Abstract
Background/Objectives: Modern healthcare faces a growing burden of antimicrobial resistance, prominently driven by ESKAPE pathogens. These organisms—Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.—are the leading causes of healthcare-associated infections, associated with limited therapeutic options and increased morbidity. Continuous surveillance is crucial for informing empirical therapy and guiding stewardship. Methods: We perform a single-center, seven-year retrospective study (2018–2024) at a 1000-bed tertiary hospital in Warsaw, Poland. Bloodstream isolates of ESKAPE pathogens were identified according to the EUCAST guidelines. Data were analyzed by pathogen, ward, and year of isolation. Results: From 2483 positive blood cultures, 3724 ESKAPE pathogens were recovered. S. aureus and K. pneumoniae predominated, particularly in the Intensive Care Unit and Hematology ward. Resistance analysis revealed persistently high vancomycin resistance in E. faecium, variable but notable methicillin resistance in S. aureus, and frequent ESBL production in K. pneumoniae with an alarming rise in carbapenemase-producing strains, including dual NDM + OXA-48 co-producers. A. baumannii exhibited near-universal multidrug resistance. P. aeruginosa demonstrated lower resistance rates with preserved colistin susceptibility, while Enterobacter spp. remained fully carbapenem-susceptible. Linezolid retained activity against E. faecium, while colistin remained effective against A. baumannii and P. aeruginosa. Modern β-lactam/β-lactamase inhibitor combinations were active against K. pneumoniae. Conclusions: Our findings underscore the critical role of ESKAPE pathogens in bloodstream infections and highlight divergent resistance patterns across species. The emergence of carbapenemase-producing K. pneumoniae and the persistence of multidrug-resistant A. baumannii are of particular concern. Sustained surveillance, robust antimicrobial stewardship, and tailored infection control strategies remained essential to curb the evolving resistance threat in tertiary care settings.
1. Introduction
Modern healthcare is increasingly challenged by the growing threat of antimicrobial resistance, especially in hospital-acquired infections. A particularly concerning group of pathogens is collectively referred to as the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.). These organisms are known for their high potential to cause healthcare-associated infections (HAIs) and their significant resistance to available antimicrobial treatment [1].
Infections caused by ESKAPE pathogens have profound clinical and economic implications. Patients with HAIs have significantly longer hospital stays–an average of 20.3 days compared to 8.7 days for non-infected patients [2]. Specific infections, such as central line-associated bloodstream infections (CLABSI) and catheter-associated urinary tract infections (CAUTI), can prolong hospitalization by 13.4 and 8.9 days, respectively, resulting in substantial additional healthcare costs [3].
Treatment costs for patients with these infections are significantly higher. For instance, the additional cost per patient for treating CLABSI is estimated at $43,975, and for CAUTI, $31,253 [3]. Furthermore, patients in isolation due to HAIs generate daily treatment costs that are approximately €950.65 higher than those for non-infected patients [4].
These infections are also associated with increased mortality. Evidence suggests that HAIs significantly raise the risk of death, especially among patients with additional clinical risk factors such as central venous catheters, mechanical ventilation, or immunosuppression [2].
While numerous studies have documented the prevalence and resistance patterns of ESKAPE pathogens in different geographic regions, there remains a need for comprehensive, institution-specific analyses that integrate both microbiological and clinical data. Previous surveillance studies have often focused on single pathogens, specific wards, or limited patient populations, which may not capture the broader dynamics of antimicrobial resistance within a tertiary care setting. For example, a study conducted at a university hospital in Italy evaluated the epidemiology and prevalence of antimicrobial resistance in ESKAPE pathogens over five years, providing valuable insights into local resistance trends [5]. Similarly, research in Ethiopia assessed the prevalence and antimicrobial susceptibility patterns of ESKAPE pathogens among patients who developed surgical site infections, highlighting the importance of region-specific data [6].
By contrast, this study systematically examines the prevalence of all ESKAPE pathogens within a large university hospital, evaluating their antimicrobial susceptibility and resistance profiles. This dataset not only provides updated local epidemiological insights but also enables comparisons with regional and global trends, thereby informing more targeted infection control and therapeutic strategies. Understanding these patterns is crucial for optimizing antimicrobial stewardship programs and mitigating the clinical and economic impacts of hospital-acquired infections (HAIs).
The study aims to analyze the prevalence of ESKAPE pathogens in a large university hospital and evaluate their antimicrobial susceptibility and resistance. Understanding the scope of the problem is essential for developing more effective preventive and therapeutic strategies.
2. Materials and Methods
2.1. Data Collection
The study was designed as a single-center retrospective study. The project was conducted at the Microbiology Laboratory of the Military Institute of Medicine–National Research Institute in Warsaw, Poland. The Military Institute of Medicine is a 1000-bed tertiary-care university hospital and a regional trauma center.
2.2. Blood Cultures and Pathogen Detection
Whole blood samples were collected concurrently for microbiological diagnostics when sepsis was clinically suspected. By standard clinical procedures, one or more sets of blood cultures—each consisting of one anaerobic and one aerobic bottle—were obtained from each patient. Blood culture bottles were loaded into a BacT/ALERT Virtuo® system (bioMérieux, Marcy-l’Étoile, France) and incubated at 37 °C until flagged positive or for a maximum of five days. Upon a positive signal, samples were Gram-stained and subcultured onto Columbia blood agar, chocolate agar, and MacConkey agar, while anaerobic bottles were plated on Schaedler agar. Aerobic plates were incubated at 37 °C in ambient air supplemented with 5% CO2, and Schaedler agar plates were incubated anaerobically at 37 °C for 48 h.
2.3. Identification of the Pathogens
Between 2018 and 2020, microorganisms were identified using the VITEK® 2 system (bioMérieux, France). We employed GN ID Cards for fermenting and non-fermenting Gram-negative bacilli and GP ID Cards for Gram-positive bacteria. From January 2021 onward, all strains were identified by Matrix-Assisted Laser Desorption Ionization-Time Of Flight mass spectrometry (MALDI-TOF; VITEK® MS, bioMérieux, France).
Non-duplicate isolates of ESKAPE pathogens were analyzed. Duplicates were defined based on unique patients’ identifiers and blood sample collection dates (within a 28-day interval). Non-ESKAPE pathogens and fungal isolates were excluded from the analysis.
2.4. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed using the VITEK-2 automated system (bioMérieux, France), following the manufacturer’s instructions. Colistin susceptibility was determined separately by the reference broth microdilution method, using the MICRONAUT MIC-Strip colistin (MMS) assay (Merlin Diagnostika GmbH, Bornheim, Germany).
The VITEK 2 system utilized the AST N-N331 and AST-N 332 cards for Gram-negative bacteria, and the AST-P644 and AST-643 cards for Gram-positive bacteria.
Results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria.
Quality control testing included the following reference strains: Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, Klebsiella pneumoniae ATCC 700603, Klebsiella pneumoniae ATCC BAA-2814, Staphylococcus aureus NCTC 124943, and Enterococcus faecalis ATCC 51299.
2.5. Detection and Resistance Mechanisms
Phenotypic detection of ESBLs was performed with double-disk synergy tests (DDST) [7].
The presence and nature of carbapenemase determinants were assessed by molecular testing of bacterial isolates using either a polymerase chain reaction (PCR)-based platform, the GeneXpert System (Cepheid, Sunnyvale, CA, USA), or a lateral flow immunochromatographic assay (RESIST-5 OOKNV, CORIS, BioConcept, Gembloux, Belgium).
Detection of methicillin resistance in Staphylococcus aureus (MRSA) was performed using the cefoxitin disk diffusion method. A zone diameter of less than 20 mm was interpreted as resistant [8].
Vancomycin resistance was assessed using the vancomycin E-test, performed according to the manufacturer’s instructions (bioMérieux, France). Isolates with the minimum inhibitory concentration (MIC) values of ≥32 μg/mL were confirmed as vancomycin-resistant Enterococcus (VRE).
2.6. Statistics and Graphics
All data preprocessing and visualizations were performed using the Anaconda Navigator environment (version 2023.07, Anaconda Inc., Austin, TX, USA) with Jupyter Notebook (version 7.4.3. Project Jupyter, Berkeley, CA, USA). Plots were generated using Python libraries, including Matplotlib (version 3.7.2), Pandas (version 2.0.3), and Seaborn (version 0.12.2).
3. Results
Between 1 January 2018, and 31 December 2024, ESKAPE pathogens were detected in 2483 out of all analyzed samples. These samples were collected from 1460 patients, and a total of 3724 bacterial strains were isolated. Among them, there were 508 E. faecium strains, 1284 S. aureus strains, 963 K. pneumoniae strains, 461 A. baumannii strains, 328 P. aeruginosa strains, and 182 Enterococcus spp. strains.
The Intensive Care Unit (ICU) yielded the highest number of isolates (n = 888). The most frequently isolated pathogen in the ICU was S. aureus (228/888, 25.7%), K. pneumoniae (226/888, 25.6%), A. baumannii (208/888, 23.4%), and E. faecium (59/888, 6.7%). The Hematology ward accounted for 594 isolates, predominantly K. pneumoniae (170/594, 28.6%) and S. aureus (124/594, 20.9%).
Temporal and spatial trends were observed in the frequency of pathogen isolation. The COVID ward, which was operational primarily during 2020–2021, exhibited a high prevalence of E. faecium (n = 47), S. aureus (n = 31), and K. pneumoniae (n = 31), with 39 out of the E. faecium isolates identified in 2020 alone.
In the Nephrology ward, S. aureus was the predominant pathogen (n = 142), while E. faecium was less frequent (n = 31).
In the Cardiology ward, S. aureus accounted for 190 isolates, significantly outnumbering other pathogens.
Throughout the study period, several temporal patterns emerged. Acinetobacter baumannii isolates in the ICU decreased after 2020 (34 in 2020 in comparison to 11 in 2024). Klebsiella pneumoniae maintained a consistently high isolation rate in both the Hematology ward and ICU. A notable increase in E. faecium was observed in the Hematology Department after 2020, with a peak between 2022 and 2024, accounting for 73.2% of all E. faecium isolates in that Department.
Less frequently detected pathogens, such as Enterobacter spp. remained relatively low in overall frequency, with 294 isolates (7.9% of the total), most commonly isolated in the Hematology Department (n = 67).
Overall, isolates were more frequently recovered from patients located in the ICU (23.8%), with lower proportions from the Hematology (15.9%) and Cardiology (10.4%) wards. Temporal trends revealed significant fluctuation in pathogen distribution, underlying the dynamic epidemiology of antimicrobial resistance in the hospital setting. The number of isolated pathogens in each hospital department is presented in Table 1 and Figure 1.

Table 1.
The number of ESKAPE pathogens isolated in different hospital departments between 2018 and 2024.
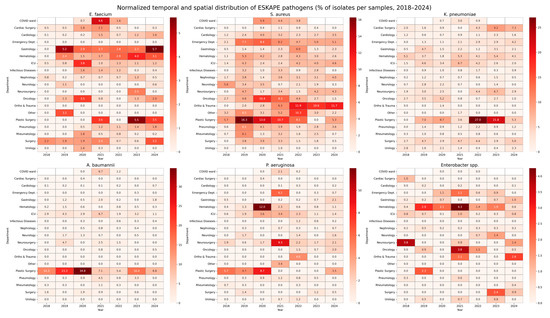
Figure 1.
Normalized temporal and spatial distribution of ESKAPE pathogens (% of isolates per samples, 2018–2024). This figure presents heatmaps illustrating the relative prevalence of ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) across hospital departments between 2018 and 2024. For each pathogen, the heatmap displays the proportion of isolates detected in a given department and year, normalized to the total number of microbiological samples collected in that department during the corresponding year. Values are expressed as percentages. Darker shades indicate a higher relative proportion of pathogen-positive samples, while lighter shades indicate lower prevalence. This visualization enables simultaneous interpretation of both the temporal dynamics and the spatial distribution of pathogens, adjusted for variations in the sampling intensity across departments.
Between 2018 and 2024, ESKAPE pathogens exhibiting antimicrobial resistance were isolated, showing variable frequencies across years and species (see Table 2 and Figure 2). The proportion of vancomycin-resistant Enterococcus faecium (VRE) ranged between 26.7% and 39.0%, with no consistent upward or downward trend.

Table 2.
Antimicrobial resistance mechanisms among ESKAPE pathogens isolated between 2018 and 2024.
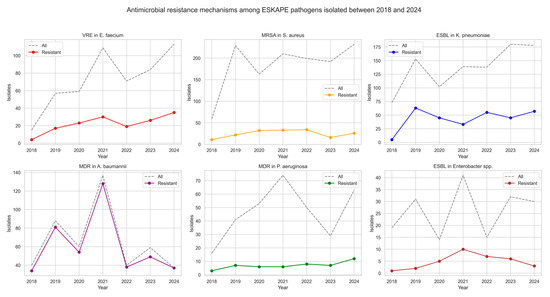
Figure 2.
Antimicrobial resistance mechanisms of ESKAPE pathogens isolated between 2018 and 2024.
Methicillin-resistant Staphylococcus aureus (MRSA) exhibited variable prevalence, peaking in 2020 (19.6%) and reaching its lowest level in 2023 (8.3%). Among K. pneumoniae isolates, ESBL production remained frequent, with annual proportions ranging from 6.8% in 2018 to 44.1% in 2020. Notably, the prevalence of carbapenemase-producing K. pneumoniae strains increased over time. NDM-type carbapenemases were most prevalent, detected in up to 16.5% of isolates in 2021. Co-production of NDM and OXA-48 enzymes emerged in 2023 (5.6%) and persisted in 2024 (6.2%), while KPC-type carbapenemases appeared sporadically. A very high prevalence of multidrug resistance (MDR) was observed in A. baumannii isolates, with rates exceeding 90% in most years and reaching 100% in 2024. In contrast, P. aeruginosa strains were less frequent, with resistance ranging from 8.1% to 24.1% annually. Among Enterobacter spp. isolates, ESBL production was observed at varying frequencies, with the highest rates in 2022 (46.7%) and 2020 (35.7%), although no consistent trend was apparent over time.
We also analyzed the antimicrobial resistance of ESKAPE pathogens. An analysis of E. faecium isolates revealed persistently high resistance to ampicillin and meropenem. Resistance to ampicillin remained at 99.6% across the study period, while imipenem resistance was 99.4%, indicating near-universal resistance to β-lactam antibiotics among the isolates. High-level gentamycin resistance (HLR) increased from 53.0% in 2018 to 77.0% in 2024, resulting in an overall resistance rate of 73.0%. Similarly, high-level streptomycin resistance averaged 65.3%, increasing from 40.0% in 2018 to over 70% in the final years of our analysis. A concerning trend was observed in glycopeptide resistance. Teicoplanin resistance rose from 7.0% in 2018 to 23.0% in 2024, while vancomycin resistance increased from 27.0% to 31.0%. Notably, no resistance to linezolid was observed in any year of the study, indicating the preserved efficacy of this agent against E. faecium in the analyzed period. The detailed data are presented in Table 3 and graphically in Figure 3.

Table 3.
Annual distribution of antimicrobial-resistant Enterococcus faecium isolates, 2018–2024.
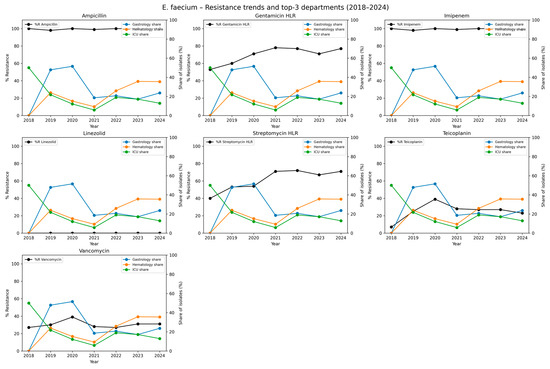
Figure 3.
Temporal trends in antimicrobial resistance of Enterococcus faecium isolates and distribution across hospital departments, 2018–2024. Each subplot represents one antimicrobial agent tested against E. faecium. The black line shows the annual percentage of resistant isolates (%R) across all departments combined. The colored lines represent the relative contribution (share of isolates, expressed as a percentage of all E. faecium isolates in a given year) of the three hospital departments with the highest cumulative number of isolates across the study period. The left y-axis corresponds to the share of isolates per department (colored lines). This dual-axis approach allows simultaneous visualization of temporal resistance trends and the spatial distribution of isolates in the clinical setting. Higher %R values indicate reduced effectiveness of a given antimicrobial, while higher departmental shares indicate wards where E. faecium was most frequently detected.
Between 2018 and 2024, resistance to most antibiotics among S. aureus isolates remained low. No resistance was detected to ceftaroline, daptomycin, linezolid, teicoplanin, tigecycline, or vancomycin in any year of the study. Resistance to oxacillin (used here as a proxy for MRSA prevalence) ranged from 8.0% to 20.0%, with a mean resistance rate of 13.6%. Resistance to ciprofloxacin and levofloxacin followed similar patterns, with each reaching a peak of 20.0% in 2020 and averaging 12.3% overall. Moderate and stable resistance levels to clindamycin and erythromycin, averaging 35.4% and 35.3%, respectively, were observed over the study period. Resistance to gentamicin remained low (mean 5.1%), with the highest recorded in 2020 (8.0%). Sporadic resistance to trimethoprim/sulfamethoxazole ranged from 2.0% to 10.0%, and remained low overall (5.5%). Resistance to rifampicin was rare (0.6%) and was detected sporadically in a small number of isolates. The detailed data are presented in Table 4 and Figure 4.

Table 4.
Annual distribution of antimicrobial-resistant Staphylococcus aureus isolates, 2018–2024.
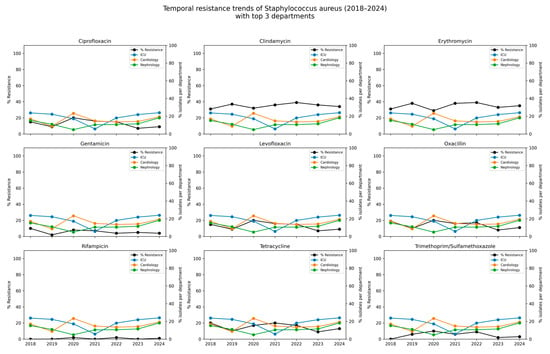
Figure 4.
Temporal resistance trends of Staphylococcus aureus (2018–2024) with spatial context of isolates from the top three hospital departments. Each subplot illustrates the temporal trend in the percentage of S. aureus isolates resistant to a given antibiotic (black line) between 2018 and 2024. In parallel, the colored lines depict the relative contribution (%) of isolates originating from the tree hospital departments with the highest cumulative number of detections (Intensive Care Unit, Cardiology ward, Hematology ward). The left axis (black line) corresponds to the % resistance to the antibiotic, while the right axis (colored lines) shows the % share of isolates from individual departments relative to the total isolates defined in a given year.
Among K. pneumoniae isolates, high and stable resistance rates were observed for amoxicillin/clavulanate, with annual resistance ranging from 71.6% to 75.6% and an overall rate of 73.6%. Similarly, high resistance was noted for ciprofloxacin (range: 42.1–83.7%, total: 54.6%), piperacillin/tazobactam (45.5–76.5%, total: 54.2%), cefuroxime (40.3–71.6%, total: 51.2%), and trimethoprim/sulfamethoxazole (41.7–56.5%, total: 46.3%). Resistance to third- and fourth-generation cephalosporins, including cefepime, cefotaxime, and ceftazidime, increased over time and exceeded 50% in recent years (total range: 45.4–47.7%). Moderate resistance was found for gentamicin (21.0–40.6%, total: 30.8%) and tobramycin (34.5–72.5%, total: 47.5%). Resistance to amikacin remained lower, with a total rate of 18.8%, though it increased from 12.7% in 2020 to 31.6% in 2023. Resistance to carbapenems also increased over the study period, reaching a rate of over 21% in 2024 (total 14.4%). Initially, resistance to newer β-lactam/β-lactamase inhibitor combinations–such as ceftazidime/avibactam, ceftolozan/tazobactam, imipenem/relebactam, and meropenem/vaborbactam, was low; however, a gradual increase was observed from 2023 to 2024, reaching a total of 12.0%. The detailed data are presented in Table 5 and Figure 5.

Table 5.
Annual distribution of antimicrobial-resistant Klebsiella pneumoniae isolates, 2018–2024.
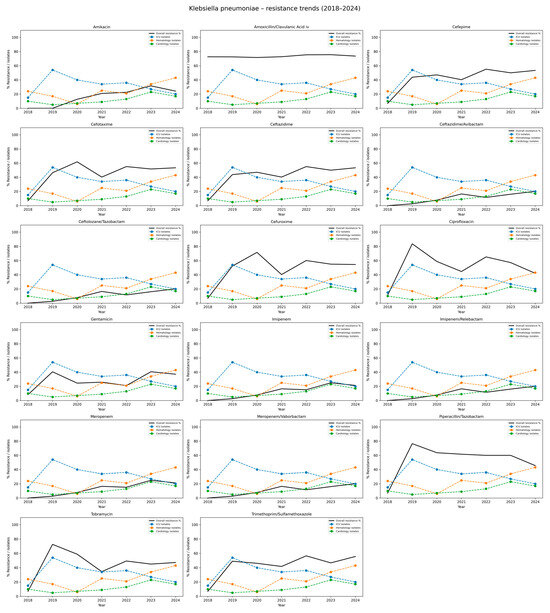
Figure 5.
Distribution of antibiotic resistance rates of Klebsiella pneumoniae isolates, 2018–2024. This table summarizes both the number of isolates tested (n) and the corresponding percentage resistance (%R) for Klebsiella pneumoniae across the years 20182024, stratified by antibiotic. Each row corresponds to an antibiotic commonly used for testing K. pneumoniae susceptibility. For each year, data are presented as n (%R), where n is the number of isolates tested and %R is the percentage resistant.
A very high level of antimicrobial resistance was observed among A. baumannii isolates over the study period (Table 6 and Figure 6). Resistance to ciprofloxacin and levofloxacin remained constant at 100% in all years. Resistance to imipenem and meropenem was also consistently high, with annual resistance rates ranging from 92.0% to 100%. Similarly, resistance to gentamicin, tobramycin, and amikacin remained above 85% in most years, reaching 100% for all three agents in 2024. Resistance to trimethoprim/sulfamethoxazole was exceptionally high, ranging from 97.5% to 100%, except for a slight decrease to 89.8% in 2023. In contrast, resistance to colistin was rare, detected in only five isolates (1.1%) throughout the entire period, indicating preserved susceptibility. These findings highlight the critical threat posed by extensively drug-resistant A. baumannii strains, which exhibit near-universal resistance to most available antibiotics except colistin.

Table 6.
Annual distribution of antimicrobial-resistant Acinetobacter baumannii isolates, 2018–2024.
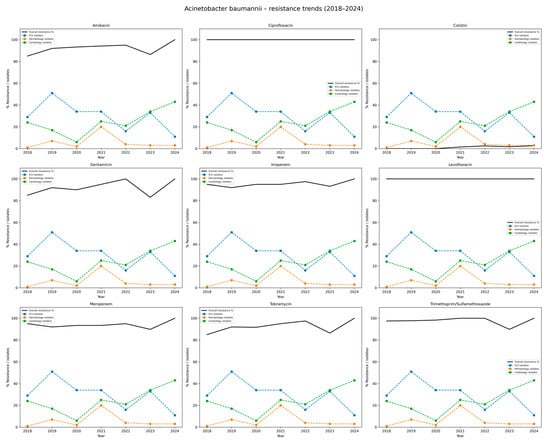
Figure 6.
Distribution of antibiotic resistance rates of Acinetobacter baumannii isolates, 2018–2024. Each subplot displays the percentage resistance of A. baumannii to a single antibiotic. The black line depicts overall resistance rates in the hospital. The colored dashed lines correspond to the departments with the highest number of isolates (here: Intensive Care Unit, Hematology ward, Cardiology ward). The Y-axis combines % resistance and isolate counts for top departments, while the X-axis shows study years (2018–2024). The figure highlights persistently high resistance rates, particularly to carbapenems and fluoroquinolones, underlying the clinical challenge posed by multidrug-resistant A. baumannii.
Table 7 presents the antibiotic resistance profiles of P. aeruginosa. The highest overall resistance was observed for ticarcillin/clavulanate, with resistance rates ranging from 26.0% to 31.0%, and a mean value of 27.3%. This pathogen also showed consistently high resistance to piperacillin/tazobactam, averaging 21.2% over the study period. Among carbapenems, resistance to imipenem increased from 18.8% in 2018 to 34.5% in 2023, with a slight decline to 22.2% in 2024. A similar trend was noted for meropenem, with resistance rising from 12.5% in 2018 to a peak of 24.1% in 2023, resulting in an average of 17.5%. Resistance levels to fluoroquinolones were variable but considerable, each averaging 16.3%. The highest resistance rate to ciprofloxacin was reported in 2022 (24.0%). Lower resistance was observed to aminoglycosides, specifically amikacin (mean 5.5%) and tobramycin (8.6%). Slightly higher but stable resistance rate to gentamicin was observed across the years, averaging 11%. Notably, no resistance to colistin was detected during the entire study period, indicating sustained susceptibility of P. aeruginosa to this last-resort antibiotic. Such data are graphically shown in Figure 7.

Table 7.
Annual distribution of antimicrobial-resistant Pseudomonas aeruginosa isolates, 2018–2024.
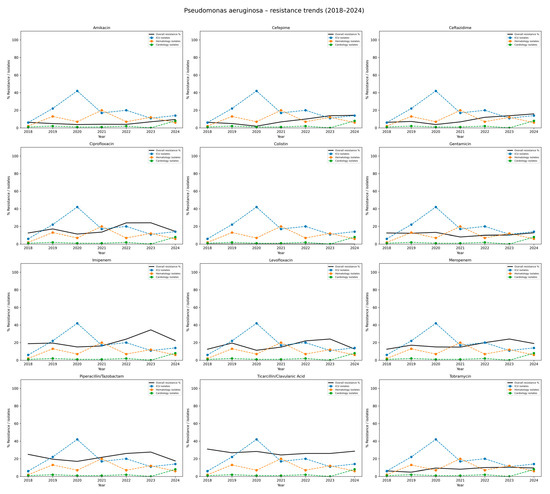
Figure 7.
Resistance trends of Pseudomonas aeruginosa isolates, 2018–2024. Each subplot illustrates the resistance rate of P. aeruginosa to a given antibiotic. The black line indicates the overall hospital resistance percentage. The colored dashed lines represent the number of isolates reported in the three hospital departments with the highest incidence (here: Intensive Care Unit, Hematology ward, Cardiology ward). The Y-axis: percentage resistance and departmental isolate counts; the X-axis: study years (2018–2024). The figure shows considerable variability across antibiotics, with worrying levels of resistance to carbapenems and fluoroquinolones, highlighting P. aeruginosa as the major multidrug-resistant pathogen.
Table 8 and Figure 8 summarize the resistance profiles of E. cloacae. Throughout the study period, isolates showed 100% resistance to amoxicillin/clavulanate, indicating that this agent is ineffective against this species in the local setting. Among cephalosporins, resistance to cefotaxime and cefuroxime was relatively high and stable, ranging from 12.9% to 46.7%, with average resistance rates above 35%. Slightly lower but still notable resistance trends to ceftazidime (ranging from 5.3% to 33.3%), and cefepime peaking at 35.7% in 2020, were observed. Resistance to fluoroquinolones, such as ciprofloxacin, increased over time, reaching 35.7% in 2020 and remaining above 20% through 2024 (mean approximately 24.3%). Aminoglycoside resistance varied depending on the specific agent: resistance to amikacin was moderate, ranging from 5.3% to 33.3%, while resistance to gentamicin and tobramycin showed fluctuating levels, reaching peaks of 28.6% and 33.3%, respectively.

Table 8.
Annual distribution of antimicrobial-resistant Enterobacter spp. isolates, 2018–2024.
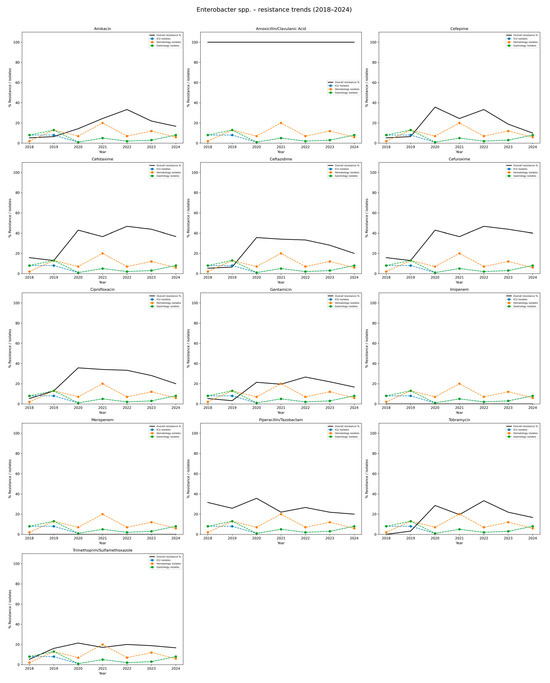
Figure 8.
Resistance trends of Enterobacter spp. isolates, 2018–2024. Each subplot shows the resistance rate of Enterobacter spp. against a specific antibiotic. The black line depicts the overall hospital resistance percentage. The colored dashed lines represent isolate counts in the three hospital departments with the highest incidence (here: Intensive Care Unit, Hematology ward, Gastrology ward). The Y-axis: percentage resistance and isolate counts; the X-axis: study years (2018–2024). This visualization highlights the increasing resistance of Enterobacter spp. to multiple β-lactams and fluoroquinolones, while carbapenem resistance remained absent during the study period.
No resistance to carbapenems was detected during the entire study period, confirming the sustained effectiveness of these agents against E. cloacae. Piperacillin/tazobactam resistance remained relatively stable, ranging from 20.0% to 35.7% (mean ca. 26.6%). Trimethoprim/sulfamethoxazole resistance fluctuated between 5.3% and 21.4%, with no consistent trend. Overall, the data reflect a complex and variable resistance pattern of E. cloacae, with consistently high resistance to β-lactam/β-lactamase inhibitor combinations and moderate levels across several other antibiotic classes, excluding carbapenems.
4. Discussion
The analysis of ESKAPE pathogens isolated from bloodstream infections between 2018 and 2024 in our center reveals significant trends in antimicrobial resistance, aligning with global patterns and underscoring the escalating threat posed by these pathogens [9,10]. ESKAPE pathogens are responsible for 42.2 to 75.6% of bloodstream infections [11,12]. Therefore, epidemiological studies concerning ESKAPE are a key element in the prevention and treatment of hospital-acquired infections.
In our study, we analyzed the frequency of infections caused by ESKAPE bacteria across various hospital departments and evaluated the dynamics of antimicrobial resistance development over a seven-year period, from 2018 to 2024, including the COVID-19 pandemic years (2020–2022).
The study material consisted of 3724 ESKAPE isolates obtained from 2483 blood samples at the Military Institute of Medicine—National Research Institute in Warsaw, the second-largest hospital in the city. Compared to other published studies on this topic, the presented dataset can be considered representative.
The most frequently isolated pathogen was S. aureus (1284 isolates; 34.5%), followed by K. pneumoniae (963; 25.8%), E. faecium (508; 13.6%), A. baumannii (461; 12.4%), P. aeruginosa (328; 8.8%), and Enterobacter spp. (182; 4.9%). The predominance of S. aureus and K. pneumoniae in our study is consistent with the findings reported by other authors [12,13,14].
The highest number of pathogens was isolated from blood samples collected from critically ill and hematological patients, which reflects global trends and is associated with department-specific risk factors, as invasive procedures and immunosuppression.
During the COVID-19 pandemic, we observed a marked and above-average increase in infections caused by E. faecium (including VRE), S. aureus, and K. pneumoniae. The conclusions of other authors were similar [15,16]. We also observed that during the pandemic period, the highest number of K. pneumoniae ESBL and K. pneumoniae NDM strains were isolated. The observations from Portugal [17] and Slovakia [18] were similar. In addition, we recorded a continuous increase in multidrug-resistant (MDR) A. baumannii isolates, with resistance rates rising from 85% in 2018 to 100% in 2024. These trends align with recent global surveillance data [19]. This rise may be attributed to the increased number of hospitalizations, reduced availability of healthcare staff, and elevated exposure of patients to infectious agents.
We observed, for the first time in 2023, the emergence of K. pneumoniae strains co-producing dual carbapenemases (NDM and OXA-48), accounting for 5.6% of K. pneumoniae isolates, with a further increase to 6.2% in 2024. This finding aligns with reports of the global emergence and local outbreaks of NDM + OXA-48–producing K. pneumoniae in recent years [20,21].
Epidemiological analysis of ESKAPE bacteria is a crucial component in guiding appropriate empirical antimicrobial therapy, which is essential for treatment efficacy and reduction in mortality. An analysis conducted from 2018 to 2020 showed that approximately 32.7% of empirical antibiotic administrations were inadequate compared to subsequent microbiological findings [22].
We also analyzed the effectiveness of antimicrobial therapy in infections caused by ESKAPE pathogens. In the treatment of E. faecium, linezolid has been proven to be the most effective option. These observations are consistent with the conclusions of Huang et al. meta-analysis [23].
We found no resistance to ceftaroline, daptomycin, linezolid, teicoplanin, vancomycin, and tigecycline in S. aureus, consistent with multiple surveillance and MIC studies reporting ≥94–100% susceptibility among clinical MRSA and MSSA isolates [24].
The restricted and controlled use of last-line agents such as linezolid, vancomycin, and daptomycin likely contributes to the sustained high susceptibility of S. aureus. In contrast, high resistance of E. faecium to ampicillin is well-documented in the literature, with studies reporting approximately 80–90% of clinical isolates being resistant, making it a characteristic feature of hospital-associated strains [25].
In clinical practice, neither ampicillin nor meropenem is routinely used to treat E. faecium infections, as their efficacy is limited. Therefore, the presence of resistance to these agents in in vitro susceptibility testing does not impact the choice of empirical therapy. These observations highlight the importance of interpreting resistance data in the context of actual antibiotic use and stewardship practices within the hospital.
The lowest resistance rate in K. pneumoniae—approximately 12%—was observed for modern β-lactam/β-lactamase inhibitor combinations, including ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam. These findings are consistent with data reported by Sader et al., who analyzed 35,360 multidrug-resistant Enterobacterales isolates collected from 75 U.S. medical centers between 2018 and 2022. In their study, ceftazidime-avibactam and meropenem-vaborbactam demonstrated susceptibility rates of 97.9%, while imipenem-relebactam showed a susceptibility rate of 93.5% [26].
Colistin remained the sole viable therapeutic option against A. baumannii, with resistance persisting at low levels (1.1–2.7%), in line with global surveillance reporting ~3–5% resistance [27]. Pseudomonas aeruginosa retained universal susceptibility to colistin throughout the study. Amikacin and cefepime showed resistance rates of 5.5% and 8.3%, respectively, which are notably lower than the typically reported rates in regional cohorts (31.1% and 44.5%) [28].
Enterobacter spp. demonstrated 100% susceptibility to carbapenems, which is comparable to susceptibility rates reported by Rhomberg et al. [29]. However, data from the CHINET surveillance network indicate that the percentage of carbapenem-resistant Enterobacter spp. reached 10%, showing an increasing trend over their 7-year study period [30].
Our study also has some limitations. The first one is its single-center design. We analyzed only isolates from blood samples, which may not fully reflect the overall spectrum of hospital-acquired infections. This study focused on the microbiological analysis of ESKAPE pathogens, including their prevalence and antimicrobial susceptibility, but did not include molecular characterization of clonal analysis. Whole-genome sequencing could provide deeper insights into resistance determinants and transmission dynamics, which will be addressed in future work.
Additionally, patient-level clinical outcomes such as mortality, length of hospital stay, and treatment success were not assessed. While these outcomes are essential for understanding the practical impact of antimicrobial resistance, they were beyond the scope of this retrospective analysis. However, despite these limitations, our data may serve as a basis for updating local empirical therapy protocols and implementing antimicrobial control strategies in a hospital setting, thereby reducing the risk of spreading multidrug-resistant bacteria. Future studies are planned to integrate molecular and clinical data, providing a more comprehensive assessment of resistance trends and their implications for patient care.
We can conclude that effective treatment of infections caused by ESKAPE pathogens relies primarily on linezolid, colistin, and modern β-lactam/β-lactamase inhibitor combinations. Among these pathogens, infections caused by A. baumannii pose the most significant therapeutic challenge due to the limited treatment options, which are restricted mainly to colistin, an antibiotic with significant toxic potential and poor tissue penetration.
5. Conclusions
Our findings confirm that multidrug-resistant ESKAPE pathogens remain a significant threat in the hospital setting. The persistence and rise in resistance mechanisms, particularly among K. pneumoniae, A. baumannii, and E. faecium, highlight the urgent need for continuous local and regional surveillance. The emergence of K. pneumonia strains co-producing dual carbapenemases (NDM + OXA-48) and the near-universal resistance of A. baumannii to most available antibiotics, except colistin, represent critical therapeutic challenges. These results emphasize the importance of stringent antimicrobial stewardship programs, infection control interventions, and the rational use of last-line agents such as linezolid, colistin, and modern β-lactam/ β-lactamase inhibitor combinations.
Future studies integrating molecular typing and patient-level clinical outcomes are essential to better understand resistance transmission dynamics, quantify the clinical impact of multidrug-resistant infections, and guide the development of more effective preventive and therapeutic strategies.
Author Contributions
Conceptualization, A.G. and A.O.-P.; methodology, A.G., K.M. and M.K.; software, A.G. and D.T.; validation, A.G., K.M., M.K. and D.T.; formal analysis, A.G., D.T., K.M., M.K., W.P., Z.R. and A.O.-P.; investigation, A.G., K.M. and M.K.; resources, A.G., K.M., M.K. and D.T.; data curation, A.G. and D.T.; writing—original draft preparation, A.G., D.T., W.P., Z.R., K.M., M.K. and A.O.-P.; writing—review and editing, A.G., D.T., W.P. and Z.R.; visualization, D.T. and A.G.; supervision, A.G., D.T., W.P., Z.R. and A.O.-P.; project administration, A.G., D.T. and A.O.-P.; funding acquisition, A.G. and A.O.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval were waived for this study because it involved only bacterial isolates obtained from patient samples during routine diagnostic procedures. No additional interventions were performed on humans, and no identifiable patient data were collected or analyzed.
Informed Consent Statement
Informed consent was waived because no intervention was involved and no patient-identifying information was included.
Data Availability Statement
The data from this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CAUTI | Catheter-associated Urinary Tract Infections |
| COVID | Coronavirus Disease |
| CLABSI | Catheter-associated Bloodstream Infections |
| DDST | Double Disk Synergy Test |
| ESBL | Extended Spectrum β-lactamases |
| ESKAPE | Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| HAI | Hospital Acquired Infections |
| HLR | High-Level Resistance |
| ICU | Intensive Care Unit |
| KPC | Klebsiella pneumoniae carbapenemase |
| MDR | Multidrug Resistant |
| MIC | Minimal Inhibitory Concentration |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MSSA | Methicillin-susceptible Staphylococcus aureus |
| NDM | New Delhi metallo-β-lactamase |
| OXA | Oxacillinase |
| PCR | Polymerase Chain Reaction |
| VRE | Vancomycin-resistant Enterococcus |
References
- Abban, M.K.; Ayerakwa, E.A.; Mosi, L.; Isawumi, A. The Burden of Hospital Acquired Infections and Antimicrobial Resistance. Heliyon 2023, 9, e20561. [Google Scholar] [CrossRef]
- Moradi, S.; Najafpour, Z.; Cheraghian, B.; Keliddar, I.; Mombeyni, R. The Extra Length of Stay, Costs, and Mortality Associated with Healthcare-Associated Infections: A Case-control Study. Health Sci. Rep. 2024, 7, e70168. [Google Scholar] [CrossRef] [PubMed]
- Sentiff, J.L.; Yenugadhati, V.; Spitz, C.; Glassman, S.; Phillips, M.; Frey, S.; Lesho, E.P. The Impact of Healthcare Associated Infections on Costs and Lengths of Stay 2019–2023. Open Forum Infect. Dis. 2023, 10, ofad500.2056. [Google Scholar] [CrossRef]
- Blatnik, P.; Bojnec, Š. Analysis of Impact of Nosocomial Infections on Cost of Patient Hospitalisation. Cent. Eur. J. Public Health 2023, 31, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Pipitò, L.; Rubino, R.; D’Agati, G.; Bono, E.; Mazzola, C.V.; Urso, S.; Zinna, G.; Distefano, S.A.; Firenze, A.; Bonura, C.; et al. Antimicrobial Resistance in ESKAPE Pathogens: A Retrospective Epidemiological Study at the University Hospital of Palermo, Italy. Antibiotics 2025, 14, 186. [Google Scholar] [CrossRef]
- Seid, M.; Bayou, B.; Aklilu, A.; Tadesse, D.; Manilal, A.; Zakir, A.; Kulyta, K.; Kebede, T.; Alodaini, H.A.; Idhayadhulla, A. Antimicrobial Resistance Patterns of WHO Priority Pathogens at General Hospital in Southern Ethiopia During the COVID-19 Pandemic, with Particular Reference to ESKAPE-Group Isolates of Surgical Site Infections. BMC Microbiol. 2025, 25, 84. [Google Scholar] [CrossRef]
- Garrec, H.; Drieux-Rouzet, L.; Golmard, J.-L.; Jarlier, V.; Robert, J. Comparison of Nine Phenotypic Methods for Detection of Extended-Spectrum Beta-Lactamase Production by Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters, versions 8.0–14.0, 2018–2024. Available online: https://www.eucast.org?clinical_breakpoints (accessed on 15 March 2025).
- Ma, J.; Song, X.; Li, M.; Yu, Z.; Cheng, W.; Yu, Z.; Zhang, W.; Zhang, Y.; Shen, A.; Sun, H.; et al. Global Spread of Carbapenem-Resistant Enterobacteriaceae: Epidemiological Features, Resistance Mechanisms, Detection and Therapy. Microbiol. Res. 2023, 266, 127249. [Google Scholar] [CrossRef]
- Ranjbar, R.; Alam, M. Antimicrobial Resistance Collaborators (2022). Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Evid. Based Nurs. 2024, 27, 16. [Google Scholar] [CrossRef]
- Marturano, J.E.; Lowery, T.J. ESKAPE Pathogens in Bloodstream Infections Are Associated With Higher Cost and Mortality but Can Be Predicted Using Diagnoses Upon Admission. Open Forum Infect. Dis. 2019, 6, ofz503. [Google Scholar] [CrossRef]
- Bereanu, A.-S.; Bereanu, R.; Mohor, C.; Vintilă, B.I.; Codru, I.R.; Olteanu, C.; Sava, M. Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania. Antibiotics 2024, 13, 400. [Google Scholar] [CrossRef]
- De Prisco, M.; Manente, R.; Santella, B.; Serretiello, E.; Dell’Annunziata, F.; Santoro, E.; Bernardi, F.F.; D’Amore, C.; Perrella, A.; Pagliano, P.; et al. Impact of ESKAPE Pathogens on Bacteremia: A Three-Year Surveillance Study at a Major Hospital in Southern Italy. Antibiotics 2024, 13, 901. [Google Scholar] [CrossRef] [PubMed]
- Hetsa, B.A.; Asante, J.; Amoako, D.G.; Abia, A.L.K.; Mbanga, J.; Essack, S.Y. Multidrug-Resistant ESKAPEEc Pathogens From Bloodstream Infections in South Africa: A Cross-Sectional Study Assessing Resistance to WHO AWaRe Antibiotics. Health Sci. Rep. 2025, 8, e70897. [Google Scholar] [CrossRef] [PubMed]
- DeVoe, C.; Segal, M.R.; Wang, L.; Stanley, K.; Madera, S.; Fan, J.; Schouest, J.; Graham-Ojo, R.; Nichols, A.; Prasad, P.A.; et al. Increased Rates of Secondary Bacterial Infections, Including Enterococcus Bacteremia, in Patients Hospitalized with Coronavirus Disease 2019 (COVID-19). Infect. Control Hosp. Epidemiol. 2022, 43, 1416–1423. [Google Scholar] [CrossRef]
- Golli, A.L.; Popa, S.G.; Ghenea, A.E.; Turcu, F.L. The Impact of the COVID-19 Pandemic on the Antibiotic Resistance of Gram-Negative Pathogens Causing Bloodstream Infections in an Intensive Care Unit. Biomedicines 2025, 13, 379. [Google Scholar] [CrossRef]
- Mendes, G.; Ramalho, J.F.; Duarte, A.; Pedrosa, A.; Silva, A.C.; Méndez, L.; Caneiras, C. First Outbreak of NDM-1-Producing Klebsiella Pneumoniae ST11 in a Portuguese Hospital Centre during the COVID-19 Pandemic. Microorganisms 2022, 10, 251. [Google Scholar] [CrossRef] [PubMed]
- Ficik, J.; Andrezál, M.; Drahovská, H.; Böhmer, M.; Szemes, T.; Liptáková, A.; Slobodníková, L. Carbapenem-Resistant Klebsiella Pneumoniae in COVID-19 Era—Challenges and Solutions. Antibiotics 2023, 12, 1285. [Google Scholar] [CrossRef]
- Yek, C.; Mancera, A.G.; Diao, G.; Walker, M.; Neupane, M.; Chishti, E.A.; Amirahmadi, R.; Richert, M.E.; Rhee, C.; Klompas, M.; et al. Impact of the COVID-19 Pandemic on Antibiotic Resistant Infection Burden in U.S. Hospitals: Retrospective Cohort Study of Trends and Risk Factors. Ann. Intern Med. 2025, 178, 796–807. [Google Scholar] [CrossRef]
- Mhango, M.; Sheehan, F.; Marmor, A.; Thirkell, C.; Kennedy, K. An Outbreak of Double Carbapenemase-Producing Klebsiella Pneumoniae, Harbouring NDM-5 and OXA-48 Genes, at a Tertiary Hospital in Canberra, Australia. Commun. Dis. Intell. 2024, 48, 1–11. [Google Scholar] [CrossRef]
- Lazar, D.S.; Nica, M.; Dascalu, A.; Oprisan, C.; Albu, O.; Codreanu, D.R.; Kosa, A.G.; Popescu, C.P.; Florescu, S.A. Carbapenem-Resistant NDM and OXA-48-like Producing K. Pneumoniae: From Menacing Superbug to a Mundane Bacteria; A Retrospective Study in a Romanian Tertiary Hospital. Antibiotics 2024, 13, 435. [Google Scholar] [CrossRef]
- Tomaszewski, D.; Guzek, A.; Rybicki, Z. The Empiric Antimicrobial Therapy: The Comparison of Initial Choice with the Results of Microbiological Investigations. An Observational Study. Pol. J. Aviat. Med. 2023, 29, 5–10. [Google Scholar] [CrossRef]
- Huang, C.; Moradi, S.; Sholeh, M.; Tabaei, F.M.; Lai, T.; Tan, B.; Meng, J.; Azizian, K. Global Trends in Antimicrobial Resistance of Enterococcus Faecium: A Systematic Review and Meta-Analysis of Clinical Isolates. Front. Pharmacol. 2025, 16, 1505674. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, Y.; Jia, P.; Zhu, Y.; Zhang, G.; Zhang, J.; Duan, S.; Kang, W.; Wang, T.; Jing, R.; et al. Global Trends of Antimicrobial Susceptibility to Ceftaroline and Ceftazidime–Avibactam: A Surveillance Study from the ATLAS Program (2012–2016). Antimicrob. Resist. Infect. Control 2020, 9, 166. [Google Scholar] [CrossRef]
- Zhang, X.; Paganelli, F.L.; Bierschenk, D.; Kuipers, A.; Bonten, M.J.M.; Willems, R.J.L.; van Schaik, W. Genome-Wide Identification of Ampicillin Resistance Determinants in Enterococcus Faecium. PLoS Genet. 2012, 8, e1002804. [Google Scholar] [CrossRef]
- Sader, H.S.; Mendes, R.E.; Duncan, L.; Kimbrough, J.H.; Carvalhaes, C.G.; Castanheira, M. Ceftazidime-Avibactam, Meropenem-Vaborbactam, and Imipenem-Relebactam Activities Against Multidrug-Resistant Enterobacterales from United States Medical Centers (2018–2022). Diagn. Microbiol. Infect. Dis. 2023, 106, 115945. [Google Scholar] [CrossRef]
- Bostanghadiri, N.; Narimisa, N.; Mirshekar, M.; Dadgar-Zankbar, L.; Taki, E.; Navidifar, T.; Darban-Sarokhalil, D. Prevalence of Colistin Resistance in Clinical Isolates of Acinetobacter baumannii: A Systematic Review and Meta-Analysis. Antimicrob. Resist. Infect. Control 2024, 13, 24. [Google Scholar] [CrossRef]
- Ramatla, T.; Nkhebenyane, J.; Lekota, K.E.; Thekisoe, O.; Monyama, M.; Achilonu, C.C.; Khasapane, G. Global Prevalence and Antibiotic Resistance Profiles of Carbapenem-Resistant Pseudomonas Aeruginosa Reported from 2014 to 2024: A Systematic Review and Meta-Analysis. Front. Microbiol. 2025, 16, 1599070. [Google Scholar] [CrossRef]
- Rhomberg, P.R.; Fritsche, T.R.; Sader, H.S.; Jones, R.N. Antimicrobial Susceptibility Pattern Comparisons Among Intensive Care Unit and General Ward Gram-Negative Isolates from the Meropenem Yearly Susceptibility Test Information Collection Program (USA). Diagn. Microbiol. Infect. Dis. 2006, 56, 57–62. [Google Scholar] [CrossRef]
- Yan, S.; Sun, Z.; Yang, Y.; Zhu, D.; Chen, Z.; on behalf of the China Antimicrobial Surveillance Network (CHINET) Study Group. Changing Antimicrobial Resistance Profile of Enterobacter spp. Isolates in Hospitals across China: A Seven-Year Analysis from the CHINET Antimicrobial Resistance Surveillance Program (2015–2021). One Health Adv. 2024, 2, 11. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).