Mutational Spectrum and Clinical Outcomes of Myelodysplastic/Myeloproliferative Neoplasms: A Single-Institution Study in Korea with Emphasis on U2AF1
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Next-Generation Sequencing for Myeloid Neoplasm-Related Genes
2.3. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
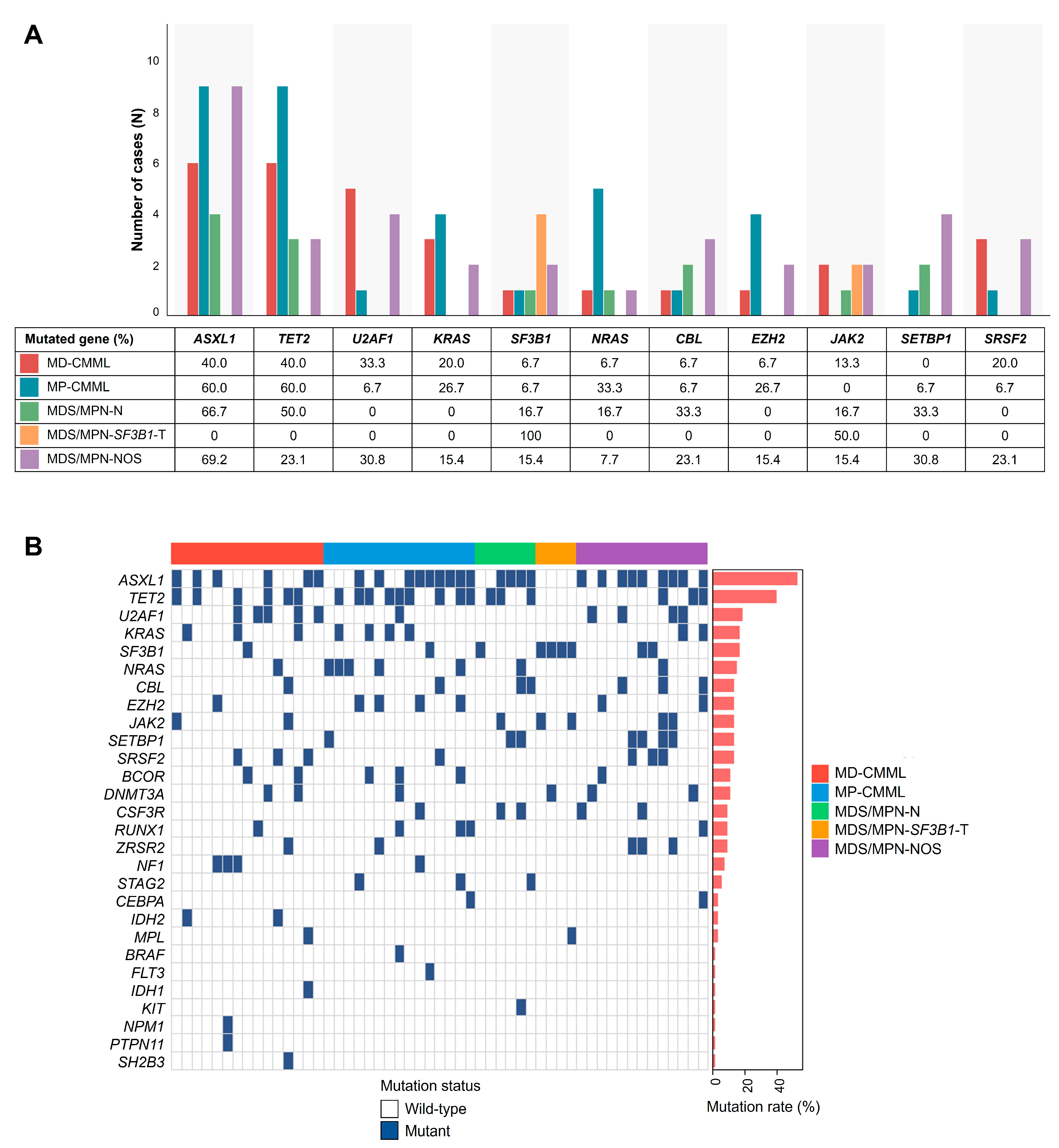
3.2. Mutational Landscape Across MDS/MPN Subtypes
3.3. Survival and Prognostic Factors in the MDS/MPN Cohort
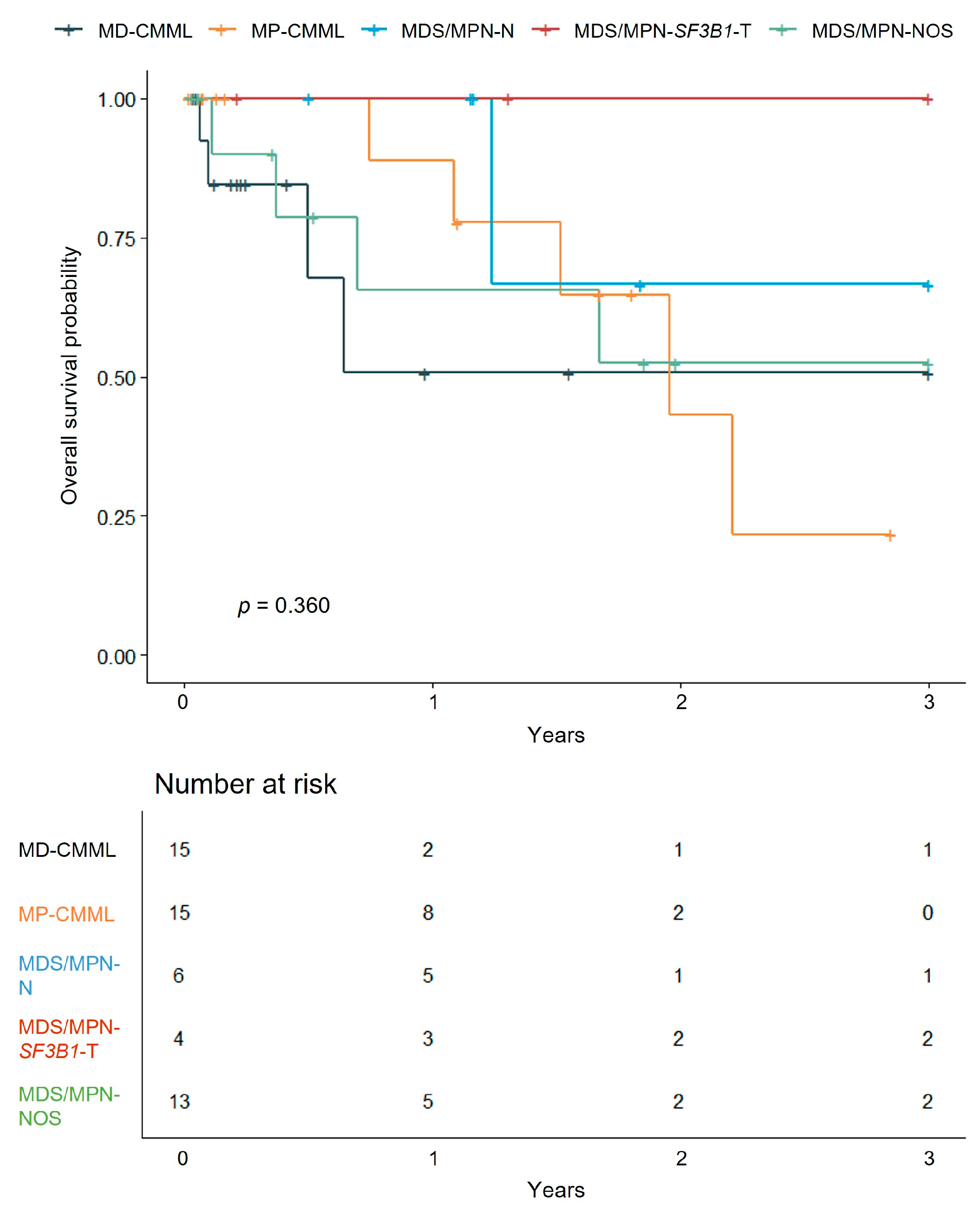
3.3.1. Survival Analysis by MDS/MPN Subtypes
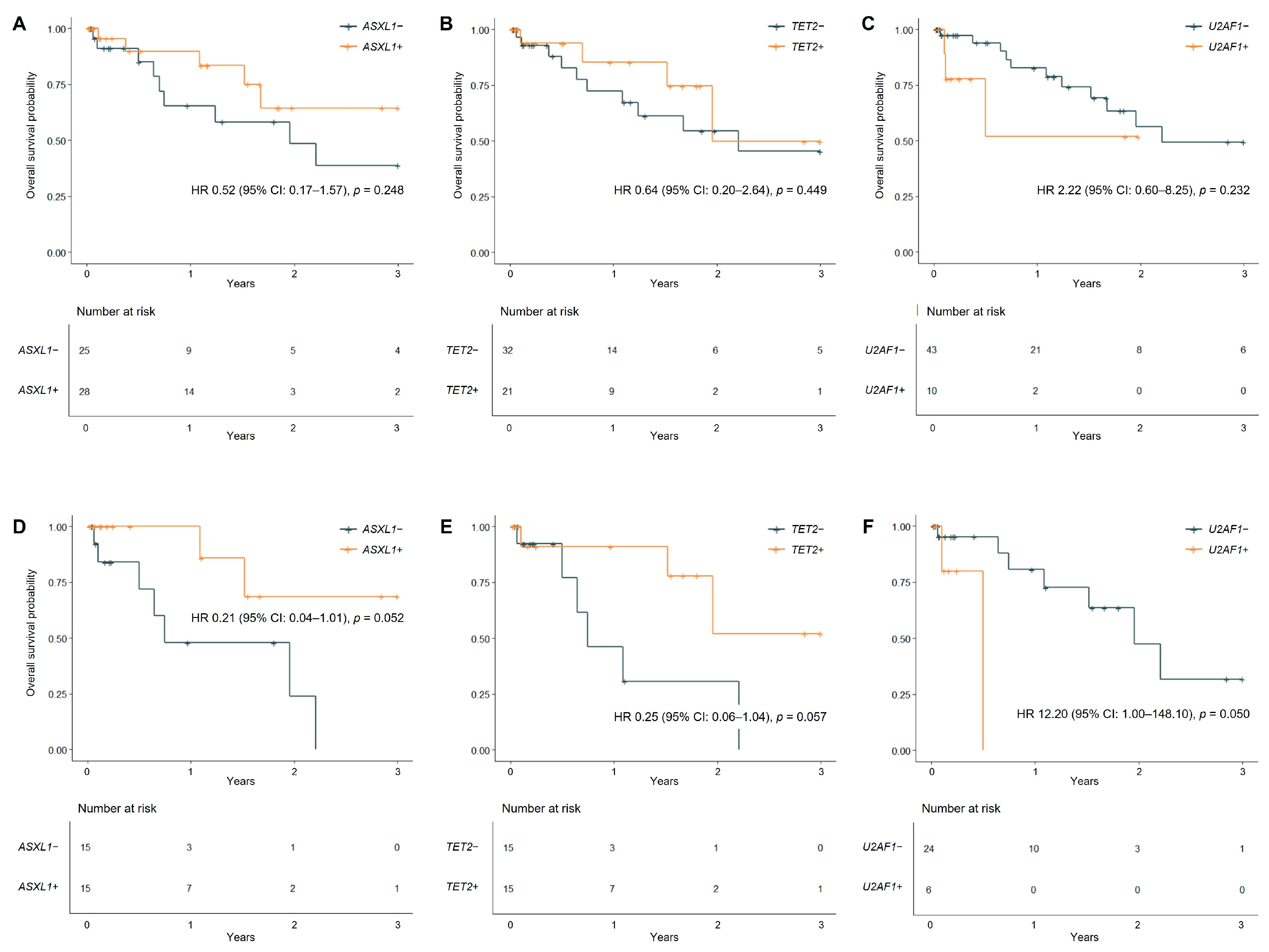
3.3.2. Prognostic Impact of Gene Mutations in MDS/MPN
3.4. Survival and Prognostic Factors in CMML
3.4.1. Prognostic Impact of Gene Mutations
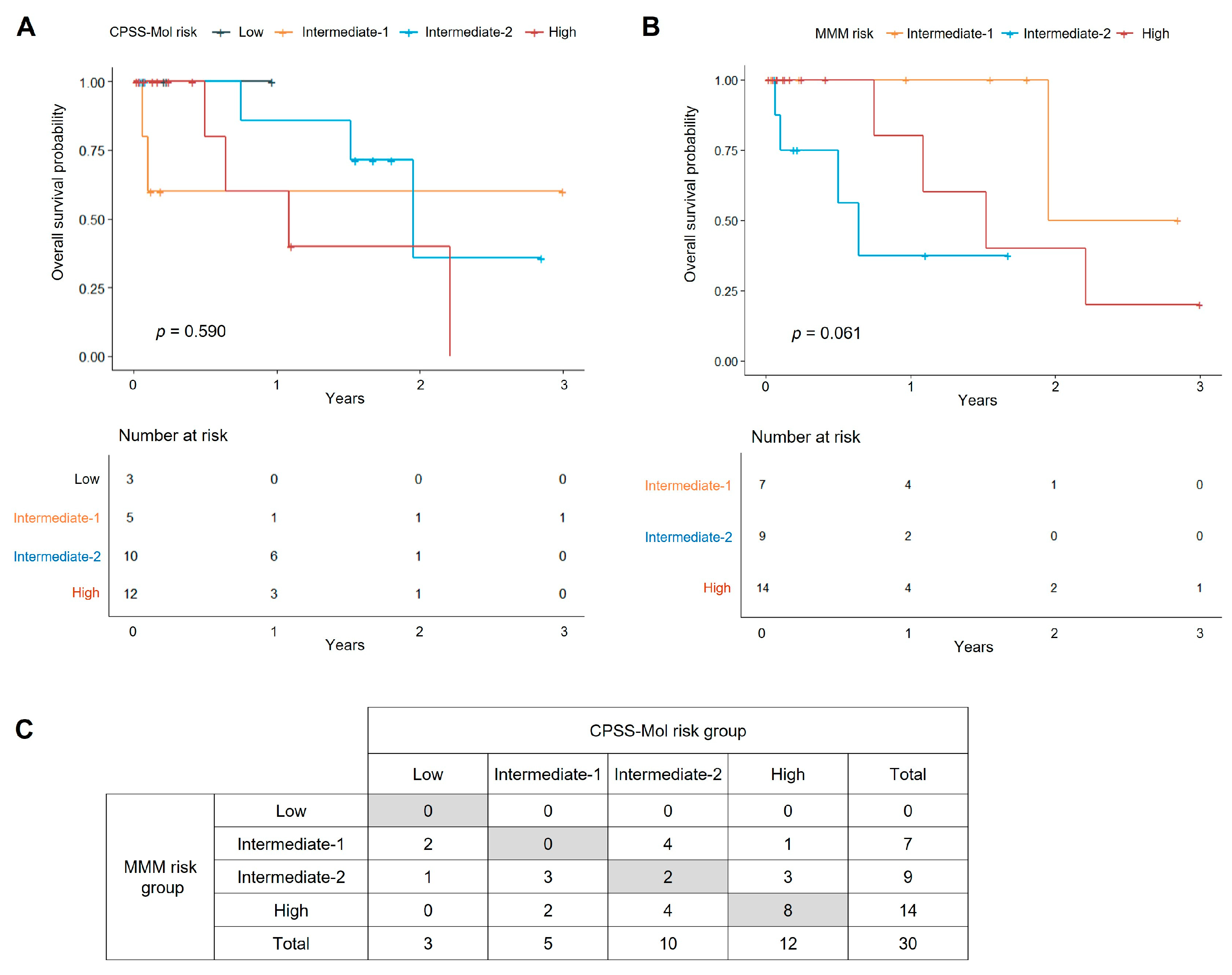
3.4.2. Survival Analysis by Prognostic Scoring Systems
3.4.3. Clinical and Molecular Prognostic Variables
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tremblay, D.; Hasserjian, R.P.; Rampal, R.K. Myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes: A practical guide to diagnosis and management. Leukemia 2025, 39, 1311–1324. [Google Scholar] [CrossRef]
- Cazzola, M.; Malcovati, L.; Invernizzi, R. Myelodysplastic/myeloproliferative neoplasms. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Fontana, D.; Elli, E.M.; Pagni, F.; Piazza, R. Myelodysplastic Syndromes/Myeloproliferative Overlap Neoplasms and Differential Diagnosis in the WHO and ICC 2022 Era: A Focused Review. Cancers 2023, 15, 3175. [Google Scholar] [CrossRef]
- Nie, Y.; Shao, L.; Zhang, H.; He, C.K.; Li, H.; Zou, J.; Chen, L.; Ji, H.; Tan, H.; Lin, Y.; et al. Mutational landscape of chronic myelomonocytic leukemia in Chinese patients. Exp. Hematol. Oncol. 2022, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Tefferi, A. Chronic myelomonocytic leukemia: 2024 update on diagnosis, risk stratification and management. Am. J. Hematol. 2024, 99, 1142–1165. [Google Scholar] [CrossRef]
- Woo, J.; Choi, D.R.; Storer, B.E.; Yeung, C.; Halpern, A.B.; Salit, R.B.; Sorror, M.L.; Woolston, D.W.; Monahan, T.; Scott, B.L.; et al. Impact of clinical, cytogenetic, and molecular profiles on long-term survival after transplantation in patients with chronic myelomonocytic leukemia. Haematologica 2020, 105, 652–660. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, H.; Wu, K.; Peng, Y.; Han, X.; Zhang, H.; Liang, L.; Chen, H.; Hu, J.; Qu, X.; et al. Knockdown of spliceosome U2AF1 significantly inhibits the development of human erythroid cells. J. Cell Mol. Med. 2019, 23, 5076–5086. [Google Scholar] [CrossRef]
- Bapat, A.; Keita, N.; Martelly, W.; Kang, P.; Seet, C.; Jacobsen, J.R.; Stoilov, P.; Hu, C.; Crooks, G.M.; Sharma, S. Myeloid Disease Mutations of Splicing Factor SRSF2 Cause G2-M Arrest and Skewed Differentiation of Human Hematopoietic Stem and Progenitor Cells. Stem Cells 2018, 36, 1663–1675. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Lasho, T.L. Genomics of myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes. Hematol. Am. Soc. Hematol. Educ. Program 2020, 2020, 450–459. [Google Scholar] [CrossRef]
- Li, F.; Qin, T.; Li, B.; Qu, S.; Pan, L.; Zhang, P.; Sun, Q.; Cai, W.; Gao, Q.; Jiao, M.; et al. Predicting survival in patients with myelodysplastic/myeloproliferative neoplasms with SF3B1 mutation and thrombocytosis. Leukemia 2024, 38, 1334–1341. [Google Scholar] [CrossRef]
- Elena, C.; Galli, A.; Such, E.; Meggendorfer, M.; Germing, U.; Rizzo, E.; Cervera, J.; Molteni, E.; Fasan, A.; Schuler, E.; et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood 2016, 128, 1408–1417. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Itzykson, R.; Lasho, T.L.; Kosmider, O.; Finke, C.M.; Hanson, C.A.; Knudson, R.A.; Ketterling, R.P.; Tefferi, A.; Solary, E. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: A two-center study of 466 patients. Leukemia 2014, 28, 2206–2212. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, T.; Spooner, M.; Khanna, S.; Hung, K.; Toop, C.R.; Kutyna, M.; Yu, J.; Brown, A.; Scott, H.; Hahn, C.; et al. RBC transfusion dependency refines the Molecular International Prognostic Scoring System for myelodysplastic syndrome. Blood Adv. 2025, 9, 4244–4247. [Google Scholar] [CrossRef] [PubMed]
- Barreyro, L.; Chlon, T.M.; Starczynowski, D.T. Chronic immune response dysregulation in MDS pathogenesis. Blood 2018, 132, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Klco, J.M.; Mullighan, C.G. Advances in germline predisposition to acute leukaemias and myeloid neoplasms. Nat. Rev. Cancer 2021, 21, 122–137. [Google Scholar] [CrossRef]
- Duncavage, E.J.; Bagg, A.; Hasserjian, R.P.; DiNardo, C.D.; Godley, L.A.; Iacobucci, I.; Jaiswal, S.; Malcovati, L.; Vannucchi, A.M.; Patel, K.P.; et al. Genomic profiling for clinical decision making in myeloid neoplasms and acute leukemia. Blood 2022, 140, 2228–2247. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Horak, P.; Griffith, M.; Danos, A.M.; Pitel, B.A.; Madhavan, S.; Liu, X.; Chow, C.; Williams, H.; Carmody, L.; Barrow-Laing, L.; et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): Joint recommendations of Clinical Genome Resource (ClinGen), Cancer Genomics Consortium (CGC), and Variant Interpretation for Cancer Consortium (VICC). Genet. Med. 2022, 24, 986–998. [Google Scholar] [CrossRef]
- Kwon, J. Diagnosis and treatment of chronic myelomonocytic leukemia. Blood Res. 2021, 56, S5–S16. [Google Scholar] [CrossRef]
- Gelsi-Boyer, V.; Brecqueville, M.; Devillier, R.; Murati, A.; Mozziconacci, M.J.; Birnbaum, D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J. Hematol. Oncol. 2012, 5, 12. [Google Scholar] [CrossRef]
- Itzykson, R.; Kosmider, O.; Renneville, A.; Gelsi-Boyer, V.; Meggendorfer, M.; Morabito, M.; Berthon, C.; Ades, L.; Fenaux, P.; Beyne-Rauzy, O.; et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J. Clin. Oncol. 2013, 31, 2428–2436. [Google Scholar] [CrossRef] [PubMed]
- Arbab Jafari, P.; Ayatollahi, H.; Sadeghi, R.; Sheikhi, M.; Asghari, A. Prognostic significance of SRSF2 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: A meta-analysis. Hematology 2018, 23, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Zhou, F.; Wang, Z.; Hua, H.; Qin, W.; Jia, Z.; Cai, X.; Chen, M.; Liu, J.; Chao, H.; et al. Mutational landscape of chronic myelomonocytic leukemia and its potential clinical significance. Int. J. Hematol. 2022, 115, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.Q.; Pan, L.J.; Qin, T.J.; Xu, Z.F.; Li, B.; Wang, H.J.; Sun, Q.; Jia, Y.J.; Li, C.W.; Cai, W.Y.; et al. Molecular features of 109 patients with chronic myelomonocytic leukemia in a single center. Zhonghua Xue Ye Xue Za Zhi 2023, 44, 373–379. [Google Scholar] [CrossRef]
- Costa, A.; Breccia, M. SOHO State of the Art Updates and Next Questions | Atypical Chronic Myeloid Leukemia: Pathogenesis, Diagnostic Challenges and Therapeutic Strategies. Clin. Lymphoma Myeloma Leuk. 2025, 25, 558–570. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Tefferi, A. Myelodysplastic syndromes with ring sideroblasts (MDS-RS) and MDS/myeloproliferative neoplasm with RS and thrombocytosis (MDS/MPN-RS-T)—“2021 update on diagnosis, risk-stratification, and management”. Am. J. Hematol. 2021, 96, 379–394. [Google Scholar] [CrossRef]
- Enjeti, A.K.; Agarwal, R.; Blombery, P.; Chee, L.; Chua, C.C.; Grigg, A.; Hamad, N.; Iland, H.; Lane, S.; Perkins, A.; et al. Panel-based gene testing in myelodysplastic/myeloproliferative neoplasm overlap syndromes: Australasian Leukaemia and Lymphoma Group (ALLG) consensus statement. Pathology 2022, 54, 389–398. [Google Scholar] [CrossRef]
- Bose, P.; Nazha, A.; Komrokji, R.S.; Patel, K.P.; Pierce, S.A.; Al-Ali, N.; Sochacki, A.; Shaver, A.; Ma, W.; Su, X.; et al. Mutational landscape of myelodysplastic/myeloproliferative neoplasm-unclassifiable. Blood 2018, 132, 2100–2103. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Lasho, T. Myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes: A focused review. Hematol. Am. Soc. Hematol. Educ. Program 2020, 2020, 460–464. [Google Scholar] [CrossRef]
- Yu, H.; Hong, J.; Shin, D.Y.; Lee, C.H. The role of ASXL1, SRSF2, and EZH2 mutations in chromatin dysregulation of myelodysplastic neoplasia and acute myeloid leukemia. Leukemia 2025, 39, 2329–2339. [Google Scholar] [CrossRef]
- Coltro, G.; Mangaonkar, A.A.; Lasho, T.L.; Finke, C.M.; Pophali, P.; Carr, R.; Gangat, N.; Binder, M.; Pardanani, A.; Fernandez-Zapico, M.; et al. Clinical, molecular, and prognostic correlates of number, type, and functional localization of TET2 mutations in chronic myelomonocytic leukemia (CMML)—A study of 1084 patients. Leukemia 2020, 34, 1407–1421. [Google Scholar] [CrossRef]
- Khan, S.; Tang, Y.; Guo, Y.; Feng, J.; Wu, H.; Song, Z.; Zhang, C.; Qin, F. Revealing the role of U2AF1 in splicing regulation and chimeric RNA dynamics. Sci. Rep. 2025, 15, 16235. [Google Scholar] [CrossRef]
| Total MDS/MPN (N = 53) | MD-CMML (N = 15) | MP-CMML (N = 15) | MDS/MPN-N (N = 6) | MDS/MPN-SF3B1-T (N = 4) | MDS/MPN-NOS (N = 13) | |
|---|---|---|---|---|---|---|
| Demographics | ||||||
| Male | 37 (69.8%) | 10 (66.7%) | 9 (60.0%) | 5 (83.3%) | 3 (75.0%) | 10 (76.9%) |
| Age, years (range) | 68 (37–87) | 69 (41–79) | 68 (37–85) | 68 (41–85) | 74 (65–82) | 65 (40–87) |
| Laboratory findings | ||||||
| WBC count (×109/L) | 18.57 (2.97–136.82) | 6.94 (2.97–12.14) | 27.07 (13.98–70.26) | 40.92 (22.30–136.82) | 4.80 (3.99–6.41) | 19.53 (4.03–71.86) |
| Hb (g/dL) | 8.2 (5.4–15.5) | 8.7 (6.0–15.5) | 8.1 (5.8–11.2) | 9.4 (7.1–12.0) | 8.4 (5.4–9.2) | 8.1 (6.6–12.1) |
| PLT count (×109/L) | 107 (6–1343) | 88 (11–625) | 93 (6–347) | 133 (29–480) | 512 (434–721) | 107 (11–1343) |
| Blast (%) | 0 (0–8) | 0 (0–4) | 1 (0–7) | 2 (0–3) | 0 (0–0) | 1 (0–8) |
| Monocyte (×109/L) | 2.31 (0.20–13.86) | 2.03 (0.78–3.98) | 6.38 (2.31–13.86) | 2.22 (1.34–4.16) | 0.42 (0.35–0.60) | 0.63 (0.20–6.47) |
| Bone marrow findings | ||||||
| Erythroid dysplasia | 29 (54.7%) | 9 (60.0%) | 8 (53.3%) | 4 (66.7%) | 4 (100%) | 5 (38.5%) |
| Granulocytic dysplasia | 40 (75.5%) | 12 (80.0%) | 12 (80.0%) | 6 (100%) | 0 (0%) | 10 (76.9%) |
| Megakaryocytic dysplasia | 45 (84.9%) | 14 (93.3%) | 14 (93.3%) | 6 (100%) | 2 (50.0%) | 9 (69.2%) |
| Myelofibrosis (grade ≥ 2) | 14 (26.4%) | 4 (26.7%) | 4 (26.7%) | 1 (16.7%) | 1 (25.0%) | 4 (30.8%) |
| Cytogenetic study | ||||||
| -7 or del(7q) | 6 (11.3%) | 4 (26.7%) | 1 (6.7%) | 0 (0%) | 0 (0%) | 1 (7.7%) |
| Trisomy 8 | 6 (11.3%) | 2 (13.3%) | 2 (13.3%) | 0 (0%) | 0 (0%) | 2 (15.4%) |
| 17p loss | 1 (1.9%) | 0 (0%) | 1 (6.7%) | 0 (0%) | 0 (0%) | 0 (0%) |
| −20 or del(20q) | 2 (3.8%) | 0 (0%) | 1 (6.7%) | 0 (0%) | 0 (0%) | 1 (7.7%) |
| Y loss | 1 (1.9%) | 1 (6.7%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Complex karyotype | 5 (9.4%) | 3 (20.0%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (15.4%) |
| NGS study | ||||||
| Number of mutated gene | 3 (0–7) | 3 (0–5) | 3 (1–7) | 3 (1–6) | 2 (1–3) | 3 (0–7) |
| ASXL1 & TET2 co-mutation | 12 (22.6%) | 3 (20.0%) | 5 (33.3%) | 2 (33.3%) | 0 (0%) | 2 (15.4%) |
| Splenomegaly | 9 (17.0%) | 3 (20.0%) | 3 (20.0%) | 2 (33.3%) | 0 (0%) | 1 (7.7%) |
| Transfusion dependency | 20 (37.7%) | 7 (46.7%) | 5 (33.3%) | 1 (16.7%) | 1 (25.0%) | 6 (46.2%) |
| Treatment regimen | ||||||
| HMA | 8 (15.1%) | 2 (13.3%) | 4 (26.7%) | 1 (16.7%) | 0 (0%) | 1 (7.7%) |
| HSCT | 5 (9.4%) | 0 (0%) | 1 (6.7%) | 3 (50.0%) | 0 (0%) | 1 (7.7%) |
| 3-year outcome | ||||||
| Progression | 8 (15.1%) | 1 (6.7%) | 5 (33.3%) | 1 (16.7%) | 0 (0%) | 1 (7.7%) |
| Death | 14 (26.4%) | 4 (26.7%) | 5 (33.3%) | 1 (16.7%) | 0 (0%) | 4 (30.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, M.-S.; Choi, D.-H.; Jang, J.H.; Jung, C.W.; Kim, H.-J.; Kim, H.-Y. Mutational Spectrum and Clinical Outcomes of Myelodysplastic/Myeloproliferative Neoplasms: A Single-Institution Study in Korea with Emphasis on U2AF1. J. Clin. Med. 2025, 14, 7074. https://doi.org/10.3390/jcm14197074
Park M-S, Choi D-H, Jang JH, Jung CW, Kim H-J, Kim H-Y. Mutational Spectrum and Clinical Outcomes of Myelodysplastic/Myeloproliferative Neoplasms: A Single-Institution Study in Korea with Emphasis on U2AF1. Journal of Clinical Medicine. 2025; 14(19):7074. https://doi.org/10.3390/jcm14197074
Chicago/Turabian StylePark, Min-Seung, Dae-Ho Choi, Jun Ho Jang, Chul Won Jung, Hee-Jin Kim, and Hyun-Young Kim. 2025. "Mutational Spectrum and Clinical Outcomes of Myelodysplastic/Myeloproliferative Neoplasms: A Single-Institution Study in Korea with Emphasis on U2AF1" Journal of Clinical Medicine 14, no. 19: 7074. https://doi.org/10.3390/jcm14197074
APA StylePark, M.-S., Choi, D.-H., Jang, J. H., Jung, C. W., Kim, H.-J., & Kim, H.-Y. (2025). Mutational Spectrum and Clinical Outcomes of Myelodysplastic/Myeloproliferative Neoplasms: A Single-Institution Study in Korea with Emphasis on U2AF1. Journal of Clinical Medicine, 14(19), 7074. https://doi.org/10.3390/jcm14197074






