Integrating New Technologies in Lipidology: A Comprehensive Review
Abstract
1. Introduction
2. Methods
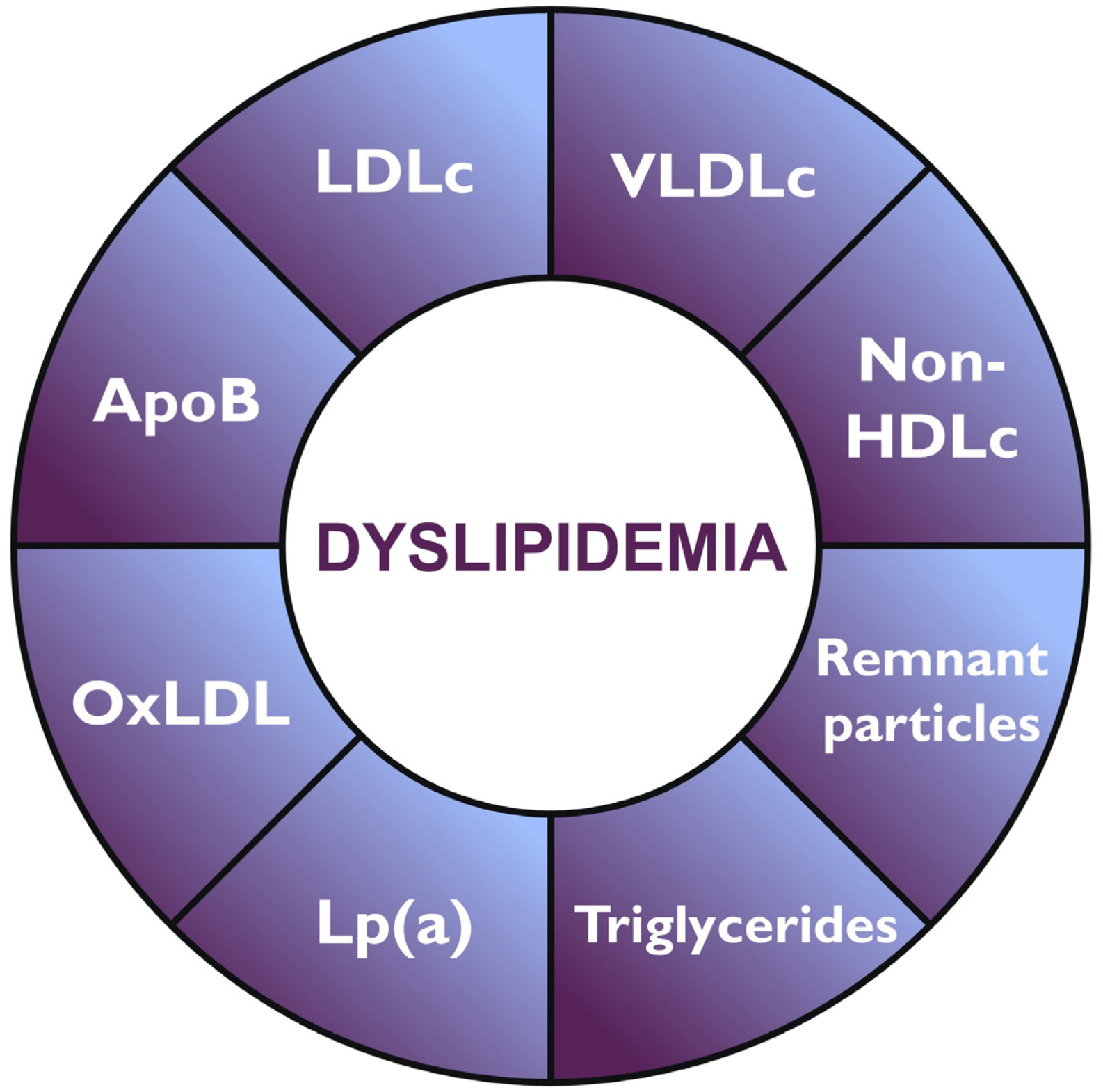
3. Advances in Lipid Profiling
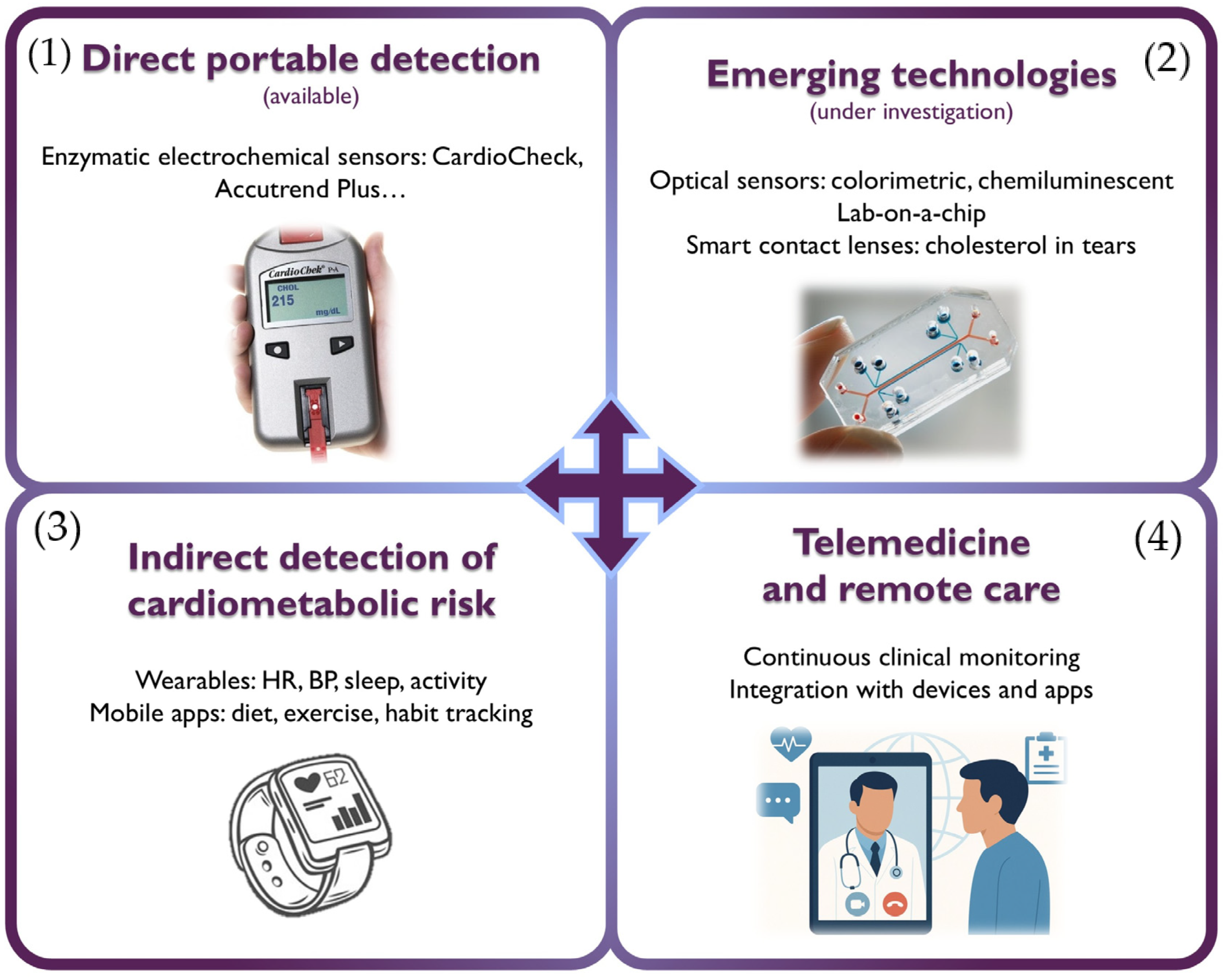
4. Novel Devices and Telemedicine in Lipidology
- -
- Enzymatic sensors: the most established technology, employing enzymes such as cholesterol oxidase (ChOx) and cholesterol esterase (ChE). These devices, found in commercial products like CardioCheck® Plus (PTS Diagnostics, Indianapolis, IN, USA) and Accutrend® Plus (Roche Diagnostics, Mannheim, Germany), enable rapid measurement of total cholesterol, HDLc, LDLc, and triglycerides from a drop of capillary blood. Although they offer high sensitivity and specificity, their performance can be affected by the instability of enzymes under environmental conditions (pH, temperature). Another limitation is their high cost.
- -
- Non-enzymatic sensors: currently under development, these systems rely on the direct oxidation of cholesterol on electrodes modified with nanomaterials such as graphene, metal oxides, or carbon nanotubes, offering greater robustness, stability, and lower potential cost.
| Technology | Detection Principle | Sample Type | Advantages | Disadvantages |
|---|---|---|---|---|
| Enzymatic electrochemical sensors | Enzymatic reaction + electrochemical signal | Capillary blood | High accuracy and specificity Commercially available | Use of unstable enzymes Requires finger-prick sampling |
| Non-enzymatic electrochemical sensors | Direct oxidation on nanomaterial-modified electrodes | Blood, serum | No enzymes required (greater stability) Potentially low cost | Not yet clinically validated Require advanced materials |
| Optical sensors (colorimetric, fluorescent) | Color or fluorescence change detected via smartphone | Capillary blood, serum | Portable and low-cost Smartphone-compatible | Not validated Sensitive to light/position Require specialized reagents |
| Chemiluminescent sensors | Light emission from chemical reaction (captured by camera) | Blood, serum | High luminescent sensitivity Accurate digital readout | In preclinical studies Require accessories (dark box) Limited to controlled environments |
| Microfluidic devices/lab-on-a-chip | Electrochemical or luminescent reaction in microchannels | Blood, serum, and biological fluids | Multi-analyte from a small volume Rapid results | High manufacturing complexity Not yet commercialized |
| Smart contact lenses | Optical detection of cholesterol in tears | Tears | Non-invasive (tear-based) Highly innovative | Still experimental Not clinically available |
5. Emerging Lipid-Lowering Therapies
5.1. RNA-Based Therapies
5.2. Gene Therapy and CRISPR-Cas9
5.3. Nanotechnology for Targeted Drug Delivery
6. Integrating Artificial Intelligence and Lipidology
6.1. Early Diagnosis of Familial Hypercholesterolemia
| Author (Year) | Cohort (s) | Algorithm (s) | Results |
|---|---|---|---|
| Myers et al. (2019) [68] | Training: EHR data from 939 clinically diagnosed individuals with FH and 83,136 controls sampled from four U.S. institutions Validation: National healthcare encounter database (170 million individuals) and an integrated healthcare delivery system dataset (174,000 individuals) | FIND HF (sequential random forest) | Internal validation: Precision 0.85, Recall 0.45, AUPRC 0.55, AUROC 0.89 External validation: 87% of the flagged individuals in the national database and 77% in the healthcare delivery system dataset were categorized as having high clinical suspicion of FH. |
| Banda et al. (2019) [71] | Training: 3.1 million patients (197 known FH patients and 6590 selected controls) Validation: Geisinger Healthcare System dataset (33,086 patients, 466 cases) | Random forest classifier | Internal validation: Positive predictive value (PPV) of 0.88 and sensitivity of 0.75 External validation: PPV of 0.85 |
| Akyea et al. (2020) [72] | Training: 3,020,832 individuals from the Clinical Practice Research Datalink (United Kingdom) Validation: 1,006,943 individuals from the same dataset | Logistic regression Random forest Gradient boosting Deep learning Ensemble model (combination) | External validation: Every model had AUC > 0.89 except for logistic regression (AUC 0.81). Ensemble learning demonstrated a high likelihood ratio (45.5) |
| Pina et al. (2020) [74] | Training: FH Gothenburg cohort (174 patients) Validation: FH-CEGP Milan cohort (364 patients) | Classification tree Gradient boosting Neural network | Internal validation: AUROCs: 0.79 (CT), 0.83 (GBM), 0.83 (NN), 0.68 (DLCN) External validation: AUROCs: 0.70 (CT), 0.78 (GBM), 0.76 (NN), 0.64 (DLCN) |
| Hesse et al. (2022) [75] | Training: Charlotte Maxeke Johannesburg Academic Hospital’s (678 patients) Validation: Groote Schuur Hospital (1376 with clinical FH and 2655 with potential FH) | Logistic regression Deep learning Random forest | External validation: AUROC 0.711 in a high-prevalence FH sample (DLCN 0.705) and 0.801 in a medium-prevalence FH sample |
| Gratton et al. (2023) [73] | Training: UK Biobank (139,779 individuals) | Least absolute shrinkage and selection operator logistic regression | Internal validation: 14 variable models with AUC of 0.77 |
| Gidding et al. (2023) [69] | Validation: Geisinger MyCode Community Health Initiative cohort (59,729 individuals) | FIND FH (random forest) Mayo Clinic (NLP) | External validation: 5.9% of 573 flagged by FIND FH and 1.9% of 10,415 by Mayo had a pathogenic or likely pathogenic (P/LP) variant. At least one algorithm identified 197 out of 280 individuals with P/LP variant (net yield: 70%) |
| Kim et al. (2024) [70] | Validation: 167,955 individuals from the Oregon Health and Science University Health Care System | FIND HF (random forest) | External validation: 121/471 flagged individuals met “likely” FH criteria and received suboptimal lipid-lowering therapies |
| Khademi et al. (2024) [76] | Training: 1591 individuals | Multi-Stage Tabular Deep Learning Network (FH-TabNet) | Internal validation: F1-score 98.2, 98.6, 87.2, and 19.2 for “Unlikely”, “Possible”, “Probable”, and “Definite” FH class prediction |
6.2. Enhanced LDLc Estimation
| Author (Year) | Cohort (s) | Algorithm (s) | Results |
|---|---|---|---|
| Singh et al. (2020) [80] | Training: 17,500 lipid profiles performed on 10,936 individuals (New York-Presbyterian Hospital/Weill Cornell Medicine) | Random forests (Weill Cornell model) | Internal validation: Correlation coefficients between estimated and measured LDLc were 0.982 (Weill Cornell model), 0.950 (Friedewald), and 0.962 (Martin–Hopkins). |
| Barakett-Hamade et al. (2021) [81] | Training: 31,922 observations from 19,279 subjects | K-nearest neighbors (LDL-KNN) | Internal validation: Intraclass correlation coefficients: 0.937 (Martin–Hopkins), 0.935 (Sampson), 0.925 (LDL-KNN), 0.894 (Friedewald), 0.869 (de Cordova), |
| Tsigalou et al. (2021) [85] | Training: 4244 records (Sismanoglio General Hospital of Komotini) Validation: 478 records (Democritus Diagnostic Center in Alexandroupolis, University Hospital of Alexandroupolis) | Linear regression (LR), Support Vector Machines (SVM), Extreme Gradient Boosting (XGB), Deep Neural Networks (DNN) | External validation. Simple ML techniques can work as well as neural networks. Standard error of the estimate (the lower, the better): - Cohort 1: 13.3 (LR), 10.1 (SVM), 15.6 (XGB), 10.1 (DNN) - Cohort 2: 20.9 (LR), 33.4 (SVM), 21 (XGB), 34.4 (DNN) |
| Ghayad et al. (2022) [82] | Training: 31,853 retrospective records (Hotel Dieu de France University Hospital) Validation: 6599 prospective records (Hotel Dieu de France University Hospital) | LDL-KNN | External validation: ICCs above the 0.9 cutoff except for low LDLc or very high triglycerides |
| Çubukçu et al. (2022) [83] | Training: 59,415 records (Başkent University) | Linear regression (LR), gradient-boosted trees, artificial neural network (ANN) | Internal validation. Better performance than classical formulas for LDLc < 70 mg/dL and TG > 177 mg/dL [F1-scores: 70.46 (ANN), 69.6 (gradient-boosted trees), 69.23 (LR), 62.71 (Martin–Hopkins), 62.38 (Friedewald)] |
| Oh et al. (2022) [84] | Training: 823,657 records (Seoul National University Hospital) | Gradient boosting (LDL-CX), neural network (LDL-CN) | Internal validation. Better correlation compared to traditional formulas [overall bias: −0.27 mg/dL (LDL-CX), −0.01 mg/dL (LDL-CN), −3.80 mg/dL (Friedewald), −2.00 mg/dL (Martin–Hopkins)] |
| Kwon et al. (2022) [86] | Training: 129,930 records (Gangnam Severance Health Check-up) Validation: 46,470 records (Korean Initiatives on Coronary Artery Calcification registry) | Deep neural network (DNN) | External validation: The DNN method had lower bias and root mean-square error than Friedewald’s, Martin’s, and Sampson equations |
6.3. Statin Use and Target LDLc Achievements
7. Digital Models and Clinical Decision Support Systems
8. Ethical, Regulatory, and Implementation Challenges
8.1. Information and Biases
8.2. Resources
8.3. Ethics
8.4. Epistemic and Ontological Considerations
8.5. Validation and Legal Frameworks
8.6. Patient Perspective
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ANGPTL3 | Angiopoietin-Like protein 3 |
| ApoB | Apolipoprotein B |
| AUC | Area Under the Curve |
| AUROC | Area Under the Receiver-Operating-Characteristic Curve |
| AUPRC | Area Under the Precision–Recall Curve |
| CDSS | Clinical Decision-Support System |
| CDS-FH | Clinical Decision-Support for Familial Hypercholesterolemia |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CT | Computed Tomography |
| CV | Cardiovascular |
| CVD | Cardiovascular Disease |
| DL | Deep Learning |
| DLCN | Dutch Lipid Clinic Network |
| EHR | Electronic Health Record |
| EMA | European Medicines Agency |
| FDA | Food and Drug Administration |
| FH | Familial Hypercholesterolemia |
| HDLc | High-Density Lipoprotein Cholesterol |
| KNN | k-Nearest Neighbours |
| LASSO | Least-Absolute-Shrinkage-and-Selection Operator |
| LDLc | Low-Density Lipoprotein Cholesterol |
| LDLR | Low-Density Lipoprotein Receptor |
| Lp(a) | Lipoprotein(a) |
| ML | Machine Learning |
| MRS | Magnetic-Resonance Spectroscopy |
| NMR | Nuclear-Magnetic Resonance |
| NLP | Natural-Language Processing |
| PCSK9 | Proprotein-Convertase Subtilisin/Kexin type 9 |
| siRNA | Small-Interfering RNA |
| TC | Total Cholesterol |
| TG | Triglycerides |
References
- Mensah, G.A.; Fuster, V.; Murray, C.J.L.; Roth, G.A. Global Burden of Cardiovascular Diseases and Risks, 1990–2022. J. Am. Coll. Cardiol. 2023, 82, 2350–2473. [Google Scholar] [CrossRef] [PubMed]
- Chong, B.; Jayabaskaran, J.; Jauhari, S.M.; Chan, S.P.; Goh, R.; Kueh, M.T.W.; Li, H.; Chin, Y.H.; Kong, G.; Anand, V.V.; et al. Global burden of cardiovascular diseases: Projections from 2025 to 2050. Eur. J. Prev. Cardiol. 2024, zwae281. [Google Scholar] [CrossRef] [PubMed]
- Ference, B.A.; Ginsberg, H.N.; Graham, I.; Ray, K.K.; Packard, C.J.; Bruckert, E.; Hegele, R.A.; Krauss, R.M.; Raal, F.J.; Schunkert, H.; et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2017, 38, 2459–2472. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, T.C.; Liu, Y.; Mohebi, R.; Miksenas, H.; Haidermota, S.; Wong, M.; Hu, X.; Cristino, J.R.; Browne, A.; Plutzky, J.; et al. Lipoprotein(a), Oxidized Phospholipids, and Coronary Artery Disease Severity and Outcomes. J. Am. Coll. Cardiol. 2023, 81, 1780–1792. [Google Scholar] [CrossRef]
- Pinto, X.; Fanlo, M.; Esteve, V.; Millan, J.; Grupo de trabajo sobre Dislipemia Aterogénica de la Sociedad Española de Arteriosclerosis. Remnant cholesterol, vascular risk, and prevention of atherosclerosis. Clin. Investig. Arterioscler. 2023, 35, 206–217. [Google Scholar] [CrossRef]
- Chong, B.; Jayabaskaran, J.; Ruban, J.; Goh, R.; Chin, Y.H.; Kong, G.; Ng, C.H.; Lin, C.; Loong, S.; Muthiah, M.D.; et al. Epicardial Adipose Tissue Assessed by Computed Tomography and Echocardiography Are Associated With Adverse Cardiovascular Outcomes: A Systematic Review and Meta-Analysis. Circ. Cardiovasc. Imaging 2023, 16, e015159. [Google Scholar] [CrossRef]
- Holmes, M.V.; Millwood, I.Y.; Kartsonaki, C.; Hill, M.R.; Bennett, D.A.; Boxall, R.; Guo, Y.; Xu, X.; Bian, Z.; Hu, R.; et al. Lipids, Lipoproteins, and Metabolites and Risk of Myocardial Infarction and Stroke. J. Am. Coll. Cardiol. 2018, 71, 620–632. [Google Scholar] [CrossRef]
- Nurmohamed, N.S.; Kraaijenhof, J.M.; Mayr, M.; Nicholls, S.J.; Koenig, W.; Catapano, A.L.; Stroes, E.S.G. Proteomics and lipidomics in atherosclerotic cardiovascular disease risk prediction. Eur. Heart J. 2023, 44, 1594–1607. [Google Scholar] [CrossRef]
- Hughes, A.; Shandhi, M.M.H.; Master, H.; Dunn, J.; Brittain, E. Wearable Devices in Cardiovascular Medicine. Circ. Res. 2023, 132, 652–670. [Google Scholar] [CrossRef]
- Naoum, I.; Saliba, W.; Aker, A.; Zafrir, B. Lipid-lowering therapy with inclisiran in the real-world setting: Initial data from a national health care service. J. Clin. Lipidol. 2024, 18, e809–e816. [Google Scholar] [CrossRef]
- Stankov, S.; Cuchel, M. Gene editing for dyslipidemias: New tools to “cut” lipids. Atherosclerosis 2023, 368, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Arrobas Velilla, T.; Guijarro, C.; Ruiz, R.C.; Pinero, M.R.; Valderrama Marcos, J.F.; Perez Perez, A.; Botana Lopez, A.M.; Lopez, A.M.; Garcia Donaire, J.A.; Obaya, J.C.; et al. Consensus document for lipid profile testing and reporting in Spanish clinical laboratories: What parameters should a basic lipid profile include? Adv. Lab. Med. 2023, 4, 138–156. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, W.C.; Otvos, J.D.; Keyes, M.J.; Pencina, M.J.; Sullivan, L.; Vasan, R.S.; Wilson, P.W.; D’Agostino, R.B. LDL Particle Number and Risk of Future Cardiovascular Disease in the Framingham Offspring Study—Implications for LDL Management. J. Clin. Lipidol. 2007, 1, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, F.; Casula, M.; Olmastroni, E. Apolipoprotein B compared with low-density lipoprotein cholesterol in the atherosclerotic cardiovascular diseases risk assessment. Pharmacol. Res. 2023, 195, 106873. [Google Scholar] [CrossRef]
- Rehman, M.B.; Tudrej, B.V. Lipoprotein(a) and risk-weighted apolipoprotein B: A novel metric for atherogenic risk. Lipids Health Dis. 2024, 23, 316. [Google Scholar] [CrossRef]
- Wu, Z.; Bagarolo, G.I.; Thoroe-Boveleth, S.; Jankowski, J. “Lipidomics”: Mass spectrometric and chemometric analyses of lipids. Adv. Drug Deliv. Rev. 2020, 159, 294–307. [Google Scholar] [CrossRef]
- Emeasoba, E.U.; Ibeson, E.; Nwosu, I.; Montemarano, N.; Shani, J.; Shetty, V.S. Clinical Relevance of Nuclear Magnetic Resonance LipoProfile. Front. Nucl. Med. 2022, 2, 960522. [Google Scholar] [CrossRef]
- Dominguez-Renedo, O.; Navarro-Cunado, A.M.; Alonso-Lomillo, M.A. Electrochemical devices for cholesterol detection. J. Pharm. Biomed. Anal. 2023, 224, 115195. [Google Scholar] [CrossRef]
- Pham, A.T.T.; Wallace, A.; Zhang, X.; Tohl, D.; Fu, H.; Chuah, C.; Reynolds, K.J.; Ramsey, C.; Tang, Y. Optical-Based Biosensors and Their Portable Healthcare Devices for Detecting and Monitoring Biomarkers in Body Fluids. Diagnostics 2021, 11, 1285. [Google Scholar] [CrossRef]
- Park, J.; Kim, J.; Kim, S.Y.; Cheong, W.H.; Jang, J.; Park, Y.G.; Na, K.; Kim, Y.T.; Heo, J.H.; Lee, C.Y.; et al. Soft, smart contact lenses with integrations of wireless circuits, glucose sensors, and displays. Sci. Adv. 2018, 4, eaap9841. [Google Scholar] [CrossRef]
- Duan, W.; Cheng, J.; Guo, J. Smartphone-based photochemical sensor for multiplex determination of glucose, uric acid, and total cholesterol in fingertip blood. Analyst 2022, 147, 3285–3290. [Google Scholar] [CrossRef] [PubMed]
- Hamaya, R.; Fukuda, H.; Takebayashi, M.; Mori, M.; Matsushima, R.; Nakano, K.; Miyake, K.; Tani, Y.; Yokokawa, H. Effects of an mHealth App (Kencom) With Integrated Functions for Healthy Lifestyles on Physical Activity Levels and Cardiovascular Risk Biomarkers: Observational Study of 12,602 Users. J. Med. Internet Res. 2021, 23, e21622. [Google Scholar] [CrossRef]
- Aomori, M.; Matsumoto, C.; Takebayashi, S.; Matsuyama, N.; Uto, Y.; Tanaka, M.; Samukawa, S.; Kato, H.; Nakajima, H.; Maeda, H. Effects of a smartphone app-based diet and physical activity program for men living with HIV who have dyslipidemia: A pilot randomized controlled trial. Jpn. J. Nurs. Sci. 2023, 20, e12535. [Google Scholar] [CrossRef] [PubMed]
- Blood, A.J.; Cannon, C.P.; Gordon, W.J.; Mailly, C.; MacLean, T.; Subramaniam, S.; Tucci, M.; Crossen, J.; Nichols, H.; Wagholikar, K.B.; et al. Results of a Remotely Delivered Hypertension and Lipid Program in More Than 10 000 Patients Across a Diverse Health Care Network. JAMA Cardiol. 2023, 8, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.C.; Kao, C.H. Telemedicine assists in the management of proatherogenic dyslipidemia and postprandial glucose variability in patients with type 2 diabetes mellitus: A cross-sectional study. Endocr. Connect. 2021, 10, 789–795. [Google Scholar] [CrossRef]
- Silverman, M.G.; Ference, B.A.; Im, K.; Wiviott, S.D.; Giugliano, R.P.; Grundy, S.M.; Braunwald, E.; Sabatine, M.S. Association between lowering LDL-C and cardiovascular risk reduction among different therapeutic interventions: A systematic review and meta-analysis. JAMA 2016, 316, 1289–1297. [Google Scholar] [CrossRef]
- Cannon, C.P.; Blazing, M.A.; Giugliano, R.P.; McCagg, A.; White, J.A.; Theroux, P.; Darius, H.; Lewis, B.S.; Ophuis, T.O.; Jukema, J.W.; et al. Ezetimibe Added to Statin Therapy after Acute Coronary Syndromes. N. Engl. J. Med. 2015, 372, 2387–2397. [Google Scholar] [CrossRef]
- Nissen, S.E.; Lincoff, A.M.; Brennan, D.; Ray, K.K.; Mason, D.; Kastelein, J.J.P.; Thompson, P.D.; Libby, P.; Cho, L.; Plutzky, J.; et al. Bempedoic Acid and Cardiovascular Outcomes in Statin-Intolerant Patients. N. Engl. J. Med. 2023, 388, 1353–1364. [Google Scholar] [CrossRef]
- Ray, K.K.; Haq, I.; Bilitou, A.; Manu, M.C.; Burden, A.; Aguiar, C.; Arca, M.; Connolly, D.L.; Eriksson, M.; Ferrières, J.; et al. Treatment gaps in the implementation of LDL cholesterol control among high- and very high-risk patients in Europe between 2020 and 2021: The multinational observational SANTORINI study. Lancet Reg. Health Eur. 2023, 29, 100624. [Google Scholar] [CrossRef]
- Catapano, A.L. Emerging therapies in dyslipidaemias. Vasc. Pharmacol. 2023, 153, 107229. [Google Scholar] [CrossRef]
- Mansfield, B.S.; Mohamed, F.; Larouche, M.; Raal, F.J. The Hurdle of Access to Emerging Therapies and Potential Solutions in the Management of Dyslipidemias. J. Clin. Med. 2024, 13, 4160. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.J.; et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Tsimikas, S.; Karwatowska-Prokopczuk, E.; Gouni-Berthold, I.; Tardif, J.C.; Baum, S.J.; Steinhagen-Thiessen, E.; Shapiro, M.D.; Stroes, E.S.; Moriarty, P.M.; Nordestgaard, B.G.; et al. Lipoprotein(a) Reduction in Persons with Cardiovascular Disease. N. Engl. J. Med. 2020, 382, 244–255. [Google Scholar] [CrossRef]
- Yeang, C.; Karwatowska-Prokopczuk, E.; Su, F.; Dinh, B.; Xia, S.; Witztum, J.L.; Tsimikas, S. Effect of Pelacarsen on Lipoprotein(a) Cholesterol and Corrected Low-Density Lipoprotein Cholesterol. J. Am. Coll. Cardiol. 2022, 79, 1035–1046. [Google Scholar] [CrossRef]
- Kosmas, C.E.; Bousvarou, M.D.; Papakonstantinou, E.J.; Tsamoulis, D.; Koulopoulos, A.; Echavarria Uceta, R.; Guzman, E.; Rallidis, L.S. Novel Pharmacological Therapies for the Management of Hyperlipoproteinemia(a). Int. J. Mol. Sci. 2023, 24, 13622. [Google Scholar] [CrossRef]
- O’Donoghue, M.L.; Rosenson, R.S.; Gencer, B.; Lopez, J.A.G.; Lepor, N.E.; Baum, S.J.; Stout, E.; Gaudet, D.; Knusel, B.; Kuder, J.F.; et al. Small Interfering RNA to Reduce Lipoprotein(a) in Cardiovascular Disease. N. Engl. J. Med. 2022, 387, 1855–1864. [Google Scholar] [CrossRef]
- Nissen, S.E.; Wang, Q.; Nicholls, S.J.; Navar, A.M.; Ray, K.K.; Schwartz, G.G.; Szarek, M.; Stroes, E.S.G.; Troquay, R.; Dorresteijn, J.A.N.; et al. Zerlasiran-A Small-Interfering RNA Targeting Lipoprotein(a): A Phase 2 Randomized Clinical Trial. JAMA 2024, 332, 1992–2002. [Google Scholar] [CrossRef]
- Blom, D.J.; Marais, A.D.; Moodley, R.; van der Merwe, N.; van Tonder, A.; Raal, F.J. RNA-based therapy in the management of lipid disorders: A review. Lipids Health Dis. 2022, 21, 41. [Google Scholar] [CrossRef]
- Ruhanen, H.; Haridas, P.A.N.; Minicocci, I.; Taskinen, J.H.; Palmas, F.; di Costanzo, A.; D’Erasmo, L.; Metso, J.; Partanen, J.; Dalli, J.; et al. ANGPTL3 deficiency alters the lipid profile and metabolism of cultured hepatocytes and human lipoproteins. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158679. [Google Scholar] [CrossRef]
- Rosenson, R.S.; Gaudet, D.; Hegele, R.A.; Ballantyne, C.M.; Nicholls, S.J.; Lucas, K.J.; San Martin, J.; Zhou, R.; Muhsin, M.; Chang, T.; et al. Zodasiran, an RNAi Therapeutic Targeting ANGPTL3, for Mixed Hyperlipidemia. N. Engl. J. Med. 2024, 391, 913–925. [Google Scholar] [CrossRef]
- Wang, X.; Musunuru, K. Angiopoietin-Like 3: From Discovery to Therapeutic Gene Editing. JACC Basic. Transl. Sci. 2019, 4, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Glass, Z.; Chen, J.; Haas, M.; Jin, X.; Zhao, X.; Rui, X.; Ye, Z.; Li, Y.; Zhang, F.; et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA 2021, 118, e2020401118. [Google Scholar] [CrossRef] [PubMed]
- U.S. National Library of Medicine. Open-Label, Phase 1b, Single-Ascending Dose and Optional re Dosing Study to Evaluate the Safety of VERVE-101 Administered to Patients with Heterozygous Familial Hypercholesterolemia, Atherosclerotic Cardiovascular Disease, and Uncontrolled Hypercholesterolemia; U.S. National Library of Medicine: Bethesda, MD, USA, 2022. Available online: https://clinicaltrials.gov/study/NCT05398029 (accessed on 8 May 2025).
- Horie, T.; Ono, K. VERVE-101: A promising CRISPR-based gene editing therapy that reduces LDL-C and PCSK9 levels in HeFH patients. Eur. Heart J. Cardiovasc. Pharmacother. 2024, 10, 89–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Centurion, F.; Misra, A.; Patel, S.; Gu, Z. Molecularly targeted nanomedicine enabled by inorganic nanoparticles for atherosclerosis diagnosis and treatment. Adv. Drug Deliv. Rev. 2023, 194, 114709. [Google Scholar] [CrossRef]
- Pang, A.S.; Dinesh, T.; Pang, N.Y.; Dinesh, V.; Pang, K.Y.; Yong, C.L.; Lee, S.J.J.; Yip, G.W.; Bay, B.H.; Srinivasan, D.K. Nanoparticles as Drug Delivery Systems for the Targeted Treatment of Atherosclerosis. Molecules 2024, 29, 2873. [Google Scholar] [CrossRef]
- Zhanga, C.-P. LDLR mRNA-Loaded Exosomes Serve as Key Vehicle in Improving Familial Hypercholesterolemia. Am. J. Biomed. Sci. Res. 2024, 22, 57–61. [Google Scholar] [CrossRef]
- Dennison Himmelfarb, C.R.; Beckie, T.M.; Allen, L.A.; Commodore-Mensah, Y.; Davidson, P.M.; Lin, G.; Lutz, B.; Spatz, E.S.; American Heart Association Council on Cardiovascular and Stroke Nursing. Shared Decision-Making and Cardiovascular Health: A Scientific Statement From the American Heart Association. Circulation 2023, 148, 912–931. [Google Scholar] [CrossRef]
- Pickering, J.W. Machine learning for decision-making in cardiology: A narrative review to aid navigating the new landscape. Rev. Esp. Cardiol. (Engl. Ed.) 2023, 76, 645–654. [Google Scholar] [CrossRef]
- Itchhaporia, D. Artificial intelligence in cardiology. Trends Cardiovasc. Med. 2022, 32, 34–41. [Google Scholar] [CrossRef]
- Howell, M.D.; Corrado, G.S.; DeSalvo, K.B. Three Epochs of Artificial Intelligence in Health Care. JAMA 2024, 331, 242–244. [Google Scholar] [CrossRef]
- D’Ascenzo, F.; De Filippo, O.; Gallone, G.; Mittone, G.; Deriu, M.A.; Iannaccone, M.; Ariza-Sole, A.; Liebetrau, C.; Manzano-Fernandez, S.; Quadri, G.; et al. Machine learning-based prediction of adverse events following an acute coronary syndrome (PRAISE): A modelling study of pooled datasets. Lancet 2021, 397, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Arshad, M.S.; Greene, S.J.; Van Spall, H.G.C.; Pandey, A.; Vemulapalli, S.; Perakslis, E.; Butler, J. Artificial intelligence and heart failure: A state-of-the-art review. Eur. J. Heart Fail. 2023, 25, 1507–1525. [Google Scholar] [CrossRef] [PubMed]
- Guha, A.; Shah, V.; Nahle, T.; Singh, S.; Kunhiraman, H.H.; Shehnaz, F.; Nain, P.; Makram, O.M.; Mahmoudi, M.; Al-Kindi, S.; et al. Artificial Intelligence Applications in Cardio-Oncology: A Comprehensive Review. Curr. Cardiol. Rep. 2025, 27, 56. [Google Scholar] [CrossRef] [PubMed]
- Kabra, R.; Israni, S.; Vijay, B.; Baru, C.; Mendu, R.; Fellman, M.; Sridhar, A.; Mason, P.; Cheung, J.W.; DiBiase, L.; et al. Emerging role of artificial intelligence in cardiac electrophysiology. Cardiovasc. Digit. Health J. 2022, 3, 263–275. [Google Scholar] [CrossRef]
- Salih, A.; Boscolo Galazzo, I.; Gkontra, P.; Lee, A.M.; Lekadir, K.; Raisi-Estabragh, Z.; Petersen, S.E. Explainable Artificial Intelligence and Cardiac Imaging: Toward More Interpretable Models. Circ. Cardiovasc. Imaging 2023, 16, e014519. [Google Scholar] [CrossRef]
- Cubukcu, H.C.; Topcu, D.I.; Yenice, S. Machine learning-based clinical decision support using laboratory data. Clin. Chem. Lab. Med. 2024, 62, 793–823. [Google Scholar] [CrossRef]
- Beheshti, S.O.; Madsen, C.M.; Varbo, A.; Nordestgaard, B.G. Worldwide Prevalence of Familial Hypercholesterolemia: Meta-Analyses of 11 Million Subjects. J. Am. Coll. Cardiol. 2020, 75, 2553–2566. [Google Scholar] [CrossRef]
- Benn, M.; Watts, G.F.; Tybjaerg-Hansen, A.; Nordestgaard, B.G. Familial hypercholesterolemia in the danish general population: Prevalence, coronary artery disease, and cholesterol-lowering medication. J. Clin. Endocrinol. Metab. 2012, 97, 3956–3964. [Google Scholar] [CrossRef]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef]
- Austin, M.A.; Hutter, C.M.; Zimmern, R.L.; Humphries, S.E. Genetic causes of monogenic heterozygous familial hypercholesterolemia: A HuGE prevalence review. Am. J. Epidemiol. 2004, 160, 407–420. [Google Scholar] [CrossRef]
- Berberich, A.J.; Hegele, R.A. The complex molecular genetics of familial hypercholesterolaemia. Nat. Rev. Cardiol. 2019, 16, 9–20. [Google Scholar] [CrossRef]
- Williams, R.R.; Hunt, S.C.; Schumacher, M.C.; Hegele, R.A.; Leppert, M.F.; Ludwig, E.H.; Hopkins, P.N. Diagnosing heterozygous familial hypercholesterolemia using new practical criteria validated by molecular genetics. Am. J. Cardiol. 1993, 72, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Gidding, S.S.; Champagne, M.A.; de Ferranti, S.D.; Defesche, J.; Ito, M.K.; Knowles, J.W.; McCrindle, B.; Raal, F.; Rader, D.; Santos, R.D.; et al. The Agenda for Familial Hypercholesterolemia: A Scientific Statement From the American Heart Association. Circulation 2015, 132, 2167–2192. [Google Scholar] [CrossRef]
- Risk of fatal coronary heart disease in familial hypercholesterolaemia. Scientific Steering Committee on behalf of the Simon Broome Register Group. BMJ 1991, 303, 893–896. [CrossRef] [PubMed]
- Nordestgaard, B.G.; Chapman, M.J.; Humphries, S.E.; Ginsberg, H.N.; Masana, L.; Descamps, O.S.; Wiklund, O.; Hegele, R.A.; Raal, F.J.; Defesche, J.C.; et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: Guidance for clinicians to prevent coronary heart disease: Consensus statement of the European Atherosclerosis Society. Eur. Heart J. 2013, 34, 3478–3490a. [Google Scholar] [CrossRef] [PubMed]
- Reeskamp, L.F.; Annink, M.E.; Schonck, W.A.M. Familial Hypercholesterolemia Detection Through Machine Learning Algorithms: A Low-Hanging Fruit. JACC Adv. 2024, 3, 101181. [Google Scholar] [CrossRef]
- Myers, K.D.; Knowles, J.W.; Staszak, D.; Shapiro, M.D.; Howard, W.; Yadava, M.; Zuzick, D.; Williamson, L.; Shah, N.H.; Banda, J.M.; et al. Precision screening for familial hypercholesterolaemia: A machine learning study applied to electronic health encounter data. Lancet Digit. Health 2019, 1, e393–e402. [Google Scholar] [CrossRef]
- Gidding, S.S.; Kirchner, H.L.; Brangan, A.; Howard, W.; Kelly, M.A.; Myers, K.D.; Morgan, K.M.; Oetjens, M.T.; Shuey, T.C.; Staszak, D.; et al. Yield of Familial Hypercholesterolemia Genetic and Phenotypic Diagnoses After Electronic Health Record and Genomic Data Screening. J. Am. Heart Assoc. 2023, 12, e030073. [Google Scholar] [CrossRef]
- Kim, K.; Faruque, S.C.; Lam, S.; Kulp, D.; He, X.; Sperling, L.S.; Eapen, D.J. Implications of Diagnosis Through a Machine Learning Algorithm on Management of People With Familial Hypercholesterolemia. JACC Adv. 2024, 3, 101184. [Google Scholar] [CrossRef]
- Banda, J.M.; Sarraju, A.; Abbasi, F.; Parizo, J.; Pariani, M.; Ison, H.; Briskin, E.; Wand, H.; Dubois, S.; Jung, K.; et al. Finding missed cases of familial hypercholesterolemia in health systems using machine learning. npj Digit. Med. 2019, 2, 23. [Google Scholar] [CrossRef]
- Akyea, R.K.; Qureshi, N.; Kai, J.; Weng, S.F. Performance and clinical utility of supervised machine-learning approaches in detecting familial hypercholesterolaemia in primary care. npj Digit. Med. 2020, 3, 142. [Google Scholar] [CrossRef] [PubMed]
- Gratton, J.; Futema, M.; Humphries, S.E.; Hingorani, A.D.; Finan, C.; Schmidt, A.F. A Machine Learning Model to Aid Detection of Familial Hypercholesterolemia. JACC Adv. 2023, 2, 100333. [Google Scholar] [CrossRef] [PubMed]
- Pina, A.; Helgadottir, S.; Mancina, R.M.; Pavanello, C.; Pirazzi, C.; Montalcini, T.; Henriques, R.; Calabresi, L.; Wiklund, O.; Macedo, M.P.; et al. Virtual genetic diagnosis for familial hypercholesterolemia powered by machine learning. Eur. J. Prev. Cardiol. 2020, 27, 1639–1646. [Google Scholar] [CrossRef]
- Hesse, R.; Raal, F.J.; Blom, D.J.; George, J.A. Familial Hypercholesterolemia Identification by Machine Learning Using Lipid Profile Data Performs as Well as Clinical Diagnostic Criteria. Circ. Genom. Precis. Med. 2022, 15, e003324. [Google Scholar] [CrossRef] [PubMed]
- Khademi, S.; Hajiakhondi, Z.; Vaseghi, G.; Sarrafzadegan, N.; Mohammadi, A. FH-TabNet: Multi-Class Familial Hypercholesterolemia Detection via a Multi-Stage Tabular Deep Learning. arXiv 2024, arXiv:2403.11032. [Google Scholar] [CrossRef]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef]
- Martin, S.S.; Blaha, M.J.; Elshazly, M.B.; Toth, P.P.; Kwiterovich, P.O.; Blumenthal, R.S.; Jones, S.R. Comparison of a novel method vs the Friedewald equation for estimating low-density lipoprotein cholesterol levels from the standard lipid profile. JAMA 2013, 310, 2061–2068. [Google Scholar] [CrossRef]
- Sampson, M.; Ling, C.; Sun, Q.; Harb, R.; Ashmaig, M.; Warnick, R.; Sethi, A.; Fleming, J.K.; Otvos, J.D.; Meeusen, J.W.; et al. A New Equation for Calculation of Low-Density Lipoprotein Cholesterol in Patients With Normolipidemia and/or Hypertriglyceridemia. JAMA Cardiol. 2020, 5, 540–548. [Google Scholar] [CrossRef]
- Singh, G.; Hussain, Y.; Xu, Z.; Sholle, E.; Michalak, K.; Dolan, K.; Lee, B.C.; van Rosendael, A.R.; Fatima, Z.; Pena, J.M.; et al. Comparing a novel machine learning method to the Friedewald formula and Martin-Hopkins equation for low-density lipoprotein estimation. PLoS ONE 2020, 15, e0239934. [Google Scholar] [CrossRef]
- Barakett-Hamade, V.; Ghayad, J.P.; McHantaf, G.; Sleilaty, G. Is Machine Learning-derived Low-Density Lipoprotein Cholesterol estimation more reliable than standard closed form equations? Insights from a laboratory database by comparison with a direct homogeneous assay. Clin. Chim. Acta 2021, 519, 220–226. [Google Scholar] [CrossRef]
- Ghayad, J.P.; Barakett-Hamade, V.; Sleilaty, G. Prospective Validation of a Machine Learning Model for Low-Density Lipoprotein Cholesterol Estimation. Lab. Med. 2022, 53, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Cubukcu, H.C.; Topcu, D.I. Estimation of Low-Density Lipoprotein Cholesterol Concentration Using Machine Learning. Lab. Med. 2022, 53, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Oh, G.C.; Ko, T.; Kim, J.H.; Lee, M.H.; Choi, S.W.; Bae, Y.S.; Kim, K.H.; Lee, H.Y. Estimation of low-density lipoprotein cholesterol levels using machine learning. Int. J. Cardiol. 2022, 352, 144–149. [Google Scholar] [CrossRef]
- Tsigalou, C.; Panopoulou, M.; Papadopoulos, C.; Karvelas, A.; Tsairidis, D.; Anagnostopoulos, K. Estimation of low-density lipoprotein cholesterol by machine learning methods. Clin. Chim. Acta 2021, 517, 108–116. [Google Scholar] [CrossRef]
- Kwon, Y.J.; Lee, H.; Baik, S.J.; Chang, H.J.; Lee, J.W. Comparison of a Machine Learning Method and Various Equations for Estimating Low-Density Lipoprotein Cholesterol in Korean Populations. Front. Cardiovasc. Med. 2022, 9, 824574. [Google Scholar] [CrossRef]
- Hidekazu, I.; Nagasawa, H.; Yamamoto, Y.; Doi, H.; Saito, M.; Ishihara, Y.; Fujita, T.; Ishida, M.; Kato, Y.; Kikuchi, R.; et al. Dataset dependency of low-density lipoprotein-cholesterol estimation by machine learning. Ann. Clin. Biochem. 2023, 60, 396–405. [Google Scholar] [CrossRef]
- Worledge, E.A.; Billups, S.J.; Titus, O.J.; Saseen, J.J. Achievement of low-density lipoprotein cholesterol thresholds in very high-risk atherosclerotic cardiovascular disease. J. Clin. Lipidol. 2025. [Google Scholar] [CrossRef]
- Bytyci, I.; Penson, P.E.; Mikhailidis, D.P.; Wong, N.D.; Hernandez, A.V.; Sahebkar, A.; Thompson, P.D.; Mazidi, M.; Rysz, J.; Pella, D.; et al. Prevalence of statin intolerance: A meta-analysis. Eur. Heart J. 2022, 43, 3213–3223. [Google Scholar] [CrossRef]
- Masi, D.; Zilich, R.; Candido, R.; Giancaterini, A.; Guaita, G.; Muselli, M.; Ponzani, P.; Santin, P.; Verda, D.; Musacchio, N.; et al. Uncovering Predictors of Lipid Goal Attainment in Type 2 Diabetes Outpatients Using Logic Learning Machine: Insights from the AMD Annals and AMD Artificial Intelligence Study Group. J. Clin. Med. 2023, 12, 4095. [Google Scholar] [CrossRef]
- Han, J.; Kim, Y.; Kang, H.J.; Seo, J.; Choi, H.; Kim, M.; Kee, G.; Park, S.; Ko, S.; Jung, H.; et al. Predicting low density lipoprotein cholesterol target attainment using machine learning in patients with coronary artery disease receiving moderate-dose statin therapy. Sci. Rep. 2025, 15, 5346. [Google Scholar] [CrossRef]
- Sarraju, A.; Zammit, A.; Ngo, S.; Witting, C.; Hernandez-Boussard, T.; Rodriguez, F. Identifying Reasons for Statin Nonuse in Patients With Diabetes Using Deep Learning of Electronic Health Records. J. Am. Heart Assoc. 2023, 12, e028120. [Google Scholar] [CrossRef] [PubMed]
- Sim, I.; Gorman, P.; Greenes, R.A.; Haynes, R.B.; Kaplan, B.; Lehmann, H.; Tang, P.C. Clinical decision support systems for the practice of evidence-based medicine. J. Am. Med. Inform. Assoc. 2001, 8, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Taheri Moghadam, S.; Sadoughi, F.; Velayati, F.; Ehsanzadeh, S.J.; Poursharif, S. The effects of clinical decision support system for prescribing medication on patient outcomes and physician practice performance: A systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2021, 21, 98. [Google Scholar] [CrossRef] [PubMed]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. npj Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef]
- Bozyel, S.; Simsek, E.; Kocyigit Burunkaya, D.; Guler, A.; Korkmaz, Y.; Seker, M.; Erturk, M.; Keser, N. Artificial Intelligence-Based Clinical Decision Support Systems in Cardiovascular Diseases. Anatol. J. Cardiol. 2024, 28, 74–86. [Google Scholar] [CrossRef]
- Kawamoto, K.; Houlihan, C.A.; Balas, E.A.; Lobach, D.F. Improving clinical practice using clinical decision support systems: A systematic review of trials to identify features critical to success. BMJ 2005, 330, 765. [Google Scholar] [CrossRef]
- Njie, G.J.; Proia, K.K.; Thota, A.B.; Finnie, R.K.C.; Hopkins, D.P.; Banks, S.M.; Callahan, D.B.; Pronk, N.P.; Rask, K.J.; Lackland, D.T.; et al. Clinical Decision Support Systems and Prevention: A Community Guide Cardiovascular Disease Systematic Review. Am. J. Prev. Med. 2015, 49, 784–795. [Google Scholar] [CrossRef]
- Kwan, J.L.; Lo, L.; Ferguson, J.; Goldberg, H.; Diaz-Martinez, J.P.; Tomlinson, G.; Grimshaw, J.M.; Shojania, K.G. Computerised clinical decision support systems and absolute improvements in care: Meta-analysis of controlled clinical trials. BMJ 2020, 370, m3216. [Google Scholar] [CrossRef]
- Zamora, A.; Fernandez de Bobadilla, F.; Carrion, C.; Vazquez, G.; Paluzie, G.; Elosua, R.; Vilaseca, M.; Martin-Urda, A.; Rivera, A.; Plana, N.; et al. Pilot study to validate a computer-based clinical decision support system for dyslipidemia treatment (HTE-DLP). Atherosclerosis 2013, 231, 401–404. [Google Scholar] [CrossRef]
- Benimetskaya, K.; Mikheenko, I.; Ponomarenko, A.; Krivosheev, Y.; Ponomarev, D.; Losik, D. A new clinical decision support system based on personalized evidence-based medicine in lipid lowering therapies. Atherosclerosis 2022, 355, 215–216. [Google Scholar] [CrossRef]
- Persson Lindell, O.; Karlsson, L.O.; Nilsson, S.; Charitakis, E.; Hagstrom, E.; Muhr, T.; Nilsson, L.; Henriksson, M.; Janzon, M. Clinical decision support for familial hypercholesterolemia (CDS-FH): Rationale and design of a cluster randomized trial in primary care. Am. Heart J. 2022, 247, 132–148. [Google Scholar] [CrossRef] [PubMed]
- Goren, A.; Santos, H.C.; Davis, T.W.; Lowe, R.B.; Monfette, M.; Meyer, M.N.; Chabris, C.F. Comparison of Clinical Decision Support Tools to Improve Pediatric Lipid Screening. J. Pediatr. 2024, 269, 113973. [Google Scholar] [CrossRef] [PubMed]
- Samadbeik, M.; Engstrom, T.; Lobo, E.H.; Kostner, K.; Austin, J.A.; Pole, J.D.; Sullivan, C. Healthcare dashboard technologies and data visualization for lipid management: A scoping review. BMC Med. Inform. Decis. Mak. 2024, 24, 352. [Google Scholar] [CrossRef] [PubMed]
- Buzancic, I.; Koh, H.J.W.; Trin, C.; Nash, C.; Ortner Hadziabdic, M.; Belec, D.; Zoungas, S.; Zomer, E.; Dalli, L.; Ademi, Z.; et al. Do clinical decision support tools improve quality of care outcomes in the primary prevention of cardiovascular disease: A systematic review and meta-analysis. Am. J. Prev. Cardiol. 2024, 20, 100855. [Google Scholar] [CrossRef]
- Groenhof, T.K.J.; Asselbergs, F.W.; Groenwold, R.H.H.; Grobbee, D.E.; Visseren, F.L.J.; Bots, M.L.; UCC-SMART Study Group. The effect of computerized decision support systems on cardiovascular risk factors: A systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2019, 19, 108. [Google Scholar] [CrossRef]
- Adusumalli, S.; Westover, J.E.; Jacoby, D.S.; Small, D.S.; VanZandbergen, C.; Chen, J.; Cavella, A.M.; Pepe, R.; Rareshide, C.A.L.; Snider, C.K.; et al. Effect of Passive Choice and Active Choice Interventions in the Electronic Health Record to Cardiologists on Statin Prescribing: A Cluster Randomized Clinical Trial. JAMA Cardiol. 2021, 6, 40–48. [Google Scholar] [CrossRef]
- Kunst, N.; Burger, E.A.; Coupe, V.M.H.; Kuntz, K.M.; Aas, E. A Guide to an Iterative Approach to Model-Based Decision Making in Health and Medicine: An Iterative Decision-Making Framework. Pharmacoeconomics 2024, 42, 363–371. [Google Scholar] [CrossRef]
- Escobar, C.; Facila, L.; Vidal-Pérez, R.; Pinedo Lapeña, A.; Vivas, D.; García Martín, A.; Manzano Fernández, S.; Gonzalez Caballero, E.; Barrios, V.; Freixa-Pamias, R. Artificial intelligence: A promising tool for the clinical cardiologist. Expert. Rev. Cardiovasc. Ther. 2025, 18, 1–15. [Google Scholar] [CrossRef]
- Hassan, M.; Kushniruk, A.; Borycki, E. Barriers to and Facilitators of Artificial Intelligence Adoption in Health Care: Scoping Review. JMIR Hum. Factors 2024, 11, e48633. [Google Scholar] [CrossRef]
- Mihan, A.; Pandey, A.; Van Spall, H.G. Mitigating the risk of artificial intelligence bias in cardiovascular care. Lancet Digit. Health 2024, 6, e749–e754. [Google Scholar] [CrossRef]
- Chen, F.; Wang, L.; Hong, J.; Jiang, J.; Zhou, L. Unmasking bias in artificial intelligence: A systematic review of bias detection and mitigation strategies in electronic health record-based models. J. Am. Med. Inform. Assoc. 2024, 31, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Khera, R.; Oikonomou, E.K.; Nadkarni, G.N.; Morley, J.R.; Wiens, J.; Butte, A.J.; Topol, E.J. Transforming Cardiovascular Care With Artificial Intelligence: From Discovery to Practice: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2024, 84, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Solanki, P.; Grundy, J.; Hussain, W. Operationalising ethics in artificial intelligence for healthcare: A framework for AI developers. AI Ethics 2023, 3, 223–240. [Google Scholar] [CrossRef]
- Hadhri, S.; Zargar, F.N.; Naeem, M.A.; Chibani, F. Bridging sustainable finance, AI, and clean technology amid economic shocks: How are they connected in median and extreme conditions? J. Environ. Manag. 2025, 391, 126375. [Google Scholar] [CrossRef]
- Rogers, W.A.; Draper, H.; Carter, S.M. Evaluation of artificial intelligence clinical applications: Detailed case analyses show value of healthcare ethics approach in identifying patient care issues. Bioethics 2021, 35, 623–633. [Google Scholar] [CrossRef]
- Hanna, M.G.; Pantanowitz, L.; Jackson, B.; Palmer, O.; Visweswaran, S.; Pantanowitz, J.; Deebajah, M.; Rashidi, H.H. Ethical and Bias Considerations in Artificial Intelligence/Machine Learning. Mod. Pathol. 2025, 38, 100686. [Google Scholar] [CrossRef]
- Arnold, M.H. Teasing out Artificial Intelligence in Medicine: An Ethical Critique of Artificial Intelligence and Machine Learning in Medicine. J. Bioeth. Inq. 2021, 18, 121–139. [Google Scholar] [CrossRef]
- Kostick-Quenet, K.; Lang, B.H.; Smith, J.; Hurley, M.; Blumenthal-Barby, J. Trust criteria for artificial intelligence in health: Normative and epistemic considerations. J. Med. Ethics 2024, 50, 544–551. [Google Scholar] [CrossRef]
- Marques, M.; Almeida, A.; Pereira, H. The Medicine Revolution Through Artificial Intelligence: Ethical Challenges of Machine Learning Algorithms in Decision-Making. Cureus 2024, 16, e69405. [Google Scholar] [CrossRef]
- Danilov, A.; Aronow, W.S. Artificial Intelligence in Cardiology: Applications and Obstacles. Curr. Probl. Cardiol. 2023, 48, 101750. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Artificial Intelligence-Enabled Device Software Functions: Lifecycle Management and Marketing Submission Recommendations; U.S. Department of Health and Human Services, Food and Drug Administration: Silver Spring, MD, USA, 2025. [Google Scholar]
- European Medicines Agency. Multi-Annual Artificial Intelligence Workplan 2023–2028: HMA-EMA Joint Big Data Steering Group; European Medicines Agency: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Moy, S.; Irannejad, M.; Manning, S.J.; Farahani, M.; Ahmed, Y.; Gao, E.; Prabhune, R.; Lorenz, S.; Mirza, R.; Klinger, C. Patient Perspectives on the Use of Artificial Intelligence in Health Care: A Scoping Review. J. Patient Cent. Res. Rev. 2024, 11, 51–62. [Google Scholar] [CrossRef] [PubMed]

| Year | CV Deaths/Year | Variation 2025–2050 | ||
|---|---|---|---|---|
| 1990 | 12.1 million | CV prevalence |  | 90.0% |
| Age-standardized cardiovascular prevalence |  | 3.6% | ||
| 2019 | 18.6 million | Total CV deaths |  | 73.4% |
| 2050 | 35.6 million | Age-standardized CV mortality rates |  | 30.5% |
| Integrating AI into Clinical Lipidology |
|---|
| Automatic laboratory alerts when values fall outside guideline-based thresholds. |
| Integrated tools combining LDLc, total cholesterol, ApoB, non-HDL, VLDLc, remnant particles, Lp(a), and glucose-triglycerides index to suggest optimal treatment strategies. |
| AI-assisted drug discovery through protein structure prediction |
| Improved models for predicting cardiovascular events in diverse populations |
| Advanced statistical modelling in clinical trials to sample size and enable automatic analysis. |
| Enhanced prediction of familial hypercholesterolemia and deeper insight into inheritance patterns, including non-Mendelian traits. |
| Risk scores for primary prevention that incorporate broader clinical and laboratory data to guide early intervention or further diagnostic evaluation. |
| Personalized dietary and physical activity recommendations for patients. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar-Cervantes, C.; Saldaña-García, J.; Torremocha-López, A.; Contreras-Lorenzo, C.; Lara-García, A.; Canales-Muñoz, L.; Martínez-González, R.; Vila-García, J.; Banach, M. Integrating New Technologies in Lipidology: A Comprehensive Review. J. Clin. Med. 2025, 14, 4984. https://doi.org/10.3390/jcm14144984
Escobar-Cervantes C, Saldaña-García J, Torremocha-López A, Contreras-Lorenzo C, Lara-García A, Canales-Muñoz L, Martínez-González R, Vila-García J, Banach M. Integrating New Technologies in Lipidology: A Comprehensive Review. Journal of Clinical Medicine. 2025; 14(14):4984. https://doi.org/10.3390/jcm14144984
Chicago/Turabian StyleEscobar-Cervantes, Carlos, Jesús Saldaña-García, Ana Torremocha-López, Cristina Contreras-Lorenzo, Alejandro Lara-García, Lucía Canales-Muñoz, Ricardo Martínez-González, Joaquín Vila-García, and Maciej Banach. 2025. "Integrating New Technologies in Lipidology: A Comprehensive Review" Journal of Clinical Medicine 14, no. 14: 4984. https://doi.org/10.3390/jcm14144984
APA StyleEscobar-Cervantes, C., Saldaña-García, J., Torremocha-López, A., Contreras-Lorenzo, C., Lara-García, A., Canales-Muñoz, L., Martínez-González, R., Vila-García, J., & Banach, M. (2025). Integrating New Technologies in Lipidology: A Comprehensive Review. Journal of Clinical Medicine, 14(14), 4984. https://doi.org/10.3390/jcm14144984








