RNA-Seq Reveals the mRNAs, miRNAs, and lncRNAs Expression Profile of Knee Joint Synovial Tissue in Osteoarthritis Patients
Abstract
1. Introduction
2. Methods and Materials
2.1. Patients and Samples
2.2. RNA Library Construction and Sequencing
2.3. Bioinformatics Analysis
2.4. Analysis of Differential Expressed miRNAs (DEmiRNAs) and Target Genes Prediction
2.5. LncRNA Transcript Assembly
2.6. LncRNA Identification, Analysis of DE mRNAs and lncRNAs, and Prediction of Target Genes
3. Results
3.1. Clinical and Biochemical Features of the Included Individuals
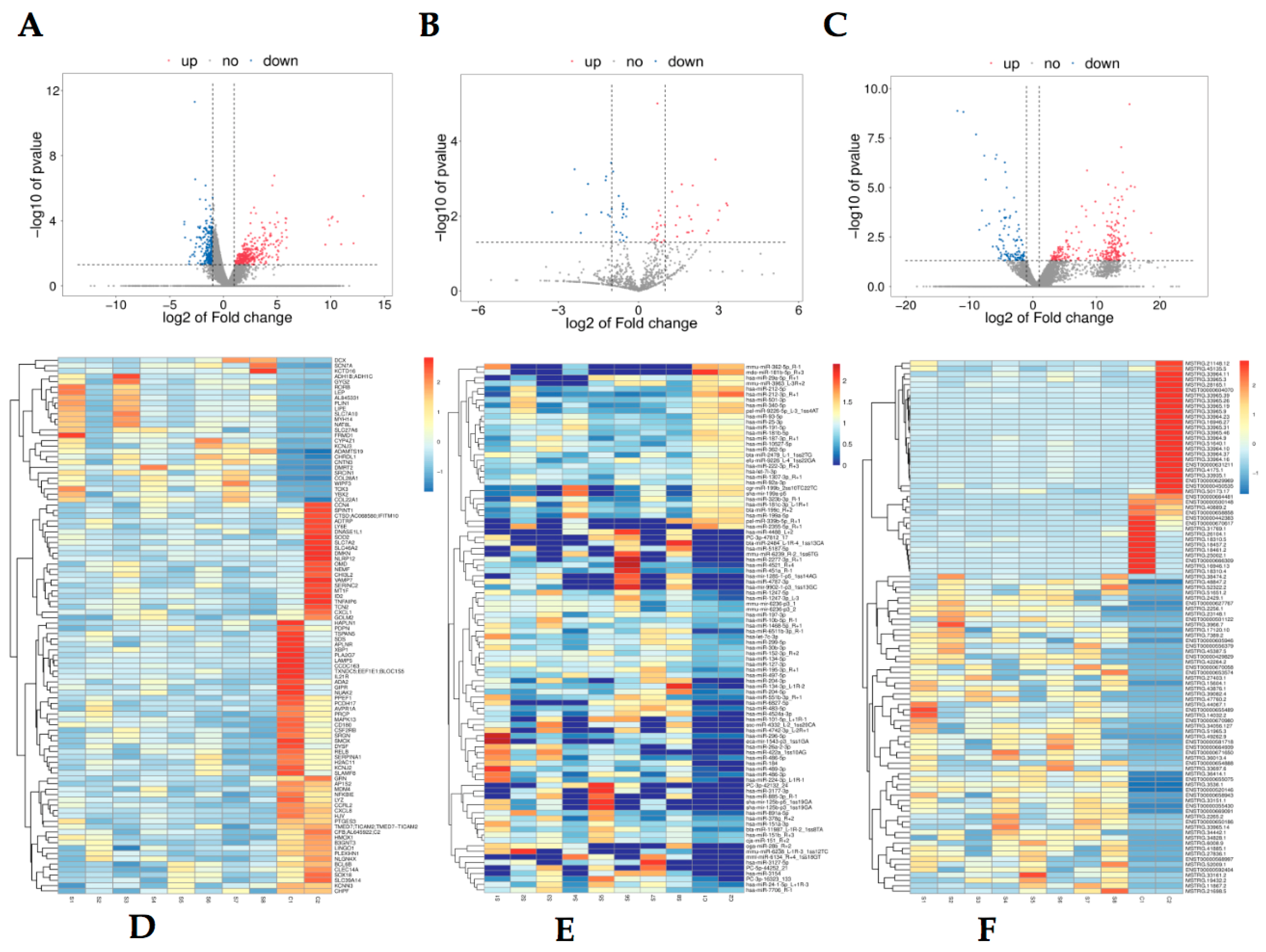
3.2. Differentially Expressed mRNA, miRNA and LncRNA Profile
3.3. GO and KEGG Enrichment Analysis of lncRNAs/miRNA Target mRNAs
3.4. Prediction of the Interaction between Differentially Expressed lncRNA-mRNA
3.5. Expression Validation of OA-Related Overlapped DEMs, lncRNAs, and mRNAs in Synovial Tissue
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACR/EULAR | American College of Rheumatol/European League Against Rheumatism |
| ALDH1A2 | aldehyde dehydrogenase 1A2 |
| ceRNAs | competing endogenous RNAs |
| CHST11 | Carbohydrate (chondroitin 4) sulfotransferase 11 |
| DE | differentially expressed |
| DEGs | differentially expressed genes |
| DEMs | differentially expressed miRNAs |
| GalNAc | N-acetylgalactosamine |
| GO | Gene Ontology |
| lncRNAs | long non-coding RNAs |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ncRNAs | non-coding RNAs |
| OA | Osteoarthritis |
| TLR4 | Toll-like receptor 4 |
| TREM1 | Triggering Receptor Expressed on Monocytes 1 |
References
- Arden, N.; Nevitt, M.C. Osteoarthritis: Epidemiology. Best Pract. Res. Clin. Rheumatol. 2006, 20, 3–25. [Google Scholar] [CrossRef]
- Cross, M.; Smith, E.; Hoy, D.; Nolte, S.; Ackerman, I.; Fransen, M.; Bridgett, L.; Williams, S.; Guillemin, F.; Hill, C.; et al. The global burden of hip and knee osteoarthritis: Estimates from the global burden of disease 2010 study. Ann. Rheum. Dis. 2014, 73, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Mahjoub, M.; Berenbaum, F.; Houard, X. Why subchondral bone in osteoarthritis? The importance of the cartilage bone Interface in osteoarthritis. Osteoporos. Int. 2012, 23 (Suppl. 8), 841–846. [Google Scholar] [CrossRef]
- Vos, T.; Flaxman, A.; Naghavi, M.; Lozano, R.; Michaud, C.; Ezzati, M. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the global burden of disease study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.; Worthington, S.; Felson, D.; Jurmain, R.; Wren, K.; Maijanen, H.; Woods, R.; Lieberman, D. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc. Natl. Acad. Sci. USA 2017, 114, 9332–9336. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Kriegsmann, J.; Hopf, T.; Jacobs, D.; Arens, N.; Krenn, V.; Schmitt-Wiedhoff, R.; Kriegsmann, M.; Heisel, C.; Biehl, C.; Thabe, H.; et al. Applications of molecular pathology in the diagnosis of joint infections. Der Orthopade 2009, 38, 531–538. [Google Scholar] [CrossRef]
- Murata, K.; Yoshitomi, H.; Tanida, S.; Ishikawa, M.; Nishitani, K.; Ito, H.; Nakamura, T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 2010, 12, R86. [Google Scholar] [CrossRef]
- Reynard, L.N.; Loughlin, J. Genetics and epigenetics of osteoarthritis. Maturitas 2012, 71, 200–204. [Google Scholar] [CrossRef]
- Ponting, C.; Oliver, P.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Harries, L.W. Long non-coding RNAs and human disease. Biochem. Soc. Trans. 2012, 40, 902–906. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Shan, Y.; Pan, Y.; Ma, J.; Jia, L. Long non-coding RNA-HOTAIR promotes osteoarthritis progression via Mir-17-5p/Fut2/Β-catenin Axis. Cell Death Dis. 2018, 9, 711. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, X.; Dai, L.; Hu, X.; Zhu, J.; Li, L.; Zhou, C.; Ao, Y. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014, 66, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Alevizos, I.; Illei, G. MicroRNAs as biomarkers in rheumatic diseases. Nat. Rev. Rheumatol. 2010, 6, 391–398. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; Mac Menamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ma, J.; Yao, J. Molecular mechanisms of the cartilage-specific microRNA-140 in osteoarthritis. Inflamm. Res. 2013, 62, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Katoh, S.; Yoshioka, H.; Senthilkumar, R.; Preethy, S.; Abraham, S. Enhanced miRNA-140 expression of osteoarthritis-affected human chondrocytes cultured in a polymer based three-dimensional (3D) matrix. Life Sci. 2021, 278, 119553. [Google Scholar] [CrossRef]
- Si, H.-B.; Zeng, Y.; Liu, S.-Y.; Zhou, Z.-K.; Chen, Y.-N.; Cheng, J.-Q.; Lu, Y.-R.; Shen., B. Intra-articular injection of microRNA-140 (miRNA-140) alleviates osteoarthritis (OA) progression by modulating extracellular matrix (ECM) homeostasis in rats. Osteoarthr. Cartil. 2017, 25, 1698–1707. [Google Scholar] [CrossRef]
- Liang, Y.; Xu, X.; Li, X.; Xiong, J.; Li, B.; Duan, L.; Wang, D.; Xia, J. Chondrocyte-Targeted MicroRNA Delivery by Engineered Exosomes toward a Cell-Free Osteoarthritis Therapy. ACS Appl. Mater. Interfaces 2020, 12, 36938–36947. [Google Scholar] [CrossRef]
- Shen, S.; Wu, Y.; Chen, J.; Xie, Z.; Huang, K.; Wang, G.; Yang, Y.; Ni, W.; Chen, Z.; Shi, P.; et al. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 2019, 78, 826–836. [Google Scholar] [CrossRef]
- Zhou, Z.-B.; Huang, G.-X.; Fu, Q.; Han, B.; Lu, J.-J.; Chen, A.-M.; Zhu, L. circRNA.33186 Contributes to the Pathogenesis of Osteoarthritis by Sponging miR-127-5p. Mol. Ther. 2019, 27, 531–541. [Google Scholar] [CrossRef]
- Hall, M.; Doherty, S.; Courtney, P.; Latief, K.; Zhang, W.; Doherty, M. Synovial pathology detected on ultrasound correlates with the severity of radiographic knee osteoarthritis more than with symptoms. Osteoarthr. Cartil. 2014, 22, 1627–1633. [Google Scholar] [CrossRef] [PubMed]
- Felson, D.T. Epidemiology of hip and knee osteoarthritis. Epidemiol. Rev. 1988, 10, 1–28. [Google Scholar] [CrossRef]
- March, L.M.; Bachmeier, C.J. Economics of osteoarthritis: A global perspective. Baillieres Clin. Rheumatol. 1997, 11, 817–834. [Google Scholar] [CrossRef]
- Rai, M.F.; Sandell, L.J. Inflammatory mediators: Tracing links between obesity and osteoarthritis. Crit. Rev. Eukaryot. Gene Expr. 2011, 21, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Mobasheri, A. Osteoarthritis year 2012 in review: Biomarkers. Osteoarthr. Cartil. 2012, 20, 1451–1464. [Google Scholar] [CrossRef]
- Lowe, C.M.; Barker, K.; Dewey, M.; Sackley, C. Effectiveness of physiotherapy exercise after knee arthroplasty for osteoarthritis: Systematic review and meta-analysis of randomised controlled trials. BMJ 2007, 335, 812. [Google Scholar] [CrossRef]
- Yau, M.; Yerges-Armstrong, L.; Liu, Y.; Lewis, C.; Duggan, D.; Renner, J.; Torner, J.; Felson, D.; McCulloch, C.; Kwoh, K.; et al. Genome-Wide Association Study of Radiographic Knee Osteoarthritis in North American Caucasians. Arthritis Rheumatol. 2017, 69, 343–351. [Google Scholar] [CrossRef]
- Bowles, J.; Knight, D.; Smith, C.; Wilhelm, D.; Richman, J.; Mamiya, S.; Yashiro, K.; Chawengsaksophak, K.; Wilson, M.; Rossant, J.; et al. Retinoid signaling determines germ cell fate in mice. Science 2006, 312, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Styrkarsdottir, U.; Thorleifsson, G.; Helgadottir, H.T.; Bomer, N.; Metrustry, S.; Bierma-Zeinstra, S. Severe osteoarthritis of the hand associates with common variants within theALDH1A2 gene and with rare variants at 1p31. Nat. Genet. 2014, 46, 498–502. [Google Scholar] [CrossRef]
- Kuai, J.; Gregory, B.; Hill, A.; Pittman, D.; Feldman, J.; Brown, T.; Carito, B.; O’Toole, M.; Ramsey, R.; Adolfsson, O.; et al. TREM-1 expression is increased in the synovium of rheumatoid arthritis patients and induces the expression of pro-inflammatory cytokines. Rheumatology 2009, 48, 1352–1358. [Google Scholar] [CrossRef]
- Lambert, C.; Dubuc, J.-E.; Montell, E.; Vergés, J.; Munaut, C.; Noël, A.; Henrotin, Y. Gene expression pattern of cells from inflamed and normal areas of osteoarthritis synovial membrane. Arthritis Rheumatol. 2014, 66, 960–968. [Google Scholar] [CrossRef]
- Sato, T.; Konomi, K.; Yamasaki, S.; Aratani, S.; Tsuchimochi, K.; Yokouchi, M.; Masuko-Hongo, K.; Yagishita, N.; Nakamura, H.; Komiya, S.; et al. Comparative analysis of gene expression profiles in intact and damaged regions of human osteoarthritic cartilage. Arthritis Rheumatol. 2006, 54, 808–817. [Google Scholar] [CrossRef]
- Rogers, E.L.; Reynard, L.N.; Loughlin, J. The role of inflammation-related genes in osteoarthritis. Osteoarthr. Cartil. 2015, 23, 1933–1938. [Google Scholar] [CrossRef] [PubMed]
- Saas, J.; Haag, J.; Rueger, D.; Chubinskaya, S.; Sohler, F.; Zimmer, R.; Bartnik, E.; Aigner, T. IL-1b, but not BMP-7 leads to a dramatic change in the gene expression pattern of human adult articular chondrocytes portraying the gene expression pattern in twodonors. Cytokine 2006, 36, 90–99. [Google Scholar] [CrossRef]
- Karlsson, C.; Dehne, T.; Lindahl, A.; Brittberg, M.; Pruss, A.; Sittinger, M.; Ringe, J. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthr. Cartil. 2010, 18, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Skrzypa, M.; Szala, D.; Gablo, N.; Czech, J.; Pajak, J.; Kopanska, M.; Trzeciak, M.; Gargasz, K.; Snela, S.; Zawlik, I. miRNA-146a-5p is upregulated in serum and cartilage samples of patients with osteoarthritis. Pol. Przegl. Chir. 2019, 91, 1–5. [Google Scholar] [CrossRef]
- Zhong, G.; Long, H.; Ma, S.; Shunhan, Y.; Li, J.; Yao, J. miRNA-335-5p relieves chondrocyte inflammation by activating autophagy in osteoarthritis. Life Sci. 2019, 226, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ahn, C.; Chun, C.-H.; Jin, E.-J. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J. Orthop. Res. 2014, 32, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Zhu, J.; Wang, B.; Wu, J.; Li, H.; Han, Y.; Xiang, D.; Chen, Y.; Li, L. LINC02288 promotes chondrocyte apoptosis and inflammation through miR-374a-3p targeting RTN3. J. Gene Med. 2021, 23, e3314. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-W.; Zhang, X.; Qian, G.-B.; Jiang, M.-J.; Wang, P.; Wang, K.-Z. Downregulation of long noncoding RNA LOC101928134 inhibits the synovial hyperplasia and cartilage destruction of osteoarthritis rats through the activation of the Janus kinase/signal transducers and activators of transcription signaling pathway by upregulating IFNA1. J. Cell Physiol. 2019, 234, 10523–10534. [Google Scholar]
- Xiao, P.; Zhu, X.; Sun, J.; Zhang, Y.; Qiu, W.; Li, J.; Wu, X. LncRNA NEAT1 regulates chondrocyte proliferation and apoptosis via targeting miR-543/PLA2G4A axis. Hum. Cell 2021, 34, 60–75. [Google Scholar] [CrossRef]
- Liu, F.; Liu, X.; Yang, Y.; Sun, Z.; Deng, S.; Jiang, Z.; Li, W.; Wu, F. NEAT1/miR-193a-3p/SOX5 axis regulates cartilage matrix degradation in human osteoarthritis. Cell Biol. Int. 2020, 44, 947–957. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, W.; Zhang, F.; Deng, Y.; Long, Z. NEAT1/miR-181c Regulates Osteopontin (OPN)-Mediated Synoviocyte Proliferation in Osteoarthritis. J. Cell. Biochem. 2017, 118, 3775–3784. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.-Y.; Yang, M.; Guo, J.; Zhang, C.; Lin, L.-L.; Liu, Y.; Wei, R.-X. Identification of the Biomarkers and Pathological Process of Osteoarthritis: Weighted Gene Co-expression Network Analysis. Front. Physiol. 2019, 10, 275. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.-P.; Chen, H.-M.; Hu, C.-C.; Chen, L.-Y.; Shih, F.-F.; Tantoh, D.M.; Lee, K.-J.; Liaw, Y.-C.; Tsai, R.-T.; Liaw, Y.-P. Interaction of Osteoarthritis and BMI on Leptin Promoter Methylation in Taiwanese Adults. Int. J. Mol. Sci. 2019, 21, 123. [Google Scholar] [CrossRef]
- Huang, Z.Y.; Luo, Z.Y.; Cai, Y.R.; Chou, C.H.; Yao, M.L.; Pei, F.X.; Kraus, V.B.; Zhou, Z.K. Single cell transcriptomics in human osteoarthritis synovium and in silico deconvoluted bulk RNA sequencing. Osteoarthr. Cartil. 2022, 30, 475–480. [Google Scholar] [CrossRef]
- Zhang, X.; Bu, Y.; Zhu, B.; Zhao, Q.; Lv, Z.; Li, B.; Liu, J. Global transcriptome analysis to identify critical genes involved in the pathology of osteoarthritis. Bone Joint Res. 2018, 7, 298–307. [Google Scholar] [CrossRef]
- Kang, L.; Dai, C.; Wang, L.; Pan, X. Potential biomarkers that discriminate rheumatoid arthritis and osteoarthritis based on the analysis and validation of datasets. BMC Musculoskelet. Disord. 2022, 23, 319. [Google Scholar] [CrossRef]



| Gender | H/W (cm)/(kg) | Age (Year) | Course (Month) | ESR (mm/h) | CRP (mg/L) | |
|---|---|---|---|---|---|---|
| OA1 | F | 155/51 | 69 | 120 | 14.3 | 4.7 |
| OA2 | F | 162/58 | 71 | 60 | 9.2 | <0.5 |
| OA3 | M | 169/71 | 75 | 96 | 3.5 | <0.5 |
| OA4 | F | 157/55 | 70 | 72 | 13.8 | 8.8 |
| OA5 | M | 162/64 | 68 | 96 | 19.7 | 5.1 |
| OA6 | M | 171/73 | 71 | 36 | 18.7 | 6.4 |
| OA7 | F | 163/62 | 73 | 48 | 24.1 | 1.3 |
| OA8 | F | 157/53 | 62 | 72 | 18.5 | 10.5 |
| cyst1 | F | 159/52 | 65 | 5 | 16.0 | 3.2 |
| cyst2 | M | 170/67 | 68 | 11 | 22.6 | 1.1 |
| Gene Name | log2 (Fold Change) | Regulation | RNA Sort |
|---|---|---|---|
| LAMP5 | −3.65 | Down | mRNA |
| HJV | −3.64 | Down | mRNA |
| CXCL8 | −2.67 | Down | mRNA |
| DMKN | −2.62 | Down | mRNA |
| PLA2G7 | −2.36 | Down | mRNA |
| NAT8L | 5.09 | Up | mRNA |
| ADH1B | 5.44 | Up | mRNA |
| CYP4Z1 | 5.78 | Up | mRNA |
| AL845331 | 5.78 | Up | mRNA |
| LEP | 5.83 | Up | mRNA |
| hsa-miR-129-5p | −4.75 | down | miRNA |
| hsa-miR-323b-3p_R-1 | −3.23 | down | miRNA |
| hsa-miR-212-3p_R+1 | −3.22 | down | miRNA |
| hsa-miR-130b-3p | −3.07 | down | miRNA |
| hsa-miR-21-3p_R+1 | −2.93 | down | miRNA |
| hsa-miR-34c-3p_1ss16GA | 4.21 | up | miRNA |
| hsa-miR-335-5p_R-1 | 4.30 | up | miRNA |
| hsa-miR-877-3p_R+1 | 4.47 | up | miRNA |
| hsa-miR-34c-5p | 5.01 | up | miRNA |
| hsa-miR-34b-3p_L-1R+1 | 6.98 | up | miRNA |
| ARRDC3-AS1 | −3.88 | Down | lncRNA |
| GUSBP11 | −3.88 | Down | lncRNA |
| MIR4435-2HG | −4.02 | Down | lncRNA |
| CKMT2-AS1 | −4.32 | Down | lncRNA |
| FP671120 | −4.39 | Down | lncRNA |
| NEAT1 | 7.72 | Up | lncRNA |
| AP000757 | 6.86 | Up | lncRNA |
| XIST | 6.84 | Up | lncRNA |
| AC060780 | 6.35 | Up | lncRNA |
| AC008691 | 5.45 | Up | lncRNA |
| mRNA | lncRNA | cis Location | p Value |
|---|---|---|---|
| SLX1A | SLX1A-SULT1A3 | 1 K | −0.70 |
| BTK | ARMCX4 | 100 K | −0.57 |
| SYNPO2 | SEC24D | 100 K | −0.50 |
| NBR1 | AC060780 | 10 K | −0.42 |
| REV3L | MFSD4B | 100 K | −0.35 |
| CSNK1A1 | CARMN | 100 K | −0.34 |
| ZFP62 | LINC00847 | 100 K | −0.31 |
| CRIP2 | MIR8071-1 | 100 K | −0.27 |
| FKBP15 | FAM225A | 100 K | −0.23 |
| DDIT3 | MBD6 | 100 K | −0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, L.; Gu, J.; Ni, Y.; Wu, J.; Zhang, D.; Gu, Y. RNA-Seq Reveals the mRNAs, miRNAs, and lncRNAs Expression Profile of Knee Joint Synovial Tissue in Osteoarthritis Patients. J. Clin. Med. 2023, 12, 1449. https://doi.org/10.3390/jcm12041449
Qiao L, Gu J, Ni Y, Wu J, Zhang D, Gu Y. RNA-Seq Reveals the mRNAs, miRNAs, and lncRNAs Expression Profile of Knee Joint Synovial Tissue in Osteoarthritis Patients. Journal of Clinical Medicine. 2023; 12(4):1449. https://doi.org/10.3390/jcm12041449
Chicago/Turabian StyleQiao, Linghui, Jun Gu, Yingjie Ni, Jianyue Wu, Dong Zhang, and Yanglin Gu. 2023. "RNA-Seq Reveals the mRNAs, miRNAs, and lncRNAs Expression Profile of Knee Joint Synovial Tissue in Osteoarthritis Patients" Journal of Clinical Medicine 12, no. 4: 1449. https://doi.org/10.3390/jcm12041449
APA StyleQiao, L., Gu, J., Ni, Y., Wu, J., Zhang, D., & Gu, Y. (2023). RNA-Seq Reveals the mRNAs, miRNAs, and lncRNAs Expression Profile of Knee Joint Synovial Tissue in Osteoarthritis Patients. Journal of Clinical Medicine, 12(4), 1449. https://doi.org/10.3390/jcm12041449




