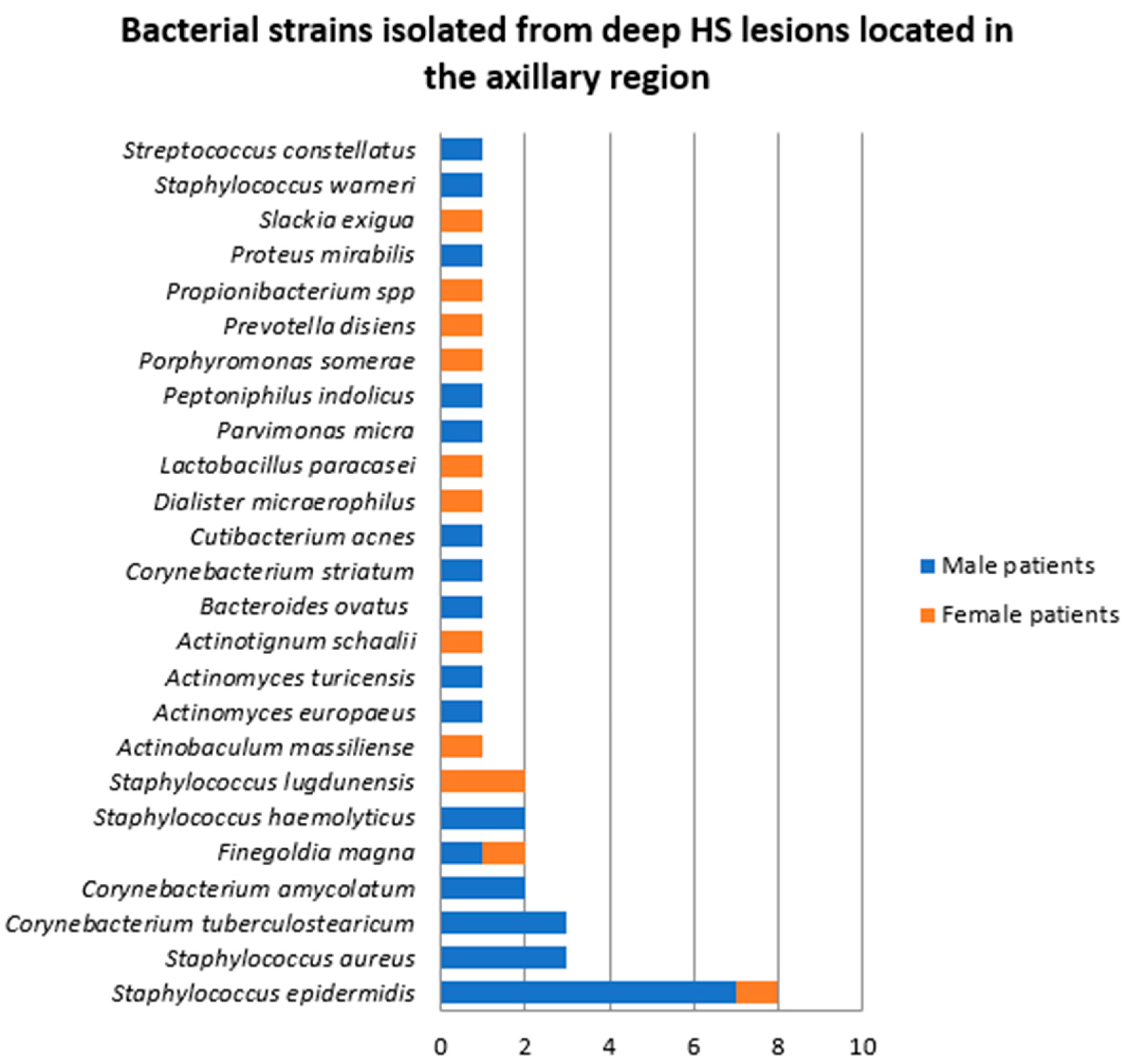
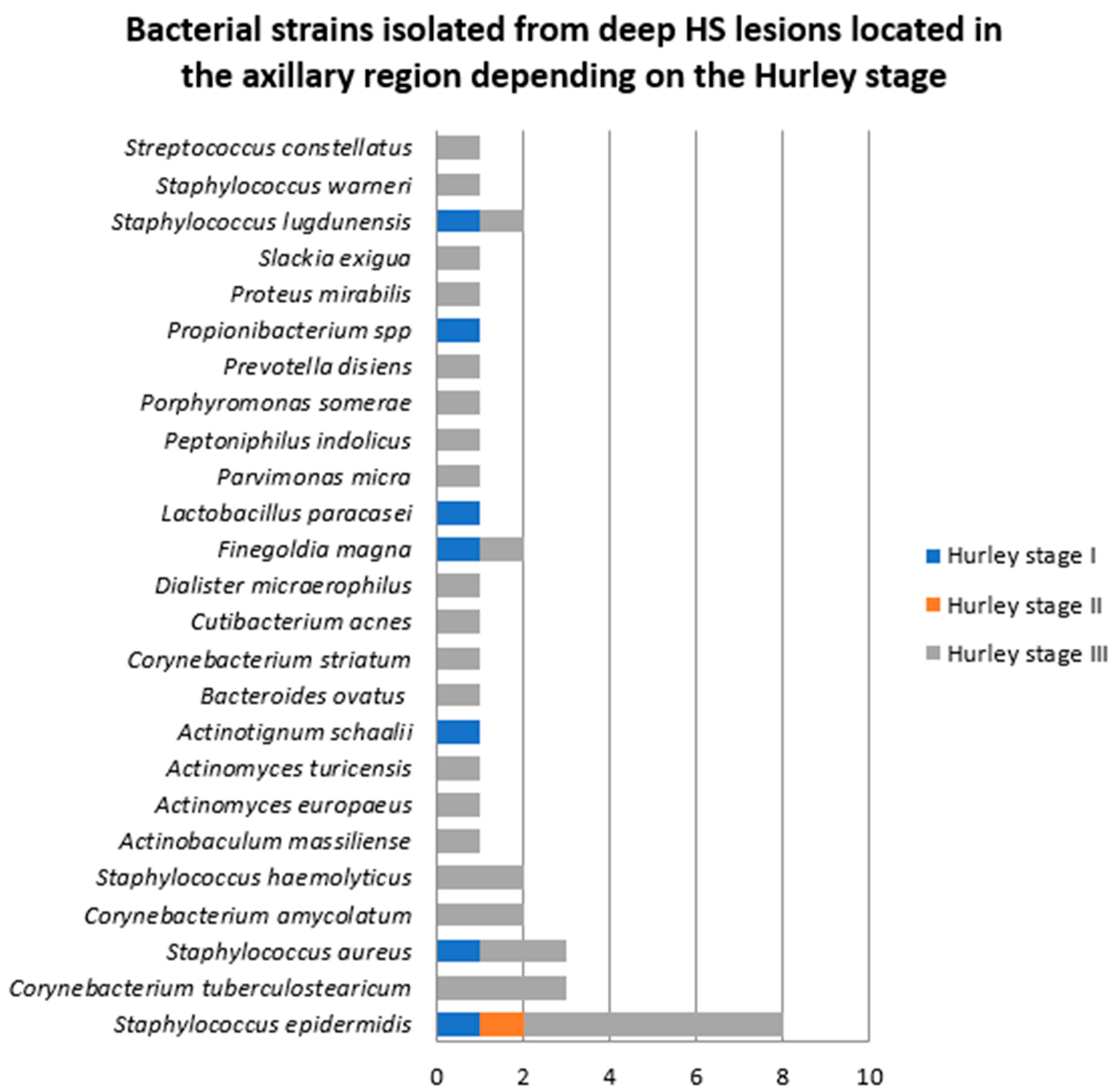
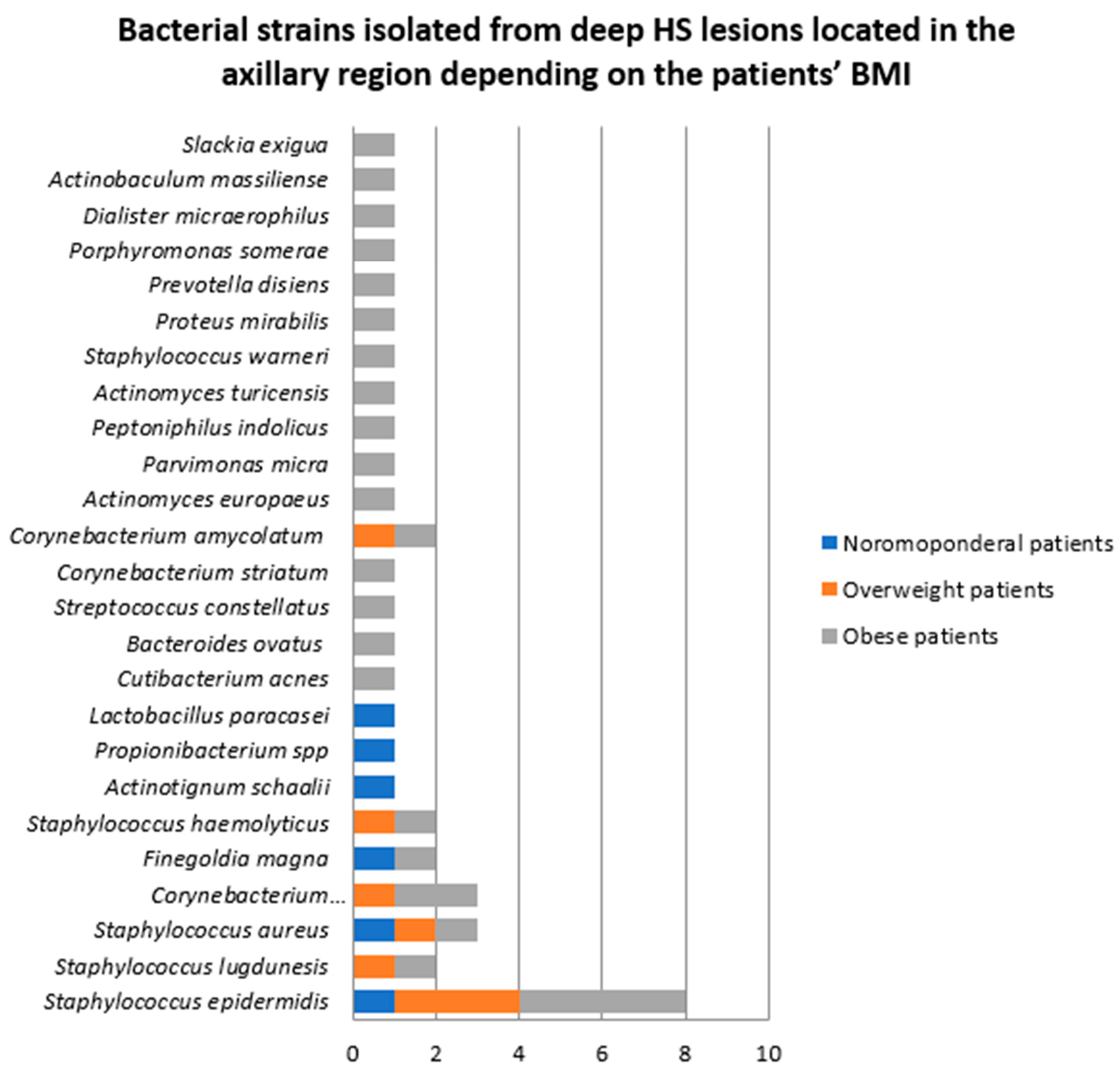
- Samples collected from deep HS lesions located in the axillary region
- 2.
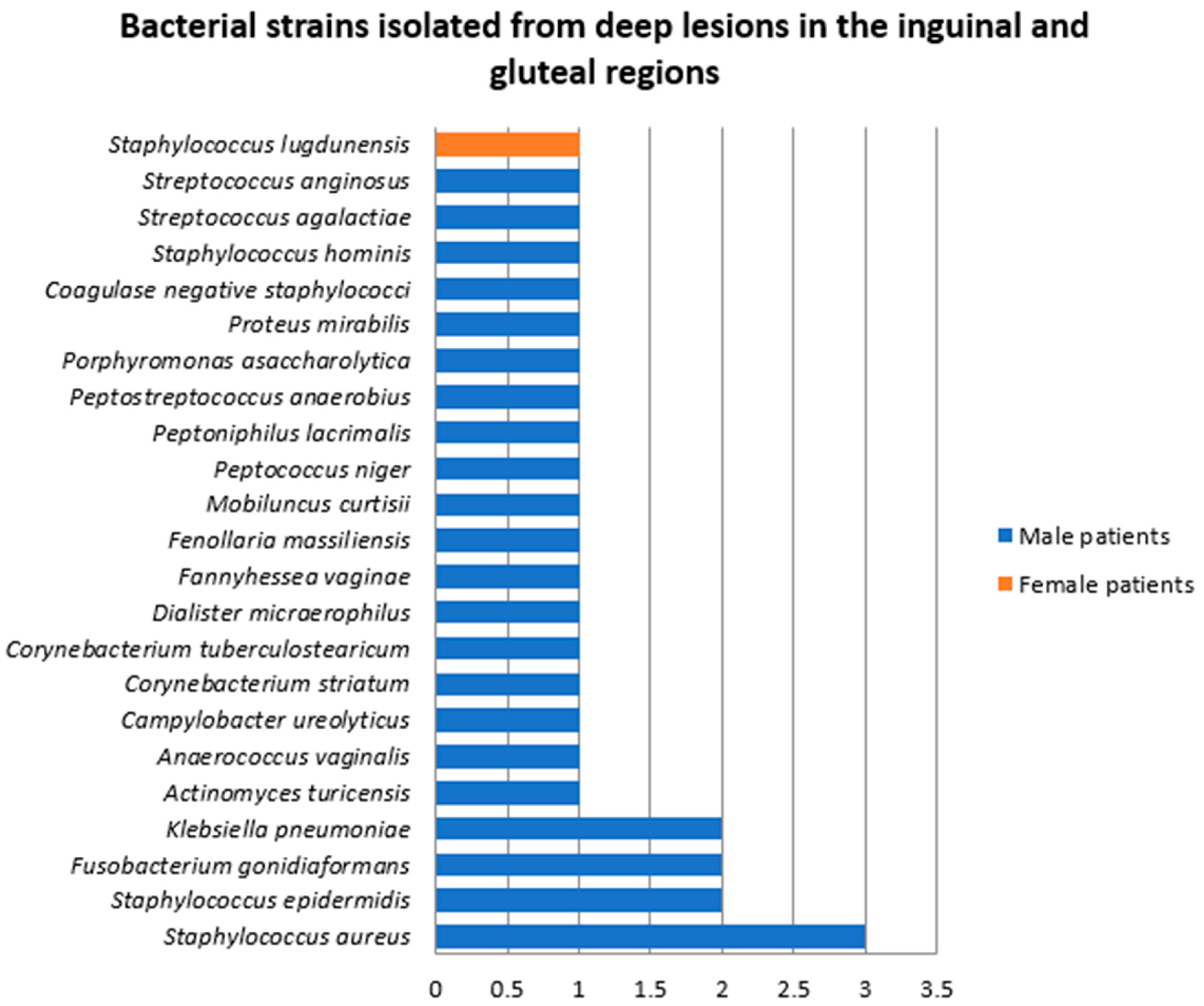
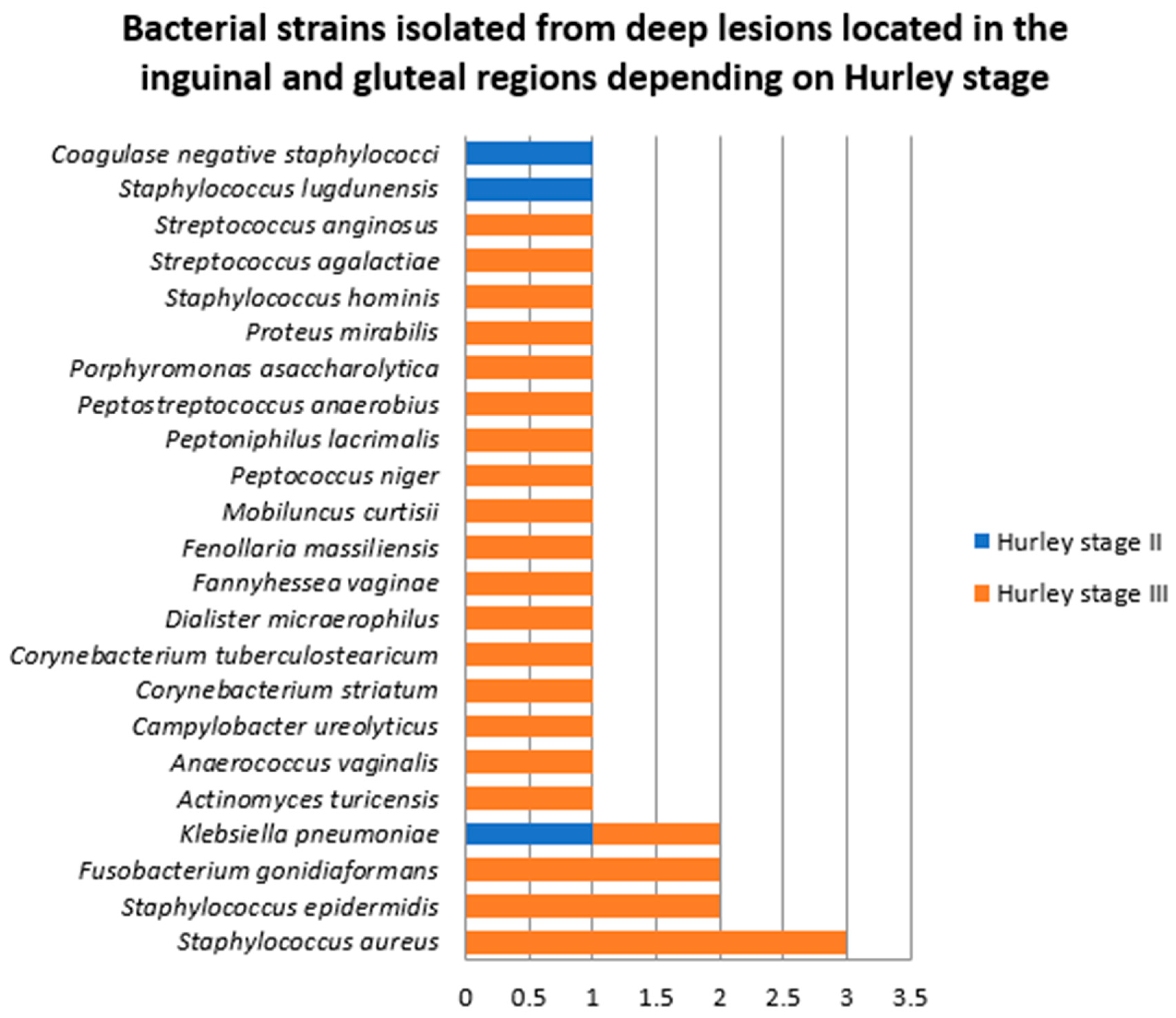
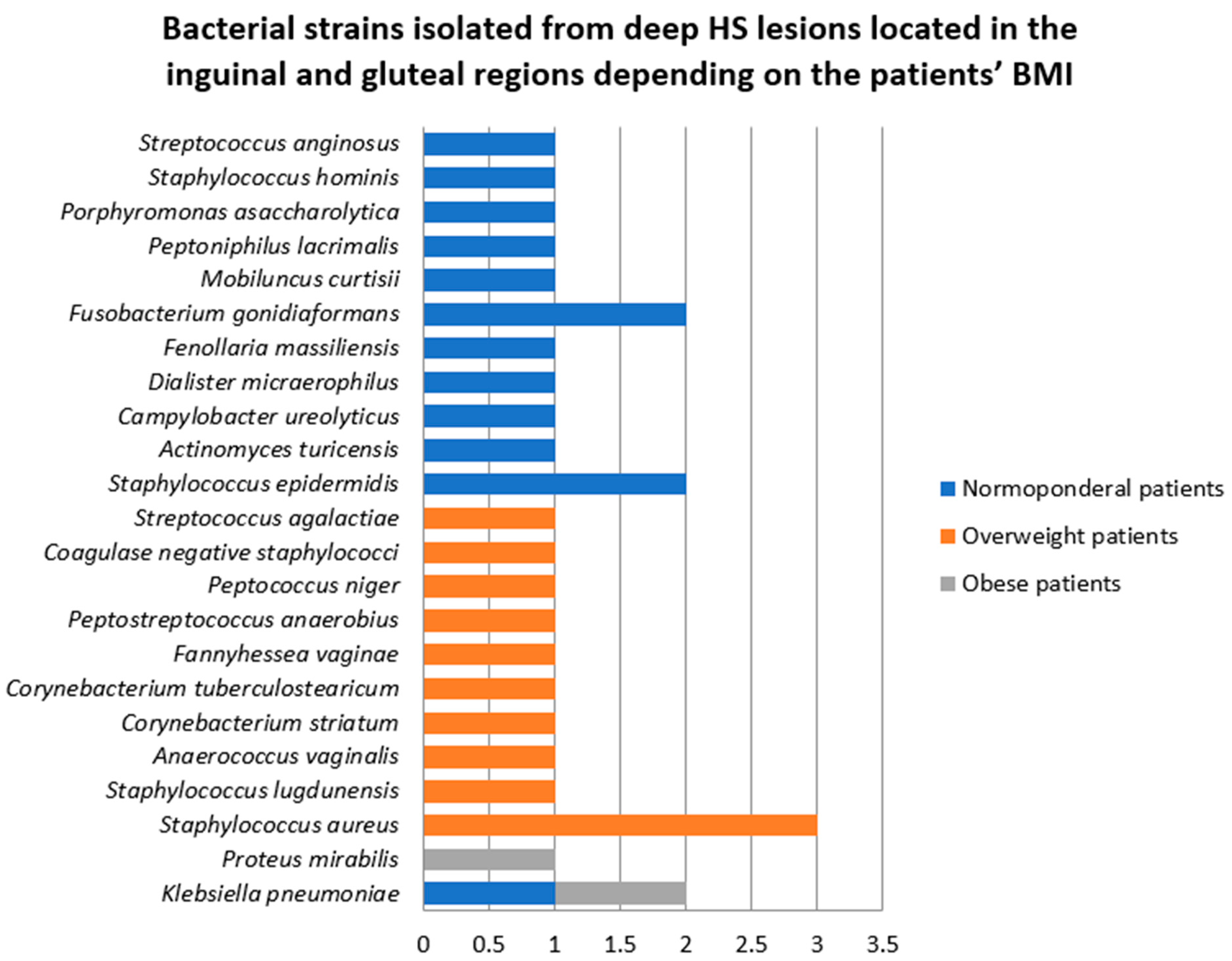
- Samples collected from deep HS lesions located in the inguinal and gluteal regions
- 3.
- Samples collected from deep HS lesions in the face and neck region
- C.
- Antibiotic resistance profile of bacterial strains isolated from HS lesions
3. Discussion
4. Materials and Methods
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HS | Hidradenitis suppurativa |
| BMI | Body mass index |
References
- Jemec, G.B. Hidradenitis suppurativa. N. Engl. J. Med. 2012, 366, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Jahns, A.C.; Killasli, H.; Nosek, D.; Lundskog, B.; Lenngren, A.; Muratova, Z.; Emtestam, L.; Alexeyev, O.A. Microbiology of hidradenitis suppurativa (acne inversa): A histological study of 27 patients. APMIS 2014, 122, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Velpeau, A. Dictionnaire de Medicine, un Repertoire des Sciences Medicales sons le Rapport, Theorique et Pratique, 2nd ed.; Nabu Press: Paris, France, 1839; p. 91. [Google Scholar]
- Verneuil, A. Etudes sur les tumeurs de la peau et quelques maladies des glandes sudoripores. Arch. Gen. Med. 1854, 4, 693–705. [Google Scholar]
- Schiefferdecker, B. Die Hautdrusen des Menschen und der Saugetierre ihre Histologishe und Rassenanatomische Bedeutung, Sowie Die Muscularis Sexualis, 1st ed.; E. Schweizerbert: Stuttgart, Germany, 1922. [Google Scholar]
- Brunsting, H.A. Hidradenitis suppurativa: Abscess of the apocrine sweat glands. Arch. Dermatol. Syph. 1939, 39, 108–120. [Google Scholar] [CrossRef]
- Cosmatos, I.; Matcho, A.; Weinstein, R.; Montgomery, M.O.; Stang, P. Analysis of patient claims data to determine the prevalence of hidradenitis suppurativa in the United States. J. Am. Acad. Dermatol. 2013, 68, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.M.; McAndrew, R.J.; Hamzavi, I. Prevalence, Risk Factors, and Comorbidities of Hidradenitis Suppurativa. Dermatol. Clin. 2016, 34, 7–16. [Google Scholar] [CrossRef] [PubMed]
- von der Werth, J.M.; Jemec, G.B. Morbidity in patients with hidradenitis suppurativa. Br. J. Dermatol. 2001, 144, 809–813. [Google Scholar] [CrossRef]
- Jemec, G.B.; Heidenheim, M.; Nielsen, N.H. Hidradenitis suppurativa-characteristics and consequences. Clin. Exp. Dermatol. 1996, 21, 419–423. [Google Scholar] [CrossRef]
- Palmer, R.A.; Keefe, M. Early-onset hidradenitis suppurativa. Clin. Exp. Dermatol. 2001, 26, 501–503. [Google Scholar] [CrossRef]
- Cucu, C.I.; Giurcaneanu, C.; Mihai, M.M.; Voiculescu, V.M.; Beiu, C.; Martin, S.; Negoita, S.; Popa, L.G.; Miron, A. Hidradenitis suppurativa in postmenopause. Acta Endocrinol. 2021, 17, 274–277. [Google Scholar] [CrossRef]
- Hurley, H.J. Axillary hyperhidrosis, apocrine bromhidrosis, hidradenitis suppurativa, and familial benign pemphigus: Surgical approach. In Dermatologic Surgery: Principles and Practice, 1st ed.; Roenigk, R.K., Roenigk, H.H., Eds.; Marcel Dekker: New York, NY, USA, 1989; pp. 729–739. [Google Scholar]
- Pink, A.E.; Simpson, M.A.; Desai, N.; Trembath, R.C.; Barker, J.N.W. γ-Secretase mutations in hidradenitis suppurativa: New insights into disease pathogenesis. J. Investig. Dermatol. 2013, 133, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wang, P.G.; Cui, Y.; Yang, S.; Zhang, Y.H.; Lin, D.; Zhang, K.Y.; Liang, Y.H.; Sun, L.D.; Yan, K.L.; et al. Inversa acne (hidradenitis suppurativa): A case report and identification of the locus at chromosome 1p21.1-1q25.3. J. Investig. Dermatol. 2006, 126, 1302–1306. [Google Scholar] [PubMed]
- Frew, J.W.; Vekic, D.A.; Woods, J.; Cains, G.D. A systematic review and critical evaluation of reported pathogenic sequence variants in hidradenitis suppurativa. Br. J. Dermatol. 2017, 177, 987–998. [Google Scholar] [CrossRef]
- Negroni, A.; Pierdomenico, M.; Cucchiara, S.; Stronati, L. NOD2 and inflammation: Current insights. J. Inflamm. Res. 2018, 11, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.K.; Ghias, M.H.; Lowes, M.A. Pathophysiology of hidradenitis suppurativa. Semin. Cutan. Med. Surg. 2017, 36, 47–54. [Google Scholar] [CrossRef]
- Knobel, M.; O’Toole, E.A.; Smith, F.J. Keratins and skin disease. Cell Tissue Res. 2015, 360, 583–589. [Google Scholar]
- Boer, J.; Weltevreden, E.F. Hidradenitis suppurativa or acne inversa. A clinicopathological study of early lesions. Br. J. Dermatol. 1996, 135, 721–725. [Google Scholar]
- van der Zee, H.H.; de Ruiter, L.; Boer, J.; van den Broecke, D.G.; den Hollander, J.C.; Laman, J.D.; Prens, E.P. Alterations in leucocyte subsets and histomorphology in normal-appearing perilesional skin and early and chronic hidradenitis suppurativa lesions. Br. J. Dermatol. 2012, 166, 98–106. [Google Scholar]
- Brook, I.; Frazier, E.H. Aerobic and anaerobic microbiology of axillary hidradenitis suppurativa. J. Med. Microbiol. 1999, 48, 103–105. [Google Scholar]
- Guet-Revillet, H.; Coignard-Biehler, H.; Jais, J.P.; Quesne, G.; Frapy, E.; Poirée, S.; Le Guern, A.S.; Le Flèche-Matéos, A.; Hovnanian, A.; Consigny, P.H.; et al. Bacterial pathogens associated with hidradenitis suppurativa, France. Emerg. Infect. Dis. 2014, 20, 1990–1998. [Google Scholar] [CrossRef]
- Nikolakis, G.; Join-Lambert, O.; Karagiannidis, I.; Guet-Revillet, H.; Zouboulis, C.C.; Nassif, A. Bacteriology of hidradenitis suppurativa/acne inversa: A review. J. Am. Acad. Dermatol. 2015, 73, S128. [Google Scholar]
- Naik, H.B.; Nassif, A.; Ramesh, M.S.; Schultz, G.; Piguet, V.; Alavi, A.; Lowes, M.A. Are Bacteria Infectious Pathogens in Hidradenitis Suppurativa? Debate at the Symposium for Hidradenitis Suppurativa Advances Meeting, November 2017. J. Investig. Dermatol. 2019, 139, 13–16. [Google Scholar] [PubMed]
- Join-Lambert, O.; Coignard, H.; Jais, J.P.; Guet-Revillet, H.; Poiree, S.; Fraitag, S.; Jullien, V.; Ribadeau-Dumas, F.; Thèze, J.; Le Guern, A.S.; et al. Efficacy of rifampin moxifloxacin-metronidazole combination therapy in hidradenitis suppurativa. Dermatology 2011, 222, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Join-Lambert, O.; Coignard-Biehler, H.; Jais, J.P.; Delage, M.; Guet-Revillet, H.; Poiree, S.; Duchatelet, S.; Jullien, V.; Hovnanian, A.; Lortholary, O.; et al. Efficacy of ertapenem in severe hidradenitis suppurativa: A pilot study in a cohort of 30 consecutive patients. J. Antimicrob. Chemother. 2016, 71, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Polkowska-Pruszyn’ska, B.; Gerkowicz, A.; Krasowska, D. The gut microbiome alterations in allergic and inflammatory skin diseases—An update. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 455–464. [Google Scholar]
- Prescott, S.L.; Larcombe, D.L.; Logan, A.C.; West, C.; Burks, W.; Caraballo, L.; Levin, M.; Etten, E.V.; Horwitz, P.; Kozyrskyj, A.; et al. The skin microbiome: Impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 2017, 10, 29. [Google Scholar]
- Radaschin, D.S.; Iancu, A.V.; Ionescu, A.M.; Gurau, G.; Niculet, E.; Bujoreanu, F.C.; Beiu, C.; Tatu, A.L.; Popa, L.G. Comparative Analysis of the Cutaneous Microbiome in Psoriasis Patients and Healthy Individuals-Insights into Microbial Dysbiosis: Final Results. Int. J. Mol. Sci. 2024, 25, 10583. [Google Scholar] [CrossRef]
- Visser, M.J.E.; Kell, D.B.; Pretorius, E. Bacterial Dysbiosis and Translocation in Psoriasis Vulgaris. Front. Cell Infect. Microbiol. 2019, 9, 7. [Google Scholar]
- Fyhrquist, N.; Muirhead, G.; Prast-Nielsen, S.; Jeanmougin, M.; Olah, P.; Skoog, T.; Jules-Clement, G.; Feld, M.; Barrientos-Somarribas, M.; Sinkko, H.; et al. Microbe-Host Interplay in Atopic Dermatitis and Psoriasis. Nat. Commun. 2019, 10, 4703. [Google Scholar]
- Radaschin, D.S.; Tatu, A.; Iancu, A.V.; Beiu, C.; Popa, L.G. The Contribution of the Skin Microbiome to Psoriasis Pathogenesis and Its Implications for Therapeutic Strategies. Medicina 2024, 60, 1619. [Google Scholar] [CrossRef]
- Carmona-Cruz, S.; Orozco-Covarrubias, L.; Sáez-de-Ocariz, M. The Human Skin Microbiome in Selected Cutaneous Diseases. Front. Cell Infect. Microbiol. 2022, 12, 834135. [Google Scholar] [CrossRef] [PubMed]
- Benzecry, V.; Grancini, A.; Guanziroli, E.; Nazzaro, G.; Barbareschi, M.; Marzano, A.V.; Muratori, S.; Veraldi, S. Hidradenitis suppurativa/acne inversa: A prospective bacteriological study and review of the literature. G. Ital. Dermatol. Venereol. 2020, 155, 459–463. [Google Scholar] [PubMed]
- Jamalpour, M.; Saki, N.; Nozari, F. Microbial profile and antibiotic susceptibility of bacteria isolated from patients with hidradenitis suppurativa. Iran. J. Dermatol. 2019, 22, 25–29. [Google Scholar]
- Nikolakis, G.; Liakou, A.I.; Bonovas, S.; Seltmann, H.; Bonitsis, N.; Join-Lambert, O.; Wild, T.; Karagiannidis, I.; Zolke-Fischer, S.; Langner, K.; et al. Bacterial colonization in hidradenitis suppurativa/acne inversa: A cross-sectional study of 50 patients and review of the literature. Acta Derm. Venereol. 2017, 97, 493–498. [Google Scholar]
- Thomas, C.; Rodby, K.A.; Thomas, J.; Shay, E.; Antony, A.K. Recalcitrant hidradenitis suppurativa: An investigation of demographics, surgical management, bacterial isolates, pharmacologic interven tion, and patient-reported health outcomes. Am. Surg. 2016, 82, 362–368. [Google Scholar] [PubMed]
- Hessam, S.; Sand, M.; Georgas, D.; Anders, A.; Bechara, F.G. Microbial profile and antimicrobial susceptibility of bacteria found in inflammatory hidradenitis suppurativa lesions. Skin Pharmacol. Physiol. 2016, 29, 161–167. [Google Scholar] [PubMed]
- Katoulis, A.C.; Koumaki, D.; Liakou, A.I.; Vrioni, G.; Koumaki, V.; Kontogiorgi, D.; Tzima, K.; Tsakris, A.; Rigopoulos, D. Aerobic and anaerobic bacteriology of hidradenitis suppurativa: A study of 22 cases. Skin Appendage Disord. 2015, 1, 55–59. [Google Scholar] [CrossRef]
- Matusiak, U.; Bieniek, A.; Szepietowski, J.C. Bacteriol ogy of hidradenitis suppurativa–which antibiotics are the treatment of choice? Acta Derm. Venereol. 2014, 94, 699–702. [Google Scholar]
- Sartorius, K.; Killasli, H.; Oprica, C.; Sullivan, A.; Lapins, J. Bacteriology of hidradenitis suppurativa exacerba tions and deep tissue cultures obtained during car bon dioxide laser treatment. Br. J. Dermatol. 2012, 166, 879–883. [Google Scholar]
- Ring, H.C.; Emtestam, L. The Microbiology of Hidradenitis Suppurativa. Dermatol. Clin. 2016, 34, 29–35. [Google Scholar]
- Jemec, G.B.; Faber, M.; Gutschik, E.; Wendelboe, P. The bacteriology of hidradenitis suppurativa. Dermatology 1996, 193, 203–206. [Google Scholar] [PubMed]
- Ring, H.C.; Riis Mikkelsen, P.; Miller, I.M.; Jenssen, H.; Fuursted, K.; Saunte, D.M.; Jemec, G.B. The bacteriology of hidradenitis suppurativa: A systematic review. Exp. Dermatol. 2015, 24, 727–731. [Google Scholar]
- Lapins, J.; Jarstrand, C.; Emtestam, L. Coagulase-negative staphylococci are the most common bacteria found in cultures from the deep portions of hidradenitis suppurativa lesions, as obtained by carbon dioxide laser surgery. Br. J. Dermatol. 1999, 140, 90. [Google Scholar]
- Riverain-Gillet, E.; Guet-Revillet, H.; Jais, J.P.; Ungeheuer, M.N.; Duchatelet, S.; Delage, M.; Lam, T.; Hovnanian, A.; Nassif, A.; Join-Lambert, O. The surface microbiome of clinically unaffected skin folds in hidradenitis suppurativa: A cross-sectional culture based and 16S rRNA gene amplicon sequencing study in 60 patients. J. Investig. Dermatol. 2020, 140, 1847–1855.e6. [Google Scholar] [PubMed]
- Ring, H.C.; Sigsgaard, V.; Thorsen, J.; Fuursted, K.; Fabricius, S.; Saunte, D.M.; Jemec, G.B. The microbiome of tunnels in hidradenitis suppurativa patients. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 1775–1780. [Google Scholar]
- Guet-Revillet, H.; Jais, J.P.; Ungeheuer, M.N.; Coignard-Biehler, H.; Duchatelet, S.; Delage, M.; Lam, T.; Hovnanian, A.; Lortholary, O.; Nassif, X.; et al. The microbiological landscape of anaerobic infections in hidradenitis suppurativa: A prospective metage nomic study. Clin. Infect. Dis. 2017, 65, 282–291. [Google Scholar] [PubMed]
- Naik, H.B.; Jo, J.H.; Paul, M.; Kong, H.H. Skin microbiota perturbations are distinct and disease severity-de pendent in hidradenitis suppurativa. J. Investig. Dermatol. 2019, 140, 922–925.e3. [Google Scholar]
- Ring, H.C.; Thorsen, J.; Saunte, D.M.; Lilje, B.; Bay, L.; Riis, P.T.; Larsen, N.; Andersen, L.O.; Nielsen, H.V.; Miller, I.M.; et al. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 2017, 153, 897–905. [Google Scholar]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar]
- Frank, K.L.; Del Pozo, J.L.; Patel, R. From clinical microbiology to infection pathogenesis: How daring to be different works for Staphylococcus lugdunensis. Clin. Microbiol. Rev. 2008, 21, 111–133. [Google Scholar]
- Brook, I. The role of anaerobic bacteria in cutaneous and soft tissue abscesses and infected cysts. Anaerobe 2007, 13, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.C.; Frew, J.W.; Krueger, J.G. A systematic review and critical appraisal of metagenomic and culture studies in hidradenitis suppurativa. Exp. Dermatol. 2021, 30, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Ring, H.C.; Thorsen, J.; Jørgensen, A.H.; Bay, L.; Bjarnsholt, T.; Fuursted, K.; Thomsen, S.F.; Jemec, G.B. Predictive Metagenomic Analysis Reveals a Role of Cutaneous Dysbiosis in the Development of Hidradenitis Suppurativa. J. Investig. Dermatol. 2020, 140, 1473–1476. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed]
- Thomi, R.; Schlapbach, C.; Yawalkar, N.; Simon, D.; Yerly, D.; Hunger, R.E. Elevated levels of the antimicrobial peptide LL-37 in hidradenitis suppurativa are associated with a Th1/Th17 immune response. Exp. Dermatol. 2018, 27, 172–177. [Google Scholar] [CrossRef]
- Hotz, C.; Boniotto, M.; Guguin, A.; Surenaud, M.; Jean-Louis, F.; Tisserand, P.; Ortonne, N.; Hersant, B.; Bosc, R.; Poli, F.; et al. Intrinsic Defect in Keratinocyte Function Leads to Inflammation in Hidradenitis Suppurativa. J. Investig. Dermatol. 2016, 136, 1768–1780. [Google Scholar] [CrossRef]
- Tricarico, P.M.; Boniotto, M.; Genovese, G.; Zouboulis, C.C.; Marzano, A.V.; Crovella, S. An Integrated Approach to Unravel Hidradenitis Suppurativa Etiopathogenesis. Front. Immunol. 2019, 10, 892. [Google Scholar] [CrossRef]
- Witte-Händel, E.; Wolk, K.; Tsaousi, A.; Irmer, M.L.; Mößner, R.; Shomroni, O.; Lingner, T.; Witte, K.; Kunkel, D.; Salinas, G.; et al. The IL-1 Pathway Is Hyperactive in Hidradenitis Suppurativa and Contributes to Skin Infiltration and Destruction. J. Investig. Dermatol. 2019, 139, 1294–1305. [Google Scholar] [CrossRef]
- Schlapbach, C.; Hänni, T.; Yawalkar, N.; Hunger, R.E. Expression of the IL-23/Th17 pathway in lesions of hidradenitis suppurativa. J. Am. Acad. Dermatol. 2011, 65, 790–798. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Bechara, F.G.; Benhadou, F.; Bettoli, V.; Bukvić Mokos, Z.; Del Marmol, V.; Dolenc-Voljč, M.; Giamarellos-Bourboulis, E.J.; Grimstad, Ø.; Guillem, P.; et al. European S2k guidelines for hidradenitis suppurativa/acne inversa part 2: Treatment. J. Eur. Acad. Dermatol. Venereol. 2024, 19. [Google Scholar] [CrossRef]
- Popa, L.G.; Giurcaneanu, C.; Mihai, M.M.; Beiu, C.; Orzan, O.A.; Negoita, S.I.; Burcea, M.; Turlea, R.I.; Enachescu, C.I. The use of cadaveric skin allografts in the management of extensive wounds. Rom. J. Leg. Med. 2021, 29, 37–44. [Google Scholar]
- Bettoli, V.; Manfredini, M.; Massoli, L.; Carillo, C.; Barozzi, A.; Amendolagine, G.; Ruina, G.; Musmeci, D.; Libanore, M.; Curtolo, A.; et al. Rates of antibiotic resistance/sensitivity in bacterial cultures of hidradenitis suppurativa patients. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 930–936. [Google Scholar] [PubMed]
- Vaienti, S.; Nazzaro, G.; Grancini, A.; Calzari, P.; Zaccaria, G.; Veraldi, S.; Vaienti, L. Lymph Node Involvement in Axillary Hidradenitis Suppurativa: A Clinical, Ultrasonographic and Bacteriological Study Conducted during Radical Surgery. J. Clin. Med. 2021, 10, 1433. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

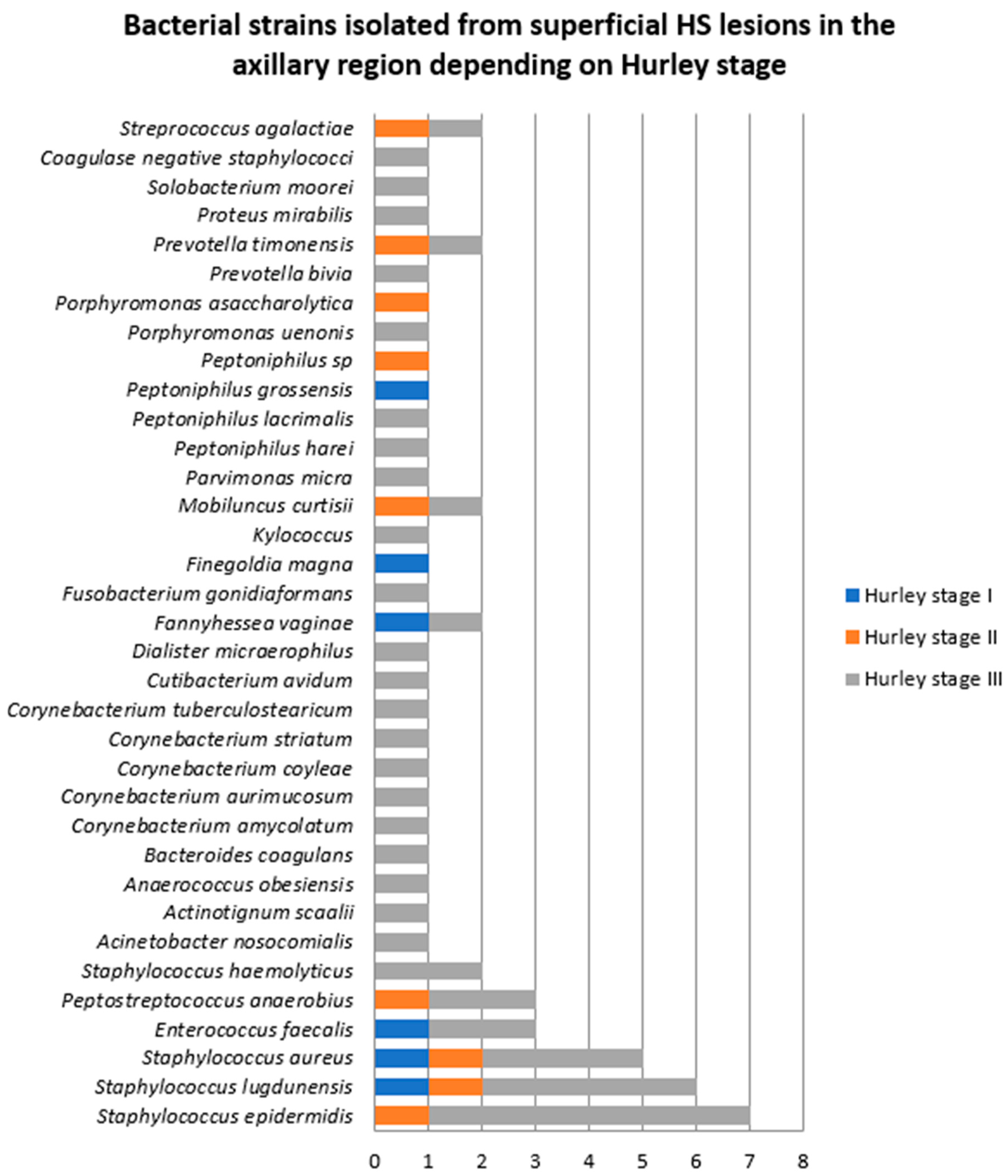
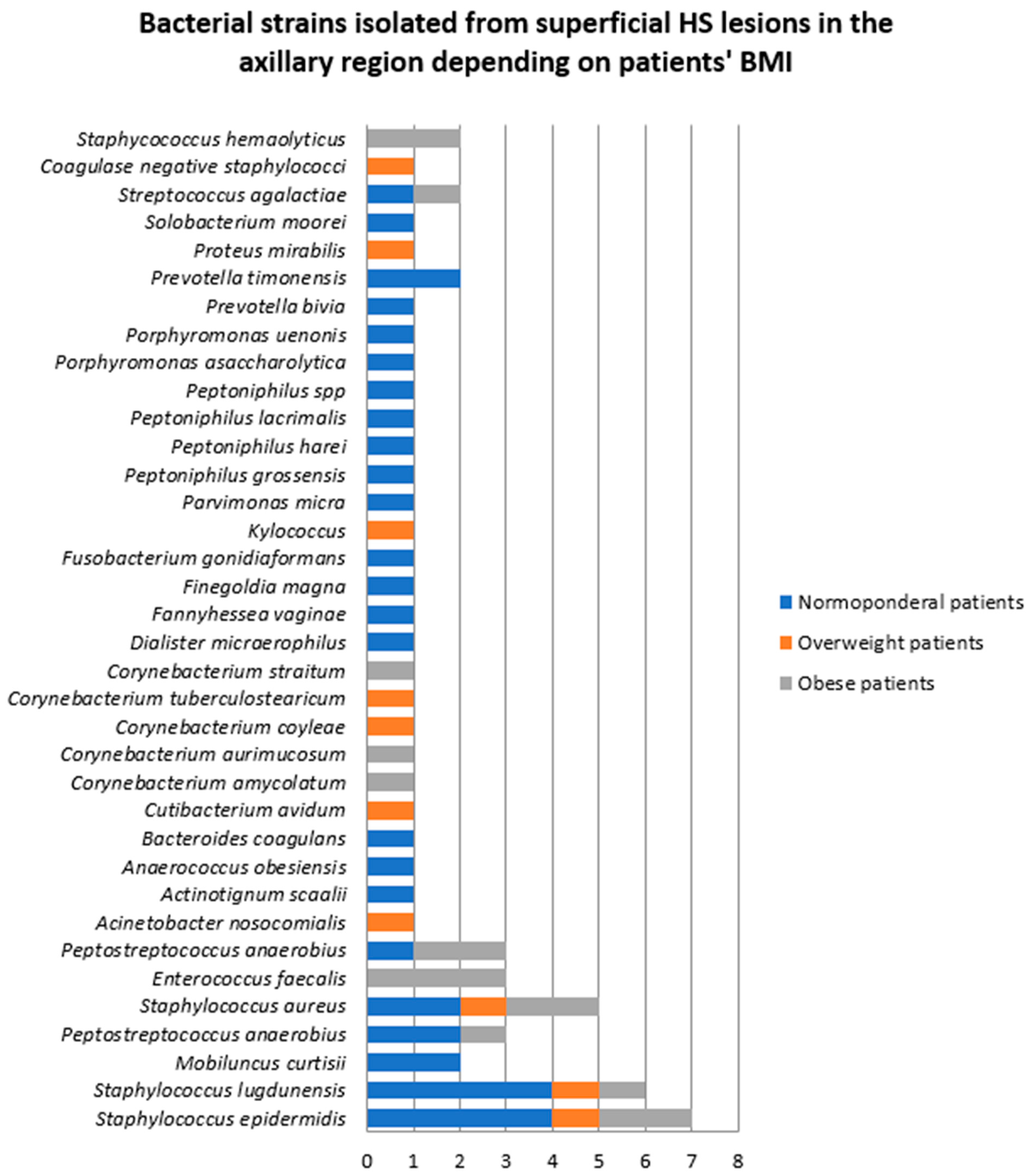

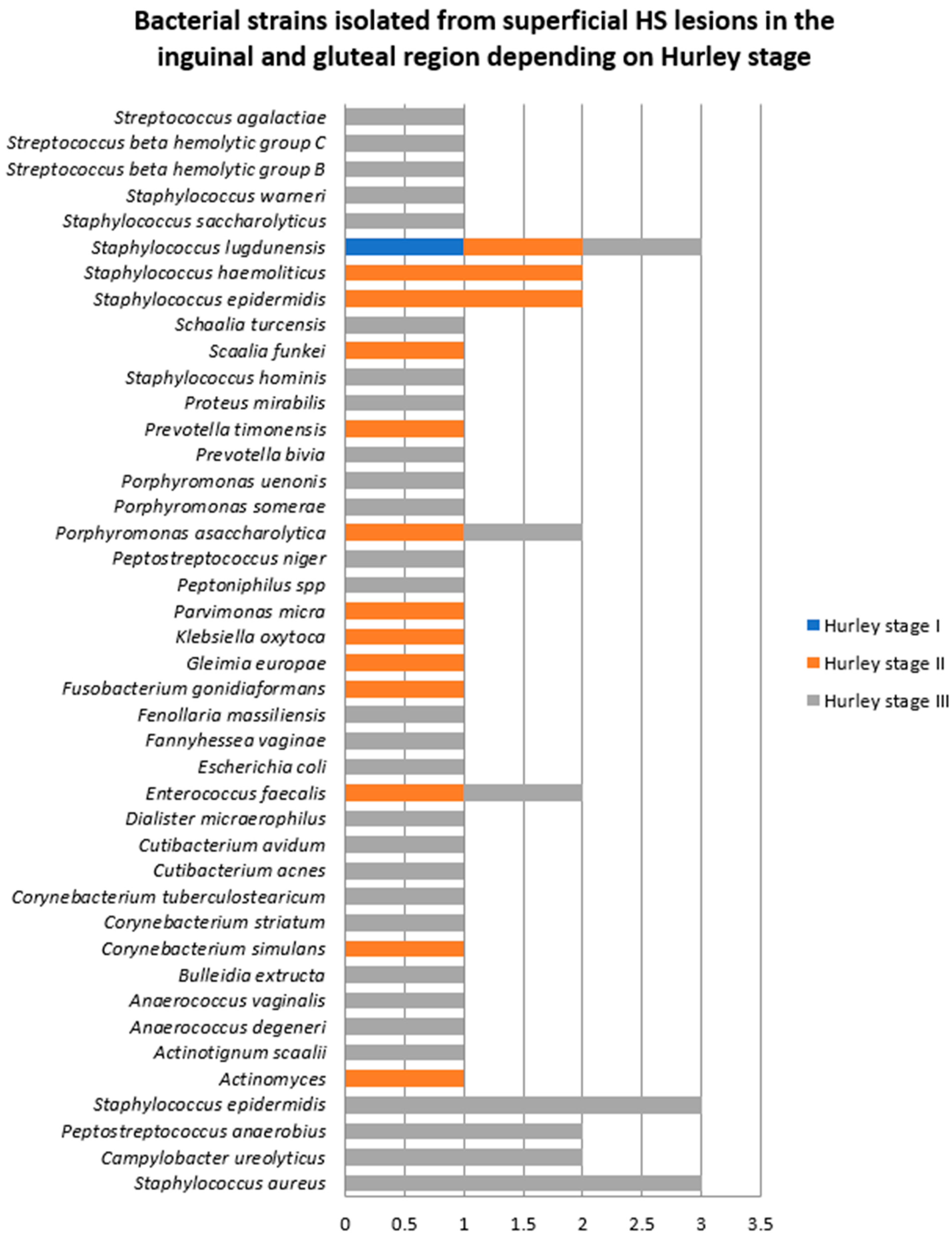
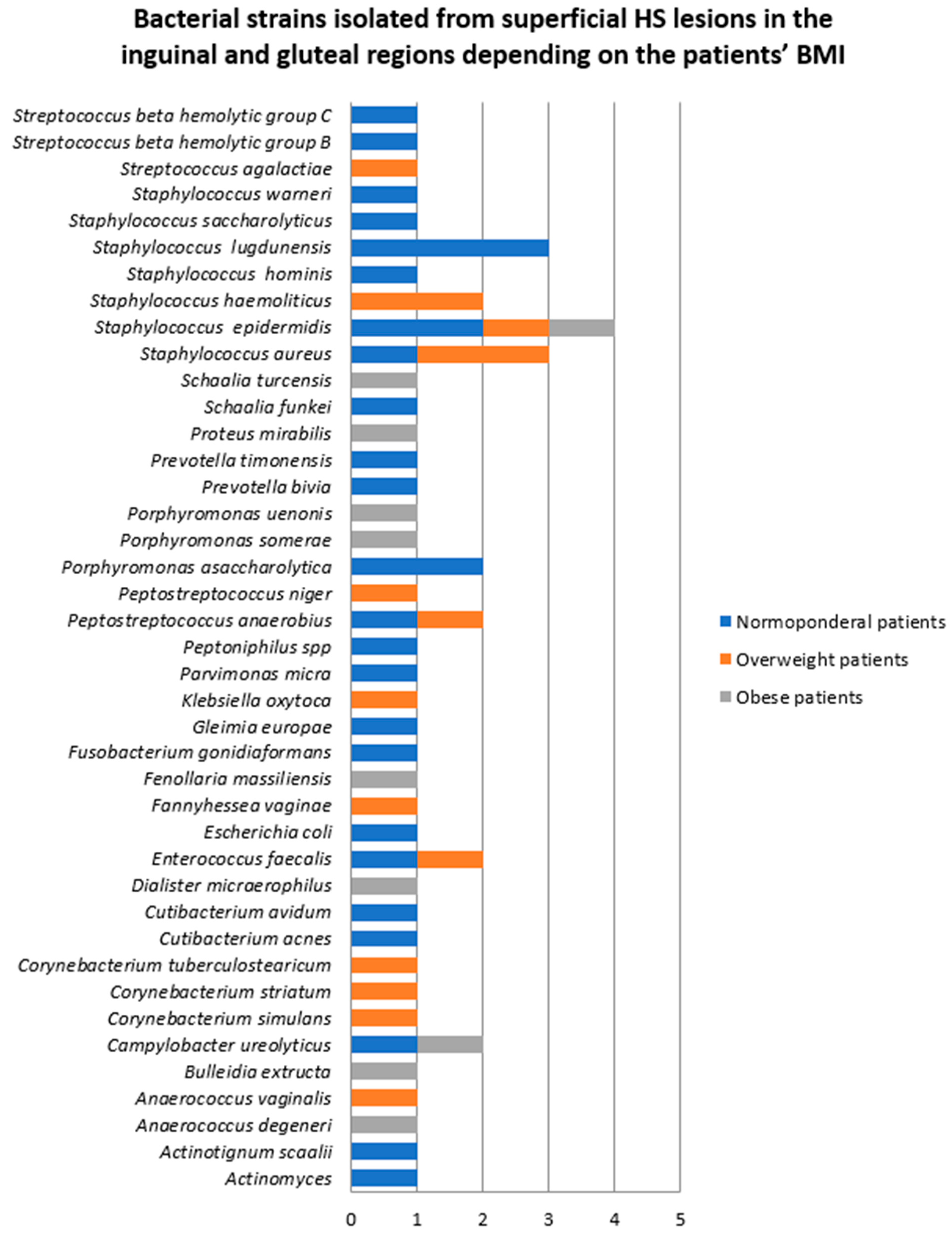

| Bacterial Strains Isolated from Superficial HS Lesions | Total | Female Patients | Male Patients |
|---|---|---|---|
| Staphylococcus epidermidis | 14 | 1 | 13 |
| Staphylococcus aureus | 10 | 1 | 9 |
| Staphylococcus lugdunensis | 9 | 4 | 5 |
| Peptoniphilus spp | 6 | 1 | 5 |
| Enterococcus faecalis | 5 | 2 | 3 |
| Proteus mirabilis | 4 | 1 | 3 |
| Staphylococcus haemolyticus | 4 | 2 | 2 |
| Porphyromonas asaccharolytica | 3 | 2 | 1 |
| Streptococcus agalactiae | 3 | 2 | 1 |
| Actinotignum scaalii | 2 | 0 | 2 |
| Campylobacter ureolyticus | 2 | 0 | 2 |
| Cutibacterium acnes | 2 | 1 | 1 |
| Cutibacterium avidum | 2 | 1 | 1 |
| Dialister micraerophilus | 2 | 0 | 2 |
| Fannyhessea vaginae | 2 | 0 | 2 |
| Fusobacterium gonidiaformans | 2 | 1 | 1 |
| Mobiluncus curtisii | 2 | 0 | 2 |
| Parvimonas micra | 2 | 1 | 1 |
| Porphyromonas uenonis | 2 | 0 | 2 |
| Prevotella bivia | 2 | 0 | 2 |
| Prevotella timonensis | 2 | 2 | 0 |
| Coagulase negative staphilococci | 2 | 1 | 1 |
| Staphylococcus hominis | 2 | 0 | 2 |
| Corynebacterium tuberculostearicum | 2 | 0 | 2 |
| Acinetobacter nosocomialis | 1 | 0 | 1 |
| Actinomyces | 1 | 0 | 1 |
| Anaerococcus degeneri | 1 | 0 | 1 |
| Anaerococcus obesiensis | 1 | 0 | 1 |
| Anaerococcus vaginalis | 1 | 0 | 1 |
| Bacteroides coagulans | 1 | 0 | 1 |
| Bulleidia extructa | 1 | 0 | 1 |
| Corynebacterium amycolatum | 1 | 0 | 1 |
| Corynebacterium aurimucosum | 1 | 0 | 1 |
| Corynebacterium coyleae | 1 | 0 | 1 |
| Corynebacterium striatum | 1 | 0 | 1 |
| Corynebacterium simulans | 1 | 0 | 1 |
| Escherichia coli | 1 | 0 | 1 |
| Fenollaria massiliensis | 1 | 0 | 1 |
| Finegoldia magna | 1 | 0 | 1 |
| Gleimia europae | 1 | 1 | 0 |
| Klebisella pneumoniae | 1 | 0 | 1 |
| Klebsiella oxytoca | 1 | 0 | 1 |
| Kylococcus | 1 | 0 | 1 |
| Peptoniphilus grossensis | 1 | 0 | 1 |
| Peptoniphilus harei | 1 | 0 | 1 |
| Peptoniphilus lacrimalis | 1 | 0 | 1 |
| Peptostreptococcus niger | 1 | 0 | 1 |
| Porphyromonas somerae | 1 | 0 | 1 |
| Staphylococcus saccharolyticus | 1 | 1 | 0 |
| Staphylococcus warneri | 1 | 1 | 0 |
| Schaalia funkei | 1 | 1 | 0 |
| Schaalia turcensis | 1 | 0 | 1 |
| Solobacterium moorei | 1 | 0 | 1 |
| Group C streptococcus | 1 | 0 | 1 |
| Peptostreptococcus anaerobius | 1 | 1 | 0 |
| Total | 120 | 28 | 92 |
| Bacterial Strains Isolated from Deep HS Lesions | Total | Female Patients | Male Patients |
|---|---|---|---|
| Staphylococcus epidermidis | 11 | 1 | 10 |
| Staphylococcus aureus | 7 | 0 | 7 |
| Corynebacterium tuberculostearicum | 6 | 0 | 6 |
| Fenollaria massiliensis | 3 | 0 | 3 |
| Staphylococcus lugdunensis | 3 | 3 | 0 |
| Actinomyces turicensis | 2 | 0 | 2 |
| Actinotignum schaalii | 2 | 2 | 0 |
| Corynebacterium amycolatum | 2 | 0 | 2 |
| Corynebacterium striatum | 2 | 0 | 2 |
| Campylobacter ureolyticus | 2 | 1 | 1 |
| Dialister micraerophilus | 2 | 0 | 2 |
| Fusobacterium gonidiaformans | 2 | 0 | 2 |
| Klebsiella pneumoniae | 2 | 0 | 2 |
| Proteus mirabilis | 2 | 0 | 2 |
| Propionibacterium spp. | 2 | 1 | 1 |
| Coagulase negative staphylococci | 2 | 1 | 1 |
| Staphylococcus haemolyticus | 2 | 0 | 2 |
| Streptococcus constellatus | 2 | 0 | 2 |
| Actinobaculum massiliense | 1 | 1 | 0 |
| Actinomyces europaeus | 1 | 0 | 1 |
| Anaerococcus vaginalis | 1 | 0 | 1 |
| Bacteroides ovatus | 1 | 0 | 1 |
| Cutibacterium acnes | 1 | 0 | 1 |
| Dialister micraerophilus | 1 | 1 | 0 |
| Enterococcus faecalis | 1 | 1 | 0 |
| Fannyhessea vaginae | 1 | 0 | 1 |
| Lactobacillus paracasei | 1 | 1 | 0 |
| Mobiluncus curtisii | 1 | 0 | 1 |
| Parvimonas micra | 1 | 0 | 1 |
| Peptococcus niger | 1 | 0 | 1 |
| Peptoniphilus indolicus | 1 | 0 | 1 |
| Peptoniphilus lacrimalis | 1 | 0 | 1 |
| Peptostreptococcus anaerobius | 1 | 0 | 1 |
| Porphyromonas asaccharolytica | 1 | 0 | 1 |
| Porphyromonas somerae | 1 | 1 | 0 |
| Prevotella disiens | 1 | 1 | 0 |
| Staphylococcus hominis | 1 | 0 | 1 |
| Staphylococcus warneri | 1 | 0 | 1 |
| Slackia exigua | 1 | 1 | 0 |
| Streptococcus agalactiae | 1 | 0 | 1 |
| Streptococcus anginosus | 1 | 0 | 1 |
| Finegoldia magna | 2 | 2 | 0 |
| Total | 81 | 18 | 63 |
| Bacterial Strain | Antibiotic Resistance |
|---|---|
| Actinotignum schaalii | Metronidazole |
| Enterococcus faecalis | Doxycycline |
| Escherichia coli | Ampicillin, Levofloxacin, Co-trimoxazole |
| Finegoldia magna | Clindamycin |
| Klebsiella pneumoniae | Co-trimoxazole |
| Propionibacterium spp. | Metronidazole |
| Proteus mirabilis | Gentamicin |
| Staphylococcus aureus | Penicillin |
| Staphylococcus aureus | Penicillin |
| Staphylococcus aureus-MRSA | Oxacillin, Erythromycin, Clindamycin |
| Staphylococcus aureus | Oxacillin, Erythromycin, Clindamycin |
| Staphylococcus aureus-MRSA, MLSBi | Penicillin, Oxacillin, Erythromycin, Clindamycin, Tetracycline |
| Coagulase negative staphylococcus | Penicillin, Erythromycin, Clindamycin |
| Coagulase negative staphylococcus | Penicillin, Erythromycin, Clindamycin, Tetracycline |
| Coagulase negative staphylococcus | Penicillin, Oxacillin, Tetracycline |
| Staphylococcus lugdunensis | Penicillin, Erythromycin, Clindamycin |
| Staphylococcus lugdunensis | Penicillin |
| Group B streptococcus | Erythromycin, Clindamicyne, Doxycycline |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cucu, C.I.; Giurcăneanu, C.; Mihai, M.M.; Andronic, T.; Ancuta, I.; Popa, M.I.; Macovei, I.S.; Popa, L.G. Unraveling the Skin Microbiome in Hidradenitis Suppurativa: Implications for Treatment and Disease Progression. J. Clin. Med. 2025, 14, 2424. https://doi.org/10.3390/jcm14072424
Cucu CI, Giurcăneanu C, Mihai MM, Andronic T, Ancuta I, Popa MI, Macovei IS, Popa LG. Unraveling the Skin Microbiome in Hidradenitis Suppurativa: Implications for Treatment and Disease Progression. Journal of Clinical Medicine. 2025; 14(7):2424. https://doi.org/10.3390/jcm14072424
Chicago/Turabian StyleCucu, Corina Ioana, Călin Giurcăneanu, Mara Madalina Mihai, Teodora Andronic, Ioan Ancuta, Mircea Ioan Popa, Ioana Sabina Macovei, and Liliana Gabriela Popa. 2025. "Unraveling the Skin Microbiome in Hidradenitis Suppurativa: Implications for Treatment and Disease Progression" Journal of Clinical Medicine 14, no. 7: 2424. https://doi.org/10.3390/jcm14072424
APA StyleCucu, C. I., Giurcăneanu, C., Mihai, M. M., Andronic, T., Ancuta, I., Popa, M. I., Macovei, I. S., & Popa, L. G. (2025). Unraveling the Skin Microbiome in Hidradenitis Suppurativa: Implications for Treatment and Disease Progression. Journal of Clinical Medicine, 14(7), 2424. https://doi.org/10.3390/jcm14072424










