Complete, Fully Automatic Detection and Classification of Benign and Malignant Breast Tumors Based on CT Images Using Artificial Intelligent and Image Processing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Breast Tumors
2.2. Research Samples and Sample Acceptance Conditions
2.3. Image Pre-Processing
2.4. Morphology
2.5. Contour Extraction Description
2.5.1. ACWE
2.5.2. GAC
2.5.3. Shi-Tomasi Corner Detection
2.6. Centroid Difference
2.7. Greedy Search Algorithm-Sequence forward Selection [40]
2.8. Support Vector Machine (SVM)
2.9. The k-Fold Cross-Validation (KCV)
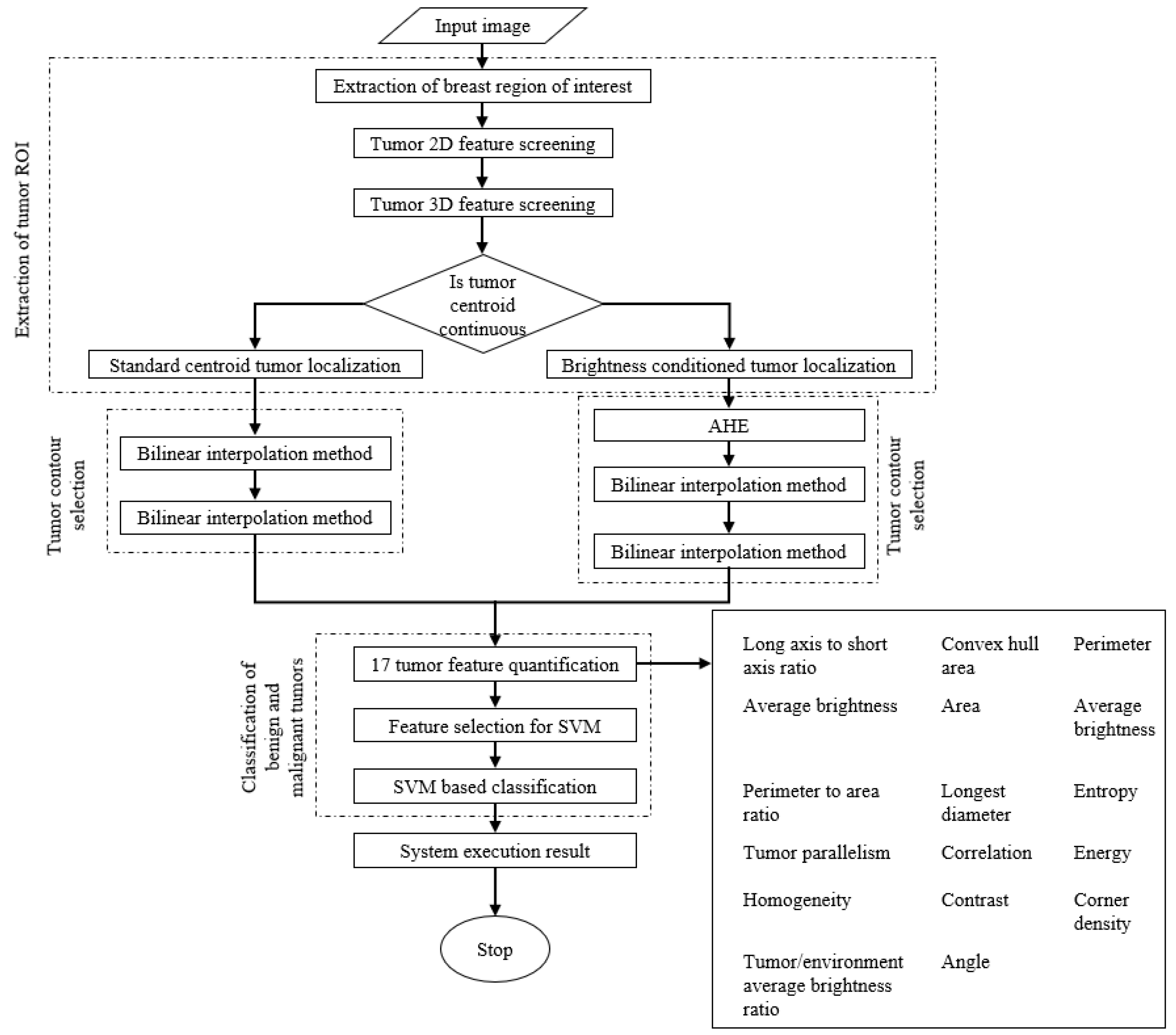
3. Results
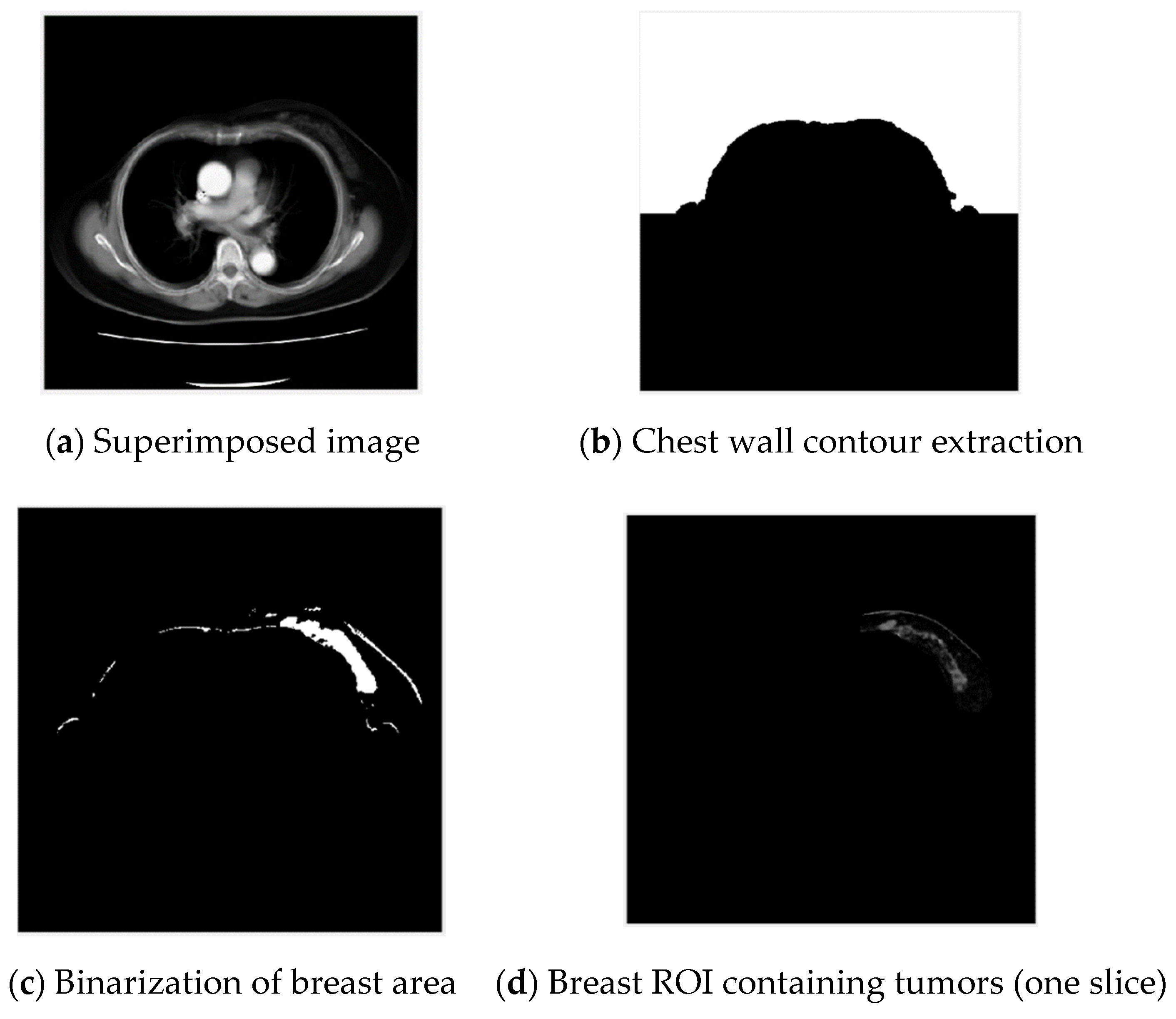
3.1. Extraction of Breast Region of Interest
3.1.1. Chest Wall Contour Extraction
3.1.2. Breast Area Distribution Analysis
3.2. Extraction of Tumor Region of Interest
Centroid Approximation-Standard Centroid Tumor Localization
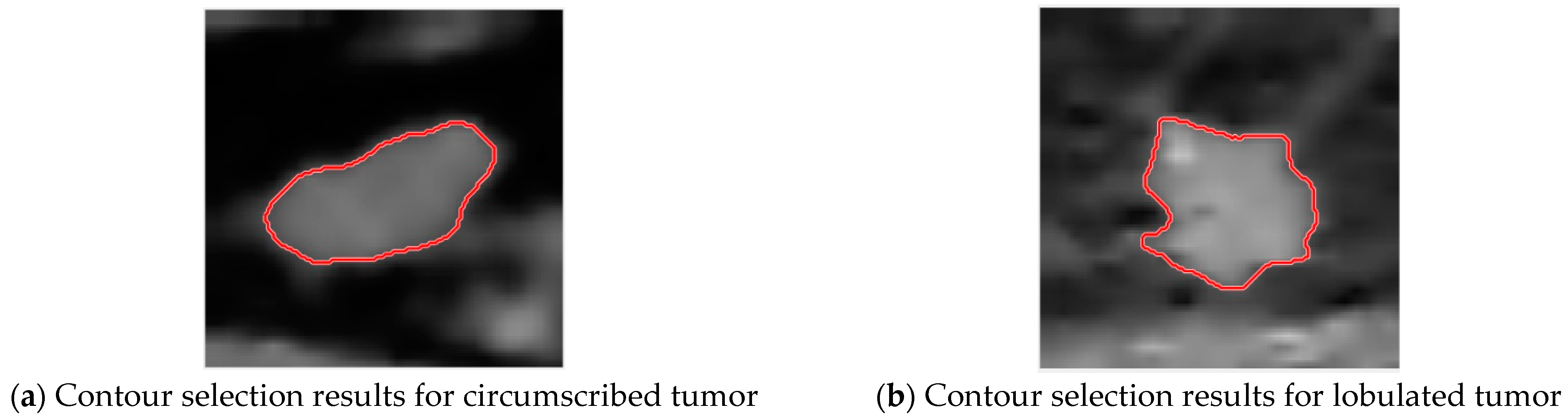
3.3. Tumor Contour Selection
3.3.1. Stable Centroid—Breast Tumor Circle Selection
3.3.2. Excessive Centroid Point Movement—Breast Tumor Circle Selection
3.4. Tumor Feature quAntification
3.5. Classification of Benign and Malignant Tumors
3.5.1. Feature Selection
3.5.2. System Effectiveness Evaluation
3.5.3. Selection of Best SVM Predictive Classifier Model
3.6. System Execution Result Evaluation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harbeck, N. Breast cancer is a systemic disease optimally treated by a multidisciplinary team. Nat. Rev. Dis. Prim. 2020, 6, 30. [Google Scholar] [CrossRef] [Green Version]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 69, 438–451. [Google Scholar] [CrossRef]
- Glick, S.J. Breast ct. Annu. Rev. Biomed. Eng. 2007, 9, 501–526. [Google Scholar] [CrossRef]
- Boone, J.M.; Lindfors, K.K. Breast CT: Potential for breast cancer screening and diagnosis. Future Oncul. 2006, 2, 351–356. [Google Scholar] [CrossRef]
- Schreier, J.; Attanasi, F.; Laaksonen, H. A full-image deep segmenter for CT images in breast cancer radiotherapy treatment. Front. Oncol. 2019, 9, 677. [Google Scholar] [CrossRef] [Green Version]
- Sreenivasu, S.V.N.; Gomathi, S.; Kumar, M.J.; Prathap, L.; Madduri, A.; Almutairi, K.; Alonazi, W.B.; Kali, D.; Jayadhas, S.A. Dense Convolutional Neural Network for Detection of Cancer from CT Images. BioMed Res. Int. 2022, 2022, 1293548. [Google Scholar] [CrossRef]
- Kocher, M.R.; Chamberlin, J.; Waltz, J.; Snoddy, M.; Stringer, N.; Stephenson, J.; Kahn, J.; Mercer, M.; Baruah, D.; Aquino, G.; et al. Tumor burden of lung metastases at initial staging in breast cancer patients detected by artificial intelligence as a prognostic tool for precision medicine. Heliyon 2022, 8, e08962. [Google Scholar] [CrossRef]
- Koh, J.; Yoon, Y.; Kim, S.; Han, K.; Kim, E.K. Deep learning for the detection of breast cancers on chest computed tomography. Clin. Breast Cancer 2022, 22, 26–31. [Google Scholar] [CrossRef]
- Dar, R.A.; Rasool, M.; Assad, A. Breast cancer detection using deep learning: Datasets, methods, and challenges ahead. Comput. Biol. Med. 2022, 149, 106073. [Google Scholar]
- Tang, J.; Rangayyan, R.M.; Xu, J.; El Naqa, I.; Yang, Y. Computer-aided detection and diagnosis of breast cancer with mammography: Recent advances. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 236–251. [Google Scholar] [CrossRef]
- Jalalian, A.; Mashohor, S.; Mahmud, R.; Karasfi, B.; Saripan, M.I.B.; Ramli, A.R.B. Foundation and methodologies in computer-aided diagnosis systems for breast cancer detection. EXCLI J. 2017, 16, 113–137. [Google Scholar]
- Lou, S.L.; Lin, H.D.; Lin, K.P.; Hoogstrate, D. Automatic breast region extraction from digital mammograms for PACS and telemammography applications. Comput. Med. Imaging Graph. 2000, 24, 205–220. [Google Scholar] [CrossRef]
- Keller, B.; Nathan, D.; Wang, Y.; Zheng, Y.; Gee, J.; Conant, E.; Kontos, D. Adaptive multi-cluster fuzzy C-means segmentation of breast parenchymal tissue in digital mammography. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Toronto, ON, Canada, 18–22 September 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 562–569. [Google Scholar] [CrossRef] [Green Version]
- Iyer, N.S.; Kandel, A.; Schneider, M. Feature-based fuzzy classification for interpretation of mammograms. Fuzzy Sets Syst. 2000, 114, 271–280. [Google Scholar] [CrossRef]
- Ertaş, G.; Gülçür, H.Ö.; Osman, O.; Uçan, O.N.; Tunacı, M.; Dursun, M. Breast MR segmentation and lesion detection with cellular neural networks and 3D template matching. Comput. Biol. Med. 2008, 38, 116–126. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern Syst. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L. A novel segmentation method for breast cancer ultrasound CAD system. In Proceedings of the 2011 International Conference on Informatics, Cybernetics, and Computer Engineering (ICCE2011), Melbourne, Australia, 19–20 November 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 307–313. [Google Scholar] [CrossRef]
- Poonguzhali, S.; Ravindran, G. A complete automatic region growing method for segmentation of masses on ultrasound images. In Proceedings of the 2006 IEEE International Conference on Biomedical and Pharmaceutical Engineering, Singapore, 11–14 December 2006; pp. 88–92. [Google Scholar]
- Osher, S.; Sethian, J. Fronts propagating with curvature dependent speed: Algorithms based on the Hamilton-Jacobi formulation. J. Comput. Phys. 1995, 118, 269–277. [Google Scholar] [CrossRef] [Green Version]
- Caselles, V.; Kimmel, R.; Sapiro, G. Geodesic active contours. Int. J. Comput. Vis. 1997, 22, 61–79. [Google Scholar] [CrossRef]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Rangayyan, R.M.; Ayres, F.J.; Desautels, J.L. Desautels A review of computer-aided diagnosis of breast cancer: Toward the detection of subtle signs. J. Frankl. Inst. 2007, 344, 312–348. [Google Scholar] [CrossRef]
- Hadjiiski, L.; Sahiner, B.; Chan, H.P. Advances in CAD for diagnosis of breast cancer. Curr. Opin. Obstet. Gynecol. 2006, 18, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Gómez, W.; Pereira, W.C.A.; Infantosi, A.F.C. Analysis of co-occurrence texture statistics as a function of gray-level quantization for classifying breast ultrasound. IEEE Trans. Med. Imaging 2012, 31, 1889–1899. [Google Scholar] [CrossRef]
- Yang, M.C.; Moon, W.K.; Wang, Y.C.F.; Bae, M.S.; Huang, C.S.; Chen, J.H.; Chang, R.F. Robust texture analysis using multi-resolution gray-scale invariant features for breast sonographic tumor diagnosis. IEEE Trans. Med. Imaging 2013, 32, 2262–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.J.; Lin, S.W.; Moon, W.K. Combining support vector machine with genetic algorithm to classify ultrasound breast tumor images. Comput. Med. Imaging Graph. 2012, 36, 627–633. [Google Scholar] [CrossRef]
- Sadoughi, F.; Kazemy, Z.; Hamedan, F.; Owji, L.; Rahmanikatigari, M.; Azadboni, T.T. Artificial intelligence methods for the diagnosis of breast cancer by image processing: A review. Breast Cancer Targets Ther. 2018, 10, 219–230. [Google Scholar] [CrossRef] [Green Version]
- D’Orsi, C.J.; Sickles, E.A.; Mendelson, E.B.; Morris, E.A. Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.P.; Hsu, H.H.; Ko, K.H.; Chu, C.M.; Chou, Y.C.; Chang, W.C.; Chang, T.H. Differentiation of malignant and benign incidental breast lesions detected by chest multidetector-row computed tomography: Added value of quantitative enhancement analysis. PLoS ONE 2016, 11, e0154569. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.C.; Hsu, H.H.; Li, C.S.; Yu, J.C.; Hsu, G.C.; Yu, C.P.; Chang, T.H.; Huang, G.S. Incidentally detected enhancing breast lesions on chest computed tomography. Korean J. Radiol. 2011, 12, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Zuiderveld, K. Contrast limited adaptive histogram equalization. Graph. Gems 1994, 474–485. Available online: https://cir.nii.ac.jp/crid/1571698601099987968 (accessed on 23 October 2022).
- Haralick, R.M.; Sternberg, S.R.; Zhuang, X. Image analysis using mathematical morphology. IEEE Trans. Pattern Anal. Mach. Intell. 1987, 4, 532–550. [Google Scholar] [CrossRef]
- Chang, F.; Chen, C.J. A component-labeling algorithm using contour tracing technique. In Proceedings of the Seventh International Conference on Document Analysis and Recognition, Edinburgh, UK, 3–6 August 2003; Volume 3, p. 741. [Google Scholar]
- Kass, M.; Witkin, A.; Terzopoulos, D. Terzopoulos. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Mumford, D.; Shah, J. Optimal approximation by piecewise smooth functions and associated variational problems. Commun. Pure Appl. Math. 1989, 42, 577–685. [Google Scholar] [CrossRef] [Green Version]
- Li, B.N.; Chui, C.K.; Chang, S.; Ong, S.H. Integrating spatial fuzzy clustering with level set methods for automated medical image segmentation. Comput. Biol. Med. 2011, 41, 1–10. [Google Scholar] [CrossRef]
- Li, C.; Kao, C.Y.; Gore, J.C.; Ding, Z. Minimization of region-scalable fitting energy for image segmentation. IEEE Trans. Image Process. 2008, 17, 1940–1949. [Google Scholar] [CrossRef] [Green Version]
- Harris, C.; Stephens, M. A combined corner and edge detector. In Proceedings of the Alvey Vision Conference, Manchester, UK, 31 August–2 September 1988; Volume 15, pp. 10–5244. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.434.4816&rep=rep1&type=pdf (accessed on 23 October 2022).
- Chickering, D.M. Optimal structure identification with greedy search. J. Mach. Learn. Res. 2002, 3, 507–554. [Google Scholar]
- Kuo, C.F.J.; Lo, W.C.; Huang, Y.R.; Tsai, H.Y.; Lee, C.L.; Wu, H.C. Automated defect inspection system for CMOS image sensor with micro multi-layer non-spherical lens module. J. Manuf. Syst. 2017, 45, 248–259. [Google Scholar] [CrossRef]
- Rodriguez, J.D.; Perez, A.; Lozano, J.A. Sensitivity analysis of k-fold cross validation in prediction error estimation. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 569–575. [Google Scholar] [CrossRef]
- Ambroise, C.; McLachlan, G.J. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl. Acad. Sci. USA 2002, 99, 6562–6566. [Google Scholar] [CrossRef] [Green Version]
- Kuo, C.F.J.; Ke, B.H.; Wu, N.Y.; Kuo, J.; Hsu, H.H. Prognostic value of tumor volume for patients with advanced lung cancer treated with chemotherapy. Comput. Methods Programs Biomed. 2017, 144, 165–177. [Google Scholar] [CrossRef]
- Kuo, C.F.J.; Lin, K.H.; Weng, W.H.; Barman, J.; Huang, C.C.; Chiu, C.W.; Lee, J.L.; Hsu, H.H. Complete fully automatic segmentation and 3-dimensional measurement of mediastinal lymph nodes for a new response evaluation criteria for solid tumors. Biocybern. Biomed. Eng. 2021, 41, 617–635. [Google Scholar] [CrossRef]
- Kuo, C.F.J.; Barman, J.; Hsieh, C.W.; Hsu, H.H. Fast fully automatic detection, classification and 3D reconstruction of pulmonary nodules in CT images by local image feature analysis. Biomed. Signal Process. Control 2021, 68, 102790. [Google Scholar] [CrossRef]
- Hussain, A.; Gordon-Dixon, A.; Almusawy, H.; Sinha, P.; Desai, A. The incidence and outcome of incidental breast lesions detected by computed tomography. Ann. R. Coll. Surg. Engl. 2010, 92, 124–126. [Google Scholar] [CrossRef] [Green Version]
- Bach, A.G.; Abbas, J.; Jasaabuu, C.; Schramm, D.; Wienke, A.; Surov, A. Comparison between incidental malignant and benign breast lesions detected by computed tomography: A systematic review. J. Med. Imaging Radiat. Oncol. 2013, 57, 529–533. [Google Scholar] [CrossRef]
- Poyraz, N.; Emlik, G.D.; Keskin, S.; Kalkan, H. Incidental breast lesions detected on computed thorax tomography. J. Breast Health 2015, 11, 163–167. [Google Scholar] [CrossRef] [Green Version]
- Kuo, H.C.; Giger, M.L.; Reiser, I.; Boone, J.M.; Lindfors, K.K.; Yang, K.; Edwards, A. Level set segmentation of breast masses in contrast-enhanced dedicated breast CT and evaluation of stopping criteria. J. Digit. Imaging 2014, 27, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Caballo, M.; Boone, J.M.; Mann, R.; Sechopoulos, I. An unsupervised automatic segmentation algorithm for breast tissue classification of dedicated breast computed tomography images. Med. Phys. 2018, 45, 2542–2559. [Google Scholar] [CrossRef] [Green Version]
- Wei, M.; Du, Y.; Wu, X.; Zhu, J. Automatic Classification of Benign and Malignant Breast Tumors in Ultrasound Image with Texture and Morphological Features. In Proceedings of the 2019 IEEE 13th International Conference on Anti-Counterfeiting, Security, and Identification (ASID), Xiamen, China, 25–27 October 2019; pp. 126–130. [Google Scholar] [CrossRef]
- Vijayarajeswari, R.; Parthasarathy, P.; Vivekanandan, S.; Basha, A.A. Classification of mammogram for early detection of breast cancer using SVM classifier and Hough transform. Measurement 2019, 146, 800–805. [Google Scholar] [CrossRef]
- AL-Dabagh, M.Z.; AL-Mukhtar, F.H. Breast Cancer Diagnostic System Based on MR images Using KPCA-Wavelet Transform and Support Vector Machine. Int. J. Adv. Eng. Res. Sci. 2017, 4, 237106. [Google Scholar] [CrossRef]
- Fujioka, T.; Kubota, K.; Mori, M.; Kikuchi, Y.; Katsuta, L.; Kasahara, M.; Oda, G.; Ishiba, T.; Nakagawa, T.; Tateishi, U. Distinction between benign and malignant breast masses at breast ultrasound using deep learning method with convolutional neural network. Jpn. J. Radiol. 2019, 37, 466–472. [Google Scholar] [CrossRef]










| Lesion Types | Types | Number | Total Number |
|---|---|---|---|
| Benign | Fibroadenoma | 69 | 89 |
| Fibrocystic change | 5 | ||
| Cyst | 5 | ||
| Breast abscess | 2 | ||
| Intraductal papilloma | 3 | ||
| Fibrotic lesion | 2 | ||
| Phyllodes tumor | 2 | ||
| Intramammary lymph node | 1 | ||
| Malignant | Invasive ductal carcinoma | 48 | 85 |
| Invasive lobular carcinoma | 10 | ||
| Mixed ductal and lobular carcinoma | 9 | ||
| Ductal carcinoma in situ | 13 | ||
| Colloid carcinoma | 5 | ||
| Molecular subtypes of malignant lesion | Total Number | Percentage (%) | |
| Luminal A | 28 | 33 | |
| Luminal B | 33 | 39 | |
| Triple negative | 12 | 14 | |
| HER2+ | 9 | 11 | |
| Unknown | 3 | 3 | |
| Lesion size | Total Number | ||
| <1 cm | 32 | ||
| >1 cm | 142 | ||
| Tumor Feature No. | AUC Value |
|---|---|
| 1. Average brightness | 0.8927 |
| 2. Convex hull area | 0.7764 |
| 3. Perimeter | 0.75 |
| 4. Average brightness | 0.7293 |
| 5. Area | 0.7252 |
| 6. Long axis to short axis ratio | 0.6906 |
| 7. Perimeter to area ratio | 0.6875 |
| 8. Longest diameter | 0.6843 |
| 9. Texture (entropy) | 0.679 |
| 10. Texture (contrast) | 0.6537 |
| 11. Tumor texture (correlation) | 0.6502 |
| 12. Texture (energy) | 0.6499 |
| 13. Texture (homogeneity) | 0.5916 |
| 14. Tumor parallelism | 0.5866 |
| 15. Corner density | 0.5759 |
| 16. Tumor/environment average brightness ratio | 0.5687 |
| 17. Angle | 0.5192 |
| Prediction | Detected as Benign Tumor (Negatives) | Detected as Malignant Tumor (Positives) | |
|---|---|---|---|
| Ground Truth | |||
| Benign tumor (Negatives) | True Negatives (TN) | False Positives (FP) | |
| Malignant tumor (Positives) | False Negatives (FN) | True Positives (TP) | |
| AUC Sort Feature Subset | Number of Selected Features | Classification Accuracy |
|---|---|---|
| model_1 | 1 | 0.7471 |
| model_2 | 2 | 0.9425 |
| model_3 | 3 | 0.954 |
| model_4 | 4 | 0.9655 |
| model_5 | 5 | 0.9655 |
| model_6 | 6 | 0.954 |
| model_7 | 7 | 0.9597 |
| model_8 | 8 | 0.9655 |
| model_9 | 9 | 0.9713 |
| model_10 | 10 | 0.9828 |
| model_11 | 11 | 0.977 |
| model_12 | 12 | 0.977 |
| model_13 | 13 | 0.9713 |
| model_14 | 14 | 0.9885 |
| model_15 | 15 | 0.9943 |
| model_16 | 16 | 0.9943 |
| model_17 | 17 | 0.9943 |
| Accuracy | AUC | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|---|
| 0.9943 | 0.9941 | 0.9882 | 1 | 1 | 0.9889 |
| Application | System Approach | Result | |
|---|---|---|---|
| This research | Machine learning in chest CT | Image processing, ACWE, GAC, and SVM |
|
| Wei [52] (2019) | Machine learning in Breast Ultrasound | Morphological features and SVM |
|
| Vijayarajeswari [53] (2019) | Machine learning in Mammography | Hough transform and SVM. |
|
| AL-Dabagh [54] (2017) | Machine learning in Breast MRI | Traditional image processing and SVM |
|
| Fujioka [55] (2019) | Deep learning in Breast Ultrasound | CNN |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, C.-F.J.; Chen, H.-Y.; Barman, J.; Ko, K.-H.; Hsu, H.-H. Complete, Fully Automatic Detection and Classification of Benign and Malignant Breast Tumors Based on CT Images Using Artificial Intelligent and Image Processing. J. Clin. Med. 2023, 12, 1582. https://doi.org/10.3390/jcm12041582
Kuo C-FJ, Chen H-Y, Barman J, Ko K-H, Hsu H-H. Complete, Fully Automatic Detection and Classification of Benign and Malignant Breast Tumors Based on CT Images Using Artificial Intelligent and Image Processing. Journal of Clinical Medicine. 2023; 12(4):1582. https://doi.org/10.3390/jcm12041582
Chicago/Turabian StyleKuo, Chung-Feng Jeffrey, Hsuan-Yu Chen, Jagadish Barman, Kai-Hsiung Ko, and Hsian-He Hsu. 2023. "Complete, Fully Automatic Detection and Classification of Benign and Malignant Breast Tumors Based on CT Images Using Artificial Intelligent and Image Processing" Journal of Clinical Medicine 12, no. 4: 1582. https://doi.org/10.3390/jcm12041582
APA StyleKuo, C.-F. J., Chen, H.-Y., Barman, J., Ko, K.-H., & Hsu, H.-H. (2023). Complete, Fully Automatic Detection and Classification of Benign and Malignant Breast Tumors Based on CT Images Using Artificial Intelligent and Image Processing. Journal of Clinical Medicine, 12(4), 1582. https://doi.org/10.3390/jcm12041582










