Predicting Survival for Veno-Arterial ECMO Using Conditional Inference Trees—A Multicenter Study
Abstract
:1. Introduction
2. Methods
2.1. Study Design
2.2. Data Collection
2.3. Statistical Analysis
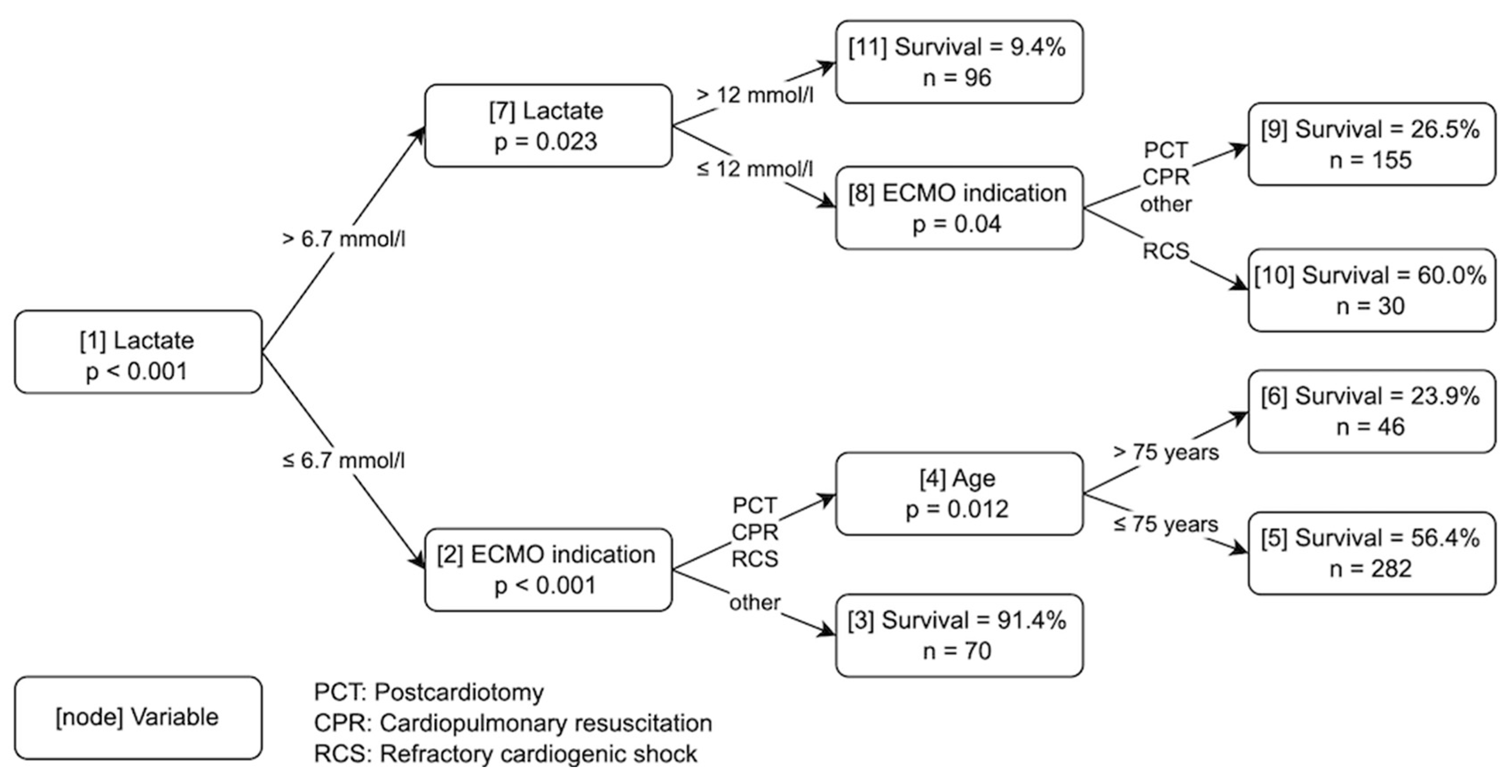
2.3.1. Model Development
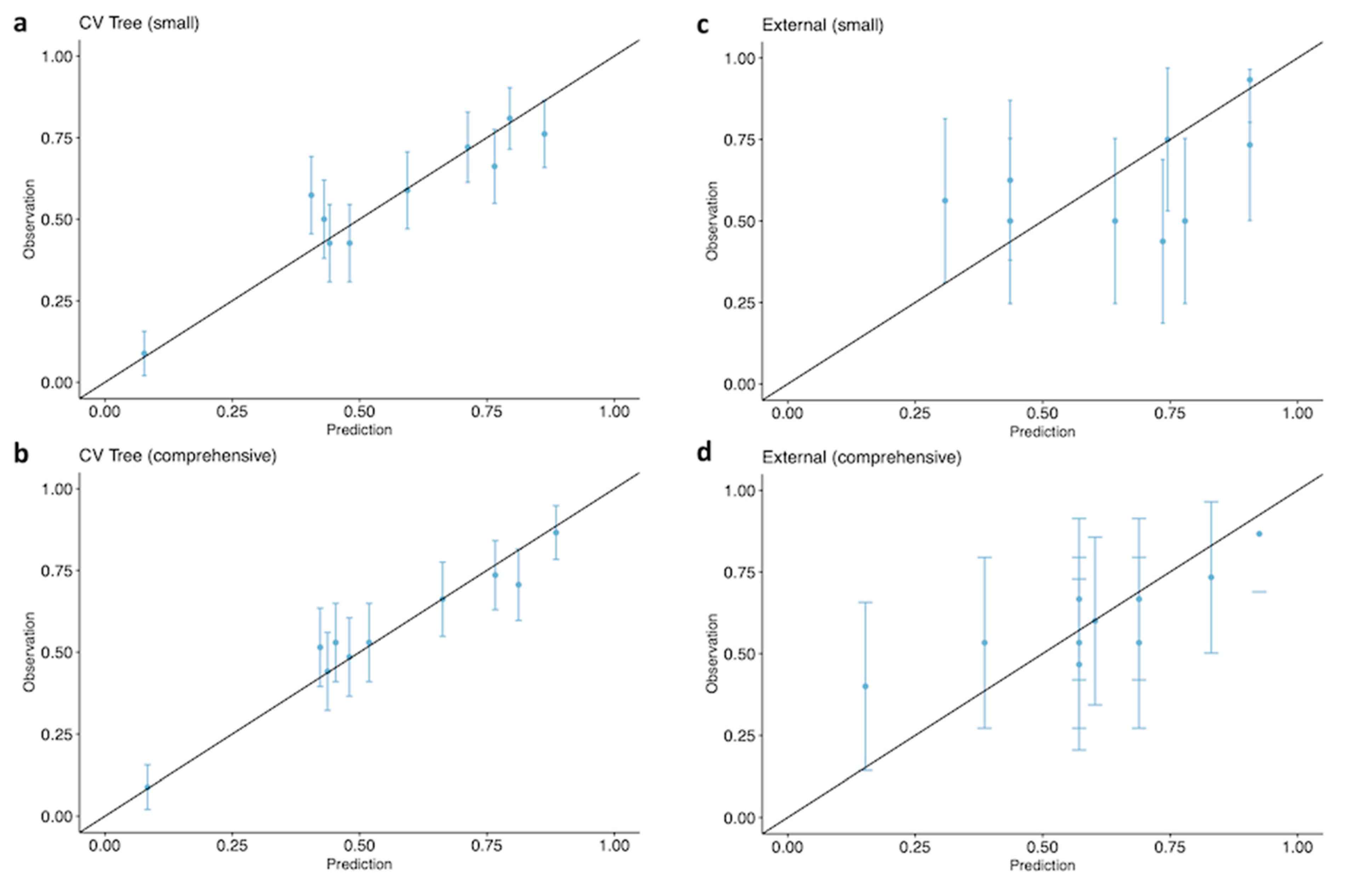
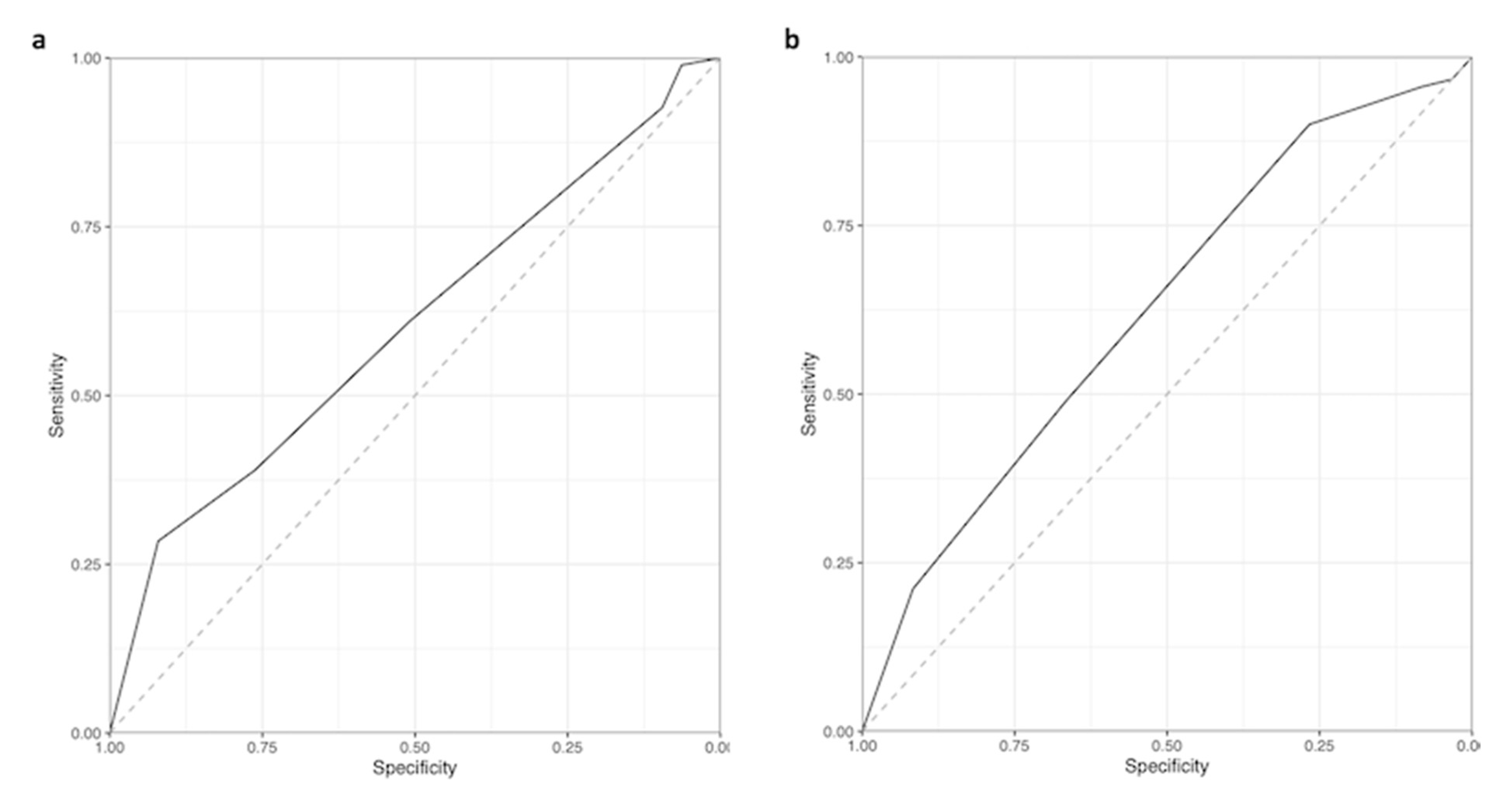
2.3.2. Model Validation
3. Results
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eckman, P.M.; Katz, J.N.; El Banayosy, A.; Bohula, E.A.; Sun, B.; van Diepen, S. Veno-Arterial Extracorporeal Membrane Oxygenation for Cardiogenic Shock: An Introduction for the Busy Clinician. Circulation 2019, 140, 2019–2037. [Google Scholar] [CrossRef] [PubMed]
- Pineton de Chambrun, M.; Bréchot, N.; Combes, A. Venoarterial extracorporeal membrane oxygenation in cardiogenic shock: Indications, mode of operation, and current evidence. Curr. Opin. Crit. Care 2019, 25, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.; Zhao, Y.; Strom, J.B.; Shen, C.; Yeh, R.W. Extracorporeal Membrane Oxygenation Use in Cardiogenic Shock: Impact of Age on In-Hospital Mortality, Length of Stay, and Costs. Crit. Care Med. 2019, 47, e214–e221. [Google Scholar] [CrossRef]
- Fernando, S.M.; Qureshi, D.; Tanuseputro, P.; Fan, E.; Munshi, L.; Rochwerg, B.; Talarico, R.; Scales, D.C.; Brodie, D.; Dhanani, S.; et al. Mortality and costs following extracorporeal membrane oxygenation in critically ill adults: A population-based cohort study. Intensive Care Med. 2019, 45, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Huang, K.Y.; Yao, C.W.; Wu, C.F.; Liang, S.J.; Li, C.H.; Tu, C.Y.; Chen, H.J. The modified SAVE score: Predicting survival using urgent veno-arterial extracorporeal membrane oxygenation within 24 hours of arrival at the emergency department. Crit. Care 2016, 20, 336. [Google Scholar] [CrossRef]
- Kowalewski, M.; Zieliński, K.; Maria Raffa, G.; Meani, P.; Lo Coco, V.; Jiritano, F.; Fina, D.; Matteucci, M.; Chiarini, G.; Willers, A.; et al. Mortality Predictors in Elderly Patients with Cardiogenic Shock on Venoarterial Extracorporeal Life Support. Analysis From the Extracorporeal Life Support Organization Registry. Crit. Care Med. 2021, 49, 7–18. [Google Scholar] [CrossRef]
- Lee, H.S.; Kim, H.S.; Lee, S.H.; Lee, S.A.; Hwang, J.J.; Park, J.B.; Kim, Y.H.; Moon, H.J.; Lee, W.S. Clinical implications of the initial SAPS II in veno-arterial extracorporeal oxygenation. J. Thorac. Dis. 2019, 11, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Muller, G.; Flecher, E.; Lebreton, G.; Luyt, C.E.; Trouillet, J.L.; Bréchot, N.; Schmidt, M.; Mastroianni, C.; Chastre, J.; Leprince, P.; et al. The ENCOURAGE mortality risk score and analysis of long-term outcomes after VA-ECMO for acute myocardial infarction with cardiogenic shock. Intensive Care Med. 2016, 42, 370–378. [Google Scholar] [CrossRef]
- Schmidt, M.; Burrell, A.; Roberts, L.; Bailey, M.; Sheldrake, J.; Rycus, P.T.; Hodgson, C.; Scheinkestel, C.; Cooper, D.J.; Thiagarajan, R.R.; et al. Predicting survival after ECMO for refractory cardiogenic shock: The survival after veno-arterial-ECMO (SAVE)-score. Eur. Heart J. 2015, 36, 2246–2256. [Google Scholar] [CrossRef]
- Wang, L.; Yang, F.; Wang, X.; Xie, H.; Fan, E.; Ogino, M.; Brodie, D.; Wang, H.; Hou, X. Predicting mortality in patients undergoing VA-ECMO after coronary artery bypass grafting: The REMEMBER score. Crit. Care 2019, 23, 11. [Google Scholar] [CrossRef]
- Wengenmayer, T.; Duerschmied, D.; Graf, E.; Chiabudini, M.; Benk, C.; Mühlschlegel, S.; Philipp, A.; Lubnow, M.; Bode, C.; Staudacher, D.L. Development and validation of a prognostic model for survival in patients treated with venoarterial extracorporeal membrane oxygenation: The PREDICT VA-ECMO score. Eur. Heart J. Acute Cardiovasc. Care 2019, 8, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Schrutka, L.; Rohmann, F.; Binder, C.; Haberl, T.; Dreyfuss, B.; Heinz, G.; Lang, I.M.; Felli, A.; Steinlechner, B.; Niessner, A.; et al. Discriminatory power of scoring systems for outcome prediction in patients with extracorporeal membrane oxygenation following cardiovascular surgery. Eur. J. Cardio-Thorac. Surg. 2019, 56, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, J.R.; Lemeshow, S.; Saulnier, F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. J. Am. Med. Assoc. 1993, 270, 2957–2963. [Google Scholar] [CrossRef]
- Abbasi, A.; Karasu, Y.; Li, C.; Sodha, N.R.; Eickhoff, C.; Ventetuolo, C.E. Machine learning to predict hemorrhage and thrombosis during extracorporeal membrane oxygenation. Crit. Care 2020, 24, 689. [Google Scholar] [CrossRef]
- Ayers, B.; Wood, K.; Gosev, I.; Prasad, S. Predicting Survival After Extracorporeal Membrane Oxygenation by Using Machine Learning. Ann. Thorac. Surg. 2020, 110, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Sahli, S.D.; Kaserer, A.; Braun, J.; Halbe, M.; Dahlem, Y.; Spahn, M.A.; Rössler, J.; Krüger, B.; Maisano, F.; Spahn, D.R.; et al. Predictors associated with mortality of extracorporeal life support therapy for acute heart failure: Single-center experience with 679 patients. J. Thorac. Dis. 2022, 14, 1960–1971. [Google Scholar] [CrossRef]
- Alba, A.C.; Foroutan, F.; Buchan, T.A.; Alvarez, J.; Kinsella, A.; Clark, K.; Zhu, A.; Lau, K.; McGuinty, C.; Aleksova, N.; et al. Mortality in patients with cardiogenic shock supported with VA ECMO: A systematic review and meta-analysis evaluating the impact of etiology on 29,289 patients. J. Heart Lung Transpl. 2021, 40, 260–268. [Google Scholar] [CrossRef]
- Guglin, M.; Zucker, M.J.; Bazan, V.M.; Bozkurt, B.; El Banayosy, A.; Estep, J.D.; Gurley, J.; Nelson, K.; Malyala, R.; Panjrath, G.S.; et al. Venoarterial ECMO for Adults: JACC Scientific Expert Panel. J. Am. Coll. Cardiol. 2019, 73, 698–716. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; Zeileis, A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J. Comput. Graph. Stat. 2006, 15, 651–674. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; van de Wiel, M.A.; Zeileis, A. A Lego System for Conditional Inference. Am. Stat. 2006, 60, 257–263. [Google Scholar] [CrossRef]
- Strobl, C.; Malley, J.; Tutz, G. An introduction to recursive partitioning: Rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol. Methods 2009, 14, 323–348. [Google Scholar] [CrossRef] [PubMed]
- Buri, M.; Tanadini, L.G.; Hothorn, T.; Curt, A. Unbiased recursive partitioning enables robust and reliable outcome prediction in acute spinal cord injury. J. Neurotrauma 2022, 39, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Tanadini, L.G.; Steeves, J.D.; Hothorn, T.; Abel, R.; Maier, D.; Schubert, M.; Weidner, N.; Rupp, R.; Curt, A. Identifying homogeneous subgroups in neurological disorders: Unbiased recursive partitioning in cervical complete spinal cord injury. Neurorehabilit. Neural Repair 2014, 28, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Hothorn, T.; Jung, H.H. RandomForest4Life: A random forest for predicting ALS disease progression. Amyotroph. Lateral Scler. Front. Degener. 2014, 15, 444–452. [Google Scholar] [CrossRef]
- Strobl, C.; Boulesteix, A.-L.; Zeileis, A.; Hothorn, T. Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinform. 2007, 8, 25. [Google Scholar] [CrossRef]
- Rufibach, K. Use of Brier score to assess binary predictions. J. Clin. Epidemiol. 2010, 63, 938–939. [Google Scholar] [CrossRef]
- Bates, S.; Hastie, T.; Tibshirani, R. Cross-validation: What does it estimate and how well does it do it? J. Am. Stat. Assoc. 2023, 1–12. [Google Scholar] [CrossRef]
- Bradley, A.A.; Schwartz, S.S.; Hashino, T. Sampling uncertainty and confidence intervals for the Brier score and Brier skill score. Weather. Forecast. 2008, 23, 992–1006. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]

| Training | |
|---|---|
| N = 679 | |
| Characteristics | |
| Age, y | 60 (49 to 69) |
| Sex, female | 187 (28%) |
| BMI, kg/m2 | 25.7 (22.9 to 29.0) [8] |
| Indication | |
| Postcardiotomy | 215 (31.7%) |
| Cardiopulmonary resuscitation | 159 (23.4%) |
| Refractory cardiogenic shock | 234 (34.5%) |
| Other | 71 (10.5%) |
| In-hospital mortality | 377 (56%) |
| Out-of-hospital resuscitation | 40 (5.9%) |
| In-hospital resuscitation | 119 (18%) |
| ROSC, min | 35 (13 to 60) |
| Comorbidities | |
| Charlson comorbidity index | 3.0 (2.0 to 5.0) [6] |
| Coronary artery disease | 326 (48%) |
| Congestive heart failure | 243 (36%) [1] |
| Peripheral vascular disease | 118 (17%) |
| Cerebrovascular event | 41 (6%) |
| Dementia | 0 (0%) |
| Obstructive pulmonary disease | 55 (8.1%) |
| Connective tissue disease | 58 (8.5%) |
| Peptic ulcer disease | 12 (1.8%) |
| Liver disease | 12 (1.8%) |
| Diabetes mellitus | 97 (14%) |
| Chronic kidney disease | 53 (7.8%) |
| Solid tumor | 50 (7.4%) |
| Leukemia | 10 (1.5%) |
| Lymphoma | 6 (0.9%) |
| AIDS | 0 (0%) |
| Blood Gas Analysis | |
| Hematocrit, mmol/L | 0.30 (0.25 to 0.38) [30] |
| Hemoglobin, g/L | 97 (82 to 124) [28] |
| Lactate, mmol/L | 5.6 (2.0 to 9.9) [83] |
| Base excess, mmol/L | −7.0 (−12.0 to −2.0) [85] |
| Bicarbonate, mmol/L | 18.4 (14.5 to 22.0) [85] |
| Carbon dioxide partial pressure, kPa | 5.11 (4.33 to 6.13) [84] |
| Oxygen partial pressure, kPa | 16 (10.0 to 30.0) [13] |
| Glucose, mmol/L | 8.8 (6.6 to 12.3) [82] |
| Laboratory Values | |
| eGFR, mL/min | 53 (37 to 74) [10] |
| Albumin, g/dL | 27 (20 to 33) [174] |
| Alkaline phosphatase, U/I | 69 (49 to 100) [196] |
| Wurzburg | Frankfurt | |
|---|---|---|
| N = 57 | N = 101 | |
| Age, y | 57 (49 to 65) | 70 (63 to 76) |
| Postcardiotomy | 2 (3.5%) | 60 (59.4%) |
| Cardiopulmonary resuscitation | 31 (54.4%) | 21 (20.8%) |
| Refractory cardiogenic shock | 18 (31.6%) | 16 (15.8%) |
| Other | 6 (10.5%) | 4 (4.0%) |
| In-hospital mortality | 39 (68%) | 56 (55%) |
| Lactate, mmol/L | 9.3 (5.3 to 14.2) | 6.0 (3.2 to 8.9) |
| eGFR, mL/min | 50 (32 to 66) [2] | 43 (32 to 60) [2] |
| Albumin, g/dL | 25 (21 to 32) [1] | 19 (15 to 24) [2] |
| Alkaline phosphatase, U/I | 64 (45 to 118) [3] | 42 (30 to 68) [5] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Braun, J.; Sahli, S.D.; Spahn, D.R.; Röder, D.; Neb, H.; Lotz, G.; Aser, R.; Wilhelm, M.J.; Kaserer, A. Predicting Survival for Veno-Arterial ECMO Using Conditional Inference Trees—A Multicenter Study. J. Clin. Med. 2023, 12, 6243. https://doi.org/10.3390/jcm12196243
Braun J, Sahli SD, Spahn DR, Röder D, Neb H, Lotz G, Aser R, Wilhelm MJ, Kaserer A. Predicting Survival for Veno-Arterial ECMO Using Conditional Inference Trees—A Multicenter Study. Journal of Clinical Medicine. 2023; 12(19):6243. https://doi.org/10.3390/jcm12196243
Chicago/Turabian StyleBraun, Julia, Sebastian D. Sahli, Donat R. Spahn, Daniel Röder, Holger Neb, Gösta Lotz, Raed Aser, Markus J. Wilhelm, and Alexander Kaserer. 2023. "Predicting Survival for Veno-Arterial ECMO Using Conditional Inference Trees—A Multicenter Study" Journal of Clinical Medicine 12, no. 19: 6243. https://doi.org/10.3390/jcm12196243
APA StyleBraun, J., Sahli, S. D., Spahn, D. R., Röder, D., Neb, H., Lotz, G., Aser, R., Wilhelm, M. J., & Kaserer, A. (2023). Predicting Survival for Veno-Arterial ECMO Using Conditional Inference Trees—A Multicenter Study. Journal of Clinical Medicine, 12(19), 6243. https://doi.org/10.3390/jcm12196243








