Abstract
Introduction: Chronic lymphocytic leukemia (CLL), the most common leukemia in Western countries, is a mature B-cell chronic lymphoproliferative disorder characterized by the accumulation of neoplastic CD5+ B lymphocytes, functionally incompetent and usually monoclonal in origin, in bone marrow, lymph nodes and blood. Diagnosis occurs predominantly in elderly patients, with a median age reported between 67 and 72 years. CLL has a heterogeneous clinical course, which can vary from indolent to, less frequently, aggressive forms. Early-stage asymptomatic CLL patients do not require immediate therapeutic intervention, but only observation; treatment is necessary for patients with advanced disease or when “active disease” is observed. The most frequent autoimmune cytopenia (AIC) is autoimmune haemolytic anaemia (AHIA). The main mechanisms underlying the appearance of AIC in CLL are not fully elucidated, the predisposition of patients with CLL to suffering autoimmune complications is variable and autoimmune cytopenia can precede, be concurrent, or follow the diagnosis of CLL. Case presentation: A 74-year-old man was admitted to the emergency room following the finding of severe macrocytic anaemia during blood tests performed that same day, in particular the patient showed a profound asthenia dating back several months. The anamnesis was silent and the patient was not taking any medications. The blood examination showed an extremely high White Blood Cell count and findings of AIHA in CLL-type mature B-cell lymphoproliferative neoplasia. Genetic investigations: Conventional karyotyping was performed and it obtained a trisomy 8 and an unbalanced translocation between the short arm of chromosome 6 and the long arm of chromosome 11, concurrent with interstitial deletions in chromosomes 6q and 11q that could not be defined in detail. Molecular cytogenetics (FISH) analyses revealed Ataxia Telangiectasia Mutated (ATM) monoallelic deletion (with loss of ATM on derivative chromosome 11) and retained signals for TP53, 13q14 and centromere 12 FISH probes. TP53 and IGHV were not mutated. Array-CGH confirmed trisomy of the entire chromosome 8 and allowed us to resolve in detail the nature of the unbalanced translocation, revealing multiple regions of genomic losses on chromosomes 6 and 11. Discussion: The present case report is an unusual CLL case with complex karyotype and refinement of all breakpoints at the gene level by the genomic array. From a genetic point of view, the case under study presented several peculiarities. Conclusions: We report the genetic findings of a CLL patient with abrupt disease onset, so far responding properly to treatments despite the presence of distinct genetic adverse traits including ATM deletion, complex karyotype and chromosome 6q chromoanagenesis event. Our report confirms that interphase FISH alone is not able to provide an overview of the whole genomic landscape in selected CLL cases and that additional techniques are required to reach an appropriate cytogenetic stratification of patients.
1. Introduction
Chronic lymphocytic leukemia (CLL), the most common leukemia in Western countries, is a mature B-cell chronic lymphoproliferative disorder characterized by the accumulation of neoplastic CD5+ B lymphocytes, functionally incompetent and usually monoclonal in origin, in bone marrow, lymph nodes and blood [,,].
Diagnosis occurs predominantly in elderly patients, with a median age reported between 67 and 72 years, where men are more frequently affected than women (ratio of 1.7:1) [].
CLL has a heterogeneous clinical course, which can vary from indolent to, less frequently, aggressive forms.
In contrast to other B-cell malignancies, immune dysregulation is a constant feature of CLL that is associated with a high prevalence of infections, autoimmune phenomena, and secondary malignancies [,,,,].
The most frequent autoimmune cytopenias (AICs) are autoimmune hemolytic anemia (AIHA, 7–10%) and immune thrombocytopenia (ITP, 1–5%), while pure red cell aplasia (<1%) and autoimmune neutropenia (0.17%) are unusual. AIHA can appear in up to a third of patients with CLL during the course of their disease, while in 10–15% of cases is already present at diagnosis [,,,,,,,].
The main mechanisms underlying the appearance of AIC in CLL are not fully elucidated and the predisposition of patients with CLL to suffering autoimmune complications is variable. For example, some studies have shown a higher incidence of AIC in patients with an advanced clinical stage, especially in those with spleen infiltration, suggesting that the spleen may act as the site of the presentation of antigens to the T cells. There are, however, some other studies in which no association was found between AIC and survival [,,,,,,,,,]. The reasons are unclear, but they could be explained with patients’ characteristics, better treatment strategies for CLL and expert management of AIC.
AIHA is defined according to autoantibody thermal characteristics [,,]. From a clinical point of view, AIHA is a heterogeneous condition caused by autoantibodies directed against the red blood cells (RBCs), with or without complement activation. The mechanisms for RBC destruction may be represented by intravascular or extravascular hemolysis.
Autoimmune cytopenia can precede, be concurrent, or follow the diagnosis of CLL. Non-active CLL and AIC should be treated according to current AIHA and ITP guidelines. However, active CLL-related AIC presents some challenges regarding its diagnosis, prognosis, and management. The diagnosis requires a high degree of suspicion as CLL-related AIC cannot manifest in their typical clinical and laboratory features; the prognosis depends on patient, disease and treatment intertwined factors. The possibility of an immune origin of cytopenia in the advanced stage should be always kept in mind [].
The diagnosis of AIHA requires the existence of anemia of abrupt onset or significant worsening of previous anemia, increased LDH with or without elevated bilirubin, low or undetectable haptoglobin, positive direct antiglobulin test (DAT) [,,]. The reticulocyte count is usually elevated, although it may be normal in some cases due to poor function of the heavily infiltrated bone marrow or treatment toxicity. The examination of a peripheral blood smear is mandatory because it usually presents spherocytosis and polychromasia [,,].
For the staging of the patient with CLL, a CT scan or an alternative imaging technique should be performed in order to evaluate the possible presence of massive or progressive splenomegaly and/or lymphadenopathies.
Treatment in CLL-associated AIHA is individualized and it depends on the presence of clinical symptoms (acuteness of the onset, grade of anemia and degree of hemolysis) and their severity, disease status and concomitant comorbidities. Anemia, if symptomatic, is an indication for therapy in both newly diagnosed and persistent AIHA [,,].
Typically elderly patients have a lower tolerance to anemia, the treatment of which is therefore frequently required. Furthermore, adverse drug reactions, as well as drug interactions and therapeutic toxicity, are more common in these patients.
RBC transfusions are generally indicated in critical cases with low hemoglobin levels (usually Hb < 6 g/dL) and/or symptomatic anemia, particularly if hemodynamically unstable.
Rituximab is a B-cell depleting monoclonal antibody with CD20 specificity that has demonstrated efficacy in many autoimmune diseases [,]. The combined therapy of rituximab and steroids, also administered in first-line treatment, shows a better response than steroid monotherapy [,,]. The addition of rituximab reduces the number of repeated steroid cycles and increases the frequency and duration of response. This is particularly useful in elderly patients with comorbidities. More recently, the targeting of Bruton’s tyrosine kinase (BTK) has markedly improved outcomes for CLL [].
Cytogenomic analyses are fundamental in the clinical management of hematological malignancies. The European recommendations [] and the IWCLL guidelines [] strongly suggest the analysis of multiple loci for the diagnosis and prognostic stratification of patients: deletions in 13q14, 11q22.3/ATM, 17p13.1/TP53, trisomy 12, TP53 mutation and IGHV mutational status. Although not mandatory, conventional cytogenetics using appropriate culture conditions identifies chromosomal abnormalities in the vast majority of cases []. A minority of cases present translocations spread across all chromosomes and chromosome bands, primarily as part of a complex karyotype [].
The present study reports a peculiar CLL case with complex karyotype and refinement of all breakpoints at the gene level by genomic array.
2. Case Presentation
A 74-year-old man was admitted to the emergency room following the finding of severe macrocytic anaemia, during blood tests performed that same day for a profound asthenia dating back several months. The anamnesis was silent and the patient was not taking any medications.
The blood examination with Automated Hematology Analyzer DASIT XN-Series showed the data reported in Table 1, whereas the analysis of the lymphocytes subpopulations and the immunophenotyping were performed respectively on AQUIOS CL and NAVIOS EX 3L 10C Beckman Coulter as in Table 2.

Table 1.
Blood examination, biochemical analysis and drugs administration evolution.

Table 2.
Comparison between the immunophenotyping on peripheral blood and bone marrow.
The microscopic examination of peripheral blood smears, prepared with the May Grünwald-Giemsa stain (obtained with automated hematology slide preparation unit SP-50; DASIT), revealed atypical lymphocytes, small mature lymphocytes with scant cytoplasm and condensed chromatin that can impart a cracked or “soccer ball” appearance, with the 6% of prolymphocytes, and Gumprecht shadows compatible with a CLL picture. The presence of numerous spherocytes and the finding of a marked polychromasia of the red blood cells correlated well with severe hemolytic anemia and with the consequent conspicuous increase in circulating reticulocytes.
All these findings have advanced the hypothesis of AIHA in lymphoproliferative neoplasia, particularly CLL.
In the patient’s staging a CT scan, an immunophenotyping on peripheral blood and a bone marrow aspiration and biopsy were carried out.
The neck-thorax-abdomen CT scan, performed with and without contrast medium, showed multiple enlarged lymph nodes with oval morphology in all the lymph node stations of the neck. No mediastinal lymphadenomegaly lymphadenomegaly was detected. Besides, bilateral axillary, retroperitoneal and inguinal pathological lymph node enlargements, as well as a homogeneous splenomegaly, were found.
The immunophenotyping showed the presence of a population of CD5+, CD19+, CD20+dim, and CD23+ B lymphocytes with clonal restriction for immunoglobulin lambda surface light-chains, which is proof of monoclonality and indicates malignancy. The immunophenotype was then consistent with CLL-type mature B-cell lymphoproliferative neoplasia (Table 2).
The bone marrow aspiration and the biopsy were performed on the right posterior superior iliac spine. The cellularity consisted, for about 20%, of an infiltrate of small lymphoid elements (Table 2) arranged in perilamellar and centrolacunar aggregates with an interstitial pattern of infiltration. Presence of plasma cells, isolated or in perisinusoidal microaggregates, without evidence of monocatenary restriction and equal to about 5% of bone marrow cellularity, was observed, as well as normo-represented, normo-maturating granulopoietic series. Erythropoietic series was overrepresented compared to granulopoietic series (myeloid series/erythroid series ratio about 1:2), with aspects of topographic dyserythropoiesis. Findings suggest medullary localization of chronic lymphatic leukemia.”
Given the severity of the hemolytic anemia and the incomplete response to corticosteroid therapy, according to ESMO 2021 guidelines, the patient was treated with Rituximab 375 mg/m2 weekly for 4 weeks, starting on the fifteenth day from the beginning of the corticosteroid therapy, after reduction of the lymphocyte count below 30,000 lymphocytes/mmc.
At discharge on the sixthteenth day from the start of the corticosteroid therapy, it was performed a biochemical reevaluation that showed a not complete recovery (Table 1).
The patient then continued with the urate-lowering and corticosteroid therapy at home. Because of an asymptomatic COVID-19 infection, the patient had to postpone the subsequent planned dose of Rituximab, but then he could complete the therapeutic scheme (Table 1).
After three months, the patient underwent revaluation. The clinical picture ameliorated with the resolution of AHIA. Due to the complete response to Rituximab and corticosteroid combined therapy and in consideration of the failure of corticosteroid therapy alone, secondary line therapy with Acalabrutinib (CalquenceTM 100 mg, 2 tabs/die) was offered to the patient (Table 1).
To these days (i.e., 8 months post patient first admission in the emergency room), the patient has carried out three treatment cycles of secondary line therapy, without significant side effects and with the maintenance of the complete response as regards the hemolytic process (Figure 1).
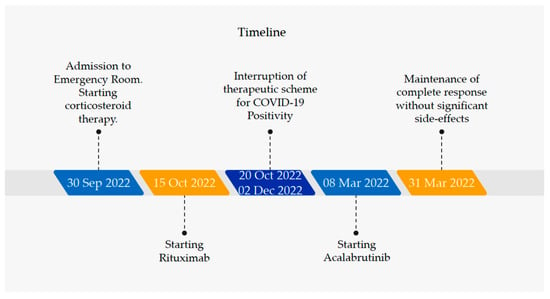
Figure 1.
Therapeutic evolution of the patient from diagnosis to complete response based on biochemical and clinical reevaluations.
3. Genetic Investigations
Conventional karyotyping was performed on peripheral blood cell cultures after 72 h of incubation with appropriate factors for the in-vitro stimulation of B Lymphocytes (ChromoLympho-B proliferation mix, EuroClone). Karyotyping was performed on Quinacrine-stained metaphases using MetaSystems Ikaros software version 5.7. As shown in Figure 2, a complex karyotype was obtained, defined by the presence of >3 numerical and/or structural abnormalities. In fact, the karyotype was characterized by trisomy 8 in 19/20 metaphases and, in all metaphases, an unbalanced translocation between the short arm of chromosome 6 and the long arm of chromosome 11 was detected, concurrent with interstitial deletions in chromosome 6q and 11q that could not be defined in detail.
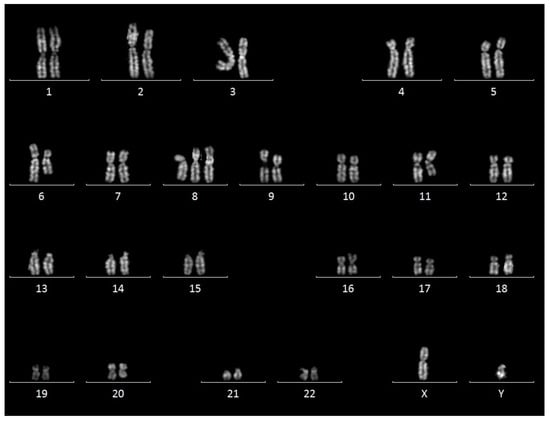
Figure 2.
Cytogenetics results. Representative karyogram showing trisomy 8 and aberrant chromosomes 6 and 11.
At first patient re-evaluation, conventional karyotyping from bone marrow cell culture was unchanged as it showed the previously reported unbalanced translocation in all metaphases and trisomy 8 in 5/20 metaphases.
In parallel, molecular cytogenetics (FISH) analyses revealed ATM monoallelic deletion (with loss of ATM on derivative chromosome 11) (Figure 3) and retained signals for TP53, 13q14 and centromere 12 FISH probes. The following FISH probes from MetaSystems were used: ATM/cep11, TP53/cep17, RB1/DLEU/LAMP, cep12.
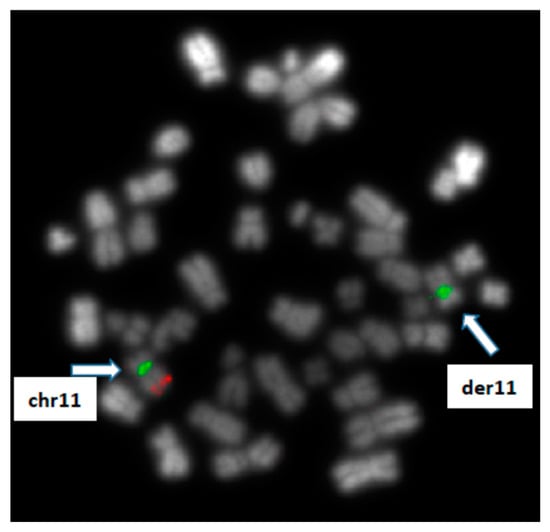
Figure 3.
Cytogenetics results: FISH analysis using probes for ATM gene (red signal) and chromosome 11 centromere (green signal) revealing a monoallelic loss of ATM on derivative chromosome 11.
Molecular analysis was performed using Sanger sequencing and did not identify mutations in the TP53 gene (exons 2–11) and in the variable region of IGH gene IgHV.
The aCGH analysis was performed with sex-matched Agilent Reference DNA using the SurePrint G3 CGH + SNP 4 × 180 K slides and the SureScan Microarray Scanner. Number of clones analyzed: 110,712 (CGH) + 59,647 (SNP). Median resolution: 25 kb. Data were analyzed with software CytoGenomics 4.0.3.12; the aberration detection method ADM-2 was applied; the Derivative LogRatio Spread (DLRSD) for this experiment was 0.17 (i.e., optimal quality). Genome positions referred to Human Genome Build 37 (hg19, Assembly February 2009). Aberrations below 50 kb and/or affecting less than five consecutive clones were not reported.
Array-CGH confirmed trisomy of the entire chromosome 8 (Figure 4A,B) and allowed to resolve in detail of the nature of the unbalanced translocation, revealing multiple regions of genomic losses on chromosomes 6 and 11.
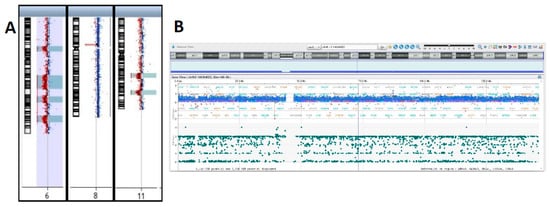
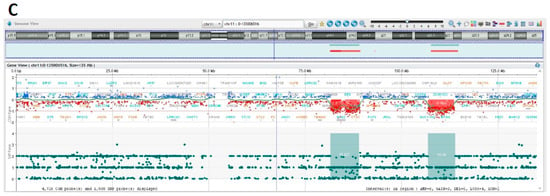
Figure 4.
Array-CGH results I. (A) Depiction of aberrant chromosomes 6, 8 and 11. Detailed CGH and SNP profiles showing. (B) Whole chromosome 8 gain. (C) Chromosome 11q deletions.
Indeed, one of the deleted regions on chromosome 11 encompasses the ATM locus that showed monoallelic deletion by FISH. The two regions of deletion on chromosome 11 are located in 11q14.1q14.2 and 11q22.3q23.2 (Figure 4C).
The exact boundaries of deletions and the corresponding gene content are listed in Table 2 and Table 3. Interestingly, chromosome 6q showed multiple regions of loss, with eighteen cumulative breakpoints, thus suggesting chromoanagenesis [,,] event as the driving force originating the derivative chromosome 6 (Figure 5, Table 3 and Table S1).

Table 3.
Genetic investigations and results.
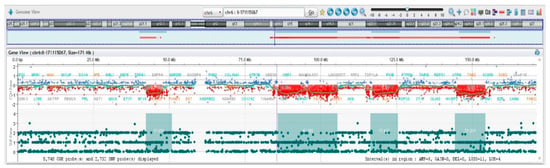
Figure 5.
Array-CGH results II. Chromosome 6 has multiple regions of imbalances, indicative of chromotripsis.
4. Discussion
From a genetic point of view, the case under study presented several peculiarities. While common in myeloid disorders, trisomy 8 is very rarely encountered in CLL at diagnosis, usually a mosaic state []. Although at mosaic state also in our patient, trisomy 8 represented a prominent cell clone. No signs of myelodysplasia were observed in the peripheral blood or bone marrow aspirate, so that we consider trisomy 8 a secondary, but very early, and positively selected event related to CLL in our patient.
We identified chromosome 11 deletions in bands 11q14.1q14.3, 11q14.3q21 and 11q22.3q23.2 (encompassing the ATM gene). As seen in our case, approximately 10–20% of CLL patient exhibit del(11)(q22q23) before treatment [,,,,], associated with loss of Ataxia Telangiectasia Mutated (ATM) serine/threonine kinase gene and representing a known marker of worse prognosis. Array-CGH showed multiple regions of deletion on chromosome 6 in our CLL case.
Chromosome 6q deletions are relatively frequent in CLL and are present in approximately 6% of the cases. In recent years, array-CGH analyses allowed to define multiple regions of deletion in 6q and no minimal critical region could be defined so far, as analyses of different case series identified distinct regions of aberration spanning from 6q16 to 6q27 [,,,,]. The association between 6q deletion and progression-free survival is conflicting in literature, probably due to the different methodologies applied (FISH, array-CGH, MLPA) and the different 6q bands investigated for prognosis [,,,]. As a result, the cytogenetic class of poor, intermediate, and good prognosis is associated with 6q deletions in literature. We anticipate that the real impact of distinct 6q deletions in patients’ prognosis and survival will benefit from a widespread application of genomic technologies for patients’ diagnosis. From this point of view, we provide a detailed description of 6q aberrations in our patient including breakpoints and gene content. The array-CGH profile of chromosome 6 is suggestive of a chromoanagenesis event.
In CLL, two-thirds of the cases show no aberrations beyond those detected by the routine FISH panel (TP53, ATM, centromere 12, 13q14) []. In fact, CLL shows a low burden of copy-number aberrations, and genomic instability is not considered a prominent feature of CLL [,].
In particular, chromoanagenesis is a rare event in CLL, reported in approximately 1–5% of cases analyzed by whole genome sequencing (WGS) or array-CGH []. Although we recognize that WGS is the only technique capable to define chromoanagenesis, such event can be inferred also by peculiar genome array profiles. Despite rarely being investigated in clinical trials, there is suggestive evidence that chromoanagenesis may confer worse survival in CLL []. In our case, the elevated number (eighteen) of breakpoints in the long arm of chromosome 6 fulfills the criteria for a bona fide chromotripsis-like event. Notably, in our patient, this event is not associated with TP53 alterations.
5. Conclusions
We report the genetic findings of a CLL patient with abrupt disease onset, so far responding properly to treatments despite the presence of distinct genetic adverse traits including ATM deletion, complex karyotype and chromosome 6q chromoanagenesis event. Our report confirms that interphase FISH alone is not able to provide an overview of the whole genomic landscape in selected CLL cases and that additional techniques are required to reach an appropriate cytogenetic stratification of patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcm12124110/s1, Table S1: Gene content of the genomic regions of deletion identified by array-CGH.
Author Contributions
M.C. and P.M. conceived the paper. The patient was treated by D.S. and A.S.; C.G., M.G., D.F. and P.M. produced genetic tests; G.P. and C.P. performed cytological assessment of sample and its immunophenotyping; S.G., M.P. and M.C. evaluated microscopic examination of the peripheral blood smear and immunophenotyping on peripheral blood; A.P. and E.V. conducted the Medline and literature review; M.C. and P.M. wrote and revised the manuscript and prepared the Table and the Figures; L.D.V. and S.G. edited and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Informed Consent Statement
Patient had provided written informed consent to participate to case presentation and for the publication of any images or potentially identifiable data included in this article.
Data Availability Statement
We state the data are available to the scientific community.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Albiol, N.; Moreno, C. Autoimmune Cytopenia in CLL: Prognosis and Management in the Era of Targeted Therapies. Cancer J. 2021, 27, 286–296. [Google Scholar] [CrossRef]
- Vitale, C.; Montalbano, M.C.; Salvetti, C.; Boccellato, E.; Griggio, V.; Boccadoro, M.; Coscia, M. Autoimmune Complications in Chronic Lymphocytic Leukemia in the Era of Targeted Drugs. Cancers 2020, 12, 282. [Google Scholar] [CrossRef] [PubMed]
- Autore, F.; Pasquale, R.; Innocenti, I.; Fresa, A.; Sora, F.; Laurenti, L. Autoimmune Hemolytic Anemia in Chronic Lymphocytic Leukemia: A Comprehensive Review. Cancers 2021, 13, 5804. [Google Scholar] [CrossRef]
- Hallek, M. Chronic lymphocytic leukemia: 2013 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2013, 88, 803–816. [Google Scholar] [CrossRef]
- Dameshek, W. Chronic lymphocytic leukemia—An accumulative disease of immunolgically incompetent lymphocytes. Blood 1967, 29, 566–584. [Google Scholar] [CrossRef]
- Hodgson, K.; Ferrer, G.; Montserrat, E.; Moreno, C. Chronic lymphocytic leukemia and autoimmunity: A systematic review. Haematologica 2011, 96, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Chiorazzi, N.; Rai, K.R.; Ferrarini, M. Chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 352, 804–815. [Google Scholar] [CrossRef]
- Strati, P.; Caligaris-Cappio, F. A matter of debate in chronic lymphocytic leukemia: Is the occurrence of autoimmune disorders an indicator of chronic lymphocytic leukemia therapy? Curr. Opin. Oncol. 2011, 23, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Jachiet, V.; Mekinian, A.; Carrat, F.; Grignano, E.; Retbi, A.; Boffa, J.-J.; Ronco, P.; Rondeau, E.; Sellam, J.; Berenbaum, F.; et al. Autoimmune manifestations associated with lymphoma: Characteristics and outcome in a multicenter retrospective cohort study. Leuk. Lymphoma 2018, 59, 1399–1405. [Google Scholar] [CrossRef]
- Mauro, F.R.; Foa, R.; Cerretti, R.; Giannarelli, D.; Coluzzi, S.; Mandelli, F.; Girelli, G. Autoimmune hemolytic anemia in chronic lymphocytic leukemia: Clinical, therapeutic, and prognostic features. Blood 2000, 95, 2786–2792. [Google Scholar] [CrossRef]
- Zent, C.S.; Ding, W.; Schwager, S.M.; Reinalda, M.S.; Hoyer, J.D.; Jelinek, D.F.; Tschumper, R.C.; Bowen, D.A.; Call, T.G.; Shanafelt, T.D.; et al. The prognostic significance of cytopenia in chronic lymphocyti leukaemia/small lymphocytic lymphoma. Br. J. Haematol. 2008, 141, 615–621. [Google Scholar] [CrossRef]
- Zent, C.S.; Ding, W.; Reinalda, M.S.; Schwager, S.M.; Hoyer, J.D.; Bowen, D.A.; Jelinek, D.F.; Tschumper, R.C.; Call, T.G.; Shanafelt, T.D.; et al. Autoimmune cytopenia in chronic lymphocytic leukemia/small lymphocytic lymphoma: Changes in clinical presentation and prognosis. Leuk. Lymphoma 2009, 50, 1261–1268. [Google Scholar] [CrossRef]
- Moreno, C.; Hodgson, K.; Ferrer, G.; Elena, M.; Filella, X.; Pereira, A.; Baumann, T.; Montserrat, E. Autoimmune cytopenia in chronic lymphocytic leukemia: Prevalence, clinical associations, and prognostic significance. Blood 2010, 116, 4771–4776. [Google Scholar] [CrossRef] [PubMed]
- Diehl, L.F.; Ketchum, L.H. Autoimmune disease and chronic lymphocytic leukemia: Autoimmune hemolytic anemia, pure red cell aplasia, and autoimmune thrombocytopenia. Semin. Oncol. 1998, 25, 80–97. [Google Scholar] [PubMed]
- Galton, D.A. The pathogenesis of chronic lymphocytic leukemia. Can. Med. Assoc. J. 1966, 94, 1005–1010. [Google Scholar]
- Barcellini, W.; Giannotta, J.A.; Fattizzo, B. Autoimmune Complications in Hematologic Neoplasms. Cancers 2021, 13, 1532. [Google Scholar] [CrossRef] [PubMed]
- Hill, Q.A.; Hill, A.; Berentsen, S. Defining autoimmune hemolytic anemia: A systematic review of the terminology used for diagnosis and treatment. Blood Adv. 2019, 3, 1897–1906. [Google Scholar] [CrossRef]
- Carli, G.; Visco, C.; Falisi, E.; Perbellini, O.; Novella, E.; Giaretta, I.; Ferrarini, I.; Sandini, A.; Alghisi, A.; Ambrosetti, A.; et al. Evans syndrome secondary to chronic lymphocytic leukaemia: Presentation, treatment, and outcome. Ann. Hematol. 2016, 95, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Maura, F.; Visco, C.; Falisi, E.; Reda, G.; Fabris, S.; Agnelli, L.; Tuana, G.; Lionetti, M.; Guercini, N.; Novella, E.; et al. B-cell receptor configuration and adverse cytogenetics are associated with autoimmune hemolytic anemia in chronic lymphocytic leukemia. Am. J. Hematol. 2013, 88, 32–36. [Google Scholar] [CrossRef]
- Vitale, C.; Salvetti, C.; Griggio, V.; Porrazzo, M.; Schiattone, L.; Zamprogna, G.; Visentin, A.; Vassallo, F.; Cassin, R.; Rigolin, G.M.; et al. Preexisting and treatment-emergent autoimmune cytopenias in patients with CLL treated with targeted drugs. Blood 2021, 137, 3507–3517. [Google Scholar] [CrossRef]
- Demir, C.; Ekinci, O. Clinical and serological autoimmune complications in chronic lymphocytic leukemia. Wien. Klin. Wochenschr. 2017, 129, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Atef, B.; Azmy, E.; Aladle, D.; Mabed, M. The prevalence and prognostic significance of autoimmune cytopenias in a cohort of Egyptian patients with chronic lymphocytic leukemia. Hematol. Oncol. Stem Cell Ther. 2019, 12, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Michalak, S.S.; Olewicz-Gawlik, A.; Rupa-Matysek, J.; Wolny-Rokicka, E.; Nowakowska, E.; Gil, L. Autoimmune hemolytic anemia: Current knowledge and perspectives. Immun. Ageing 2020, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Hill, Q.A.; Stamps, R.; Massey, E.; Grainger, J.D.; Provan, D.; Hill, A.; Haematology, T.B.S.F. The diagnosis and management of primary autoimmune haemolytic anaemia. Br. J. Haematol. 2017, 176, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Barcellini, W.; Fattizzo, B. How I treat warm autoimmune hemolytic anemia. Blood 2021, 137, 1283–1294. [Google Scholar] [CrossRef]
- Berentsen, S. How I manage patients with cold agglutinin disease. Br. J. Haematol. 2018, 181, 320–330. [Google Scholar] [CrossRef]
- Go, R.S.; Winters, J.L.; Kay, N.E. How I treat autoimmune hemolytic anemia. Blood 2017, 129, 2971–2979. [Google Scholar] [CrossRef] [PubMed]
- Dearden, C.; Wade, R.; Else, M.; Richards, S.; Milligan, D.; Hamblin, T.; Catovsky, D.; for the UK National Cancer Research Institute (NCRI) Haematological Oncology Clinical Studies Group and NCRI CLL Working Group. The prognostic significance of a positive direct antiglobulin test in chronic lymphocytic leukemia: A beneficial effect of the combination of fludarabine and cyclophosphamide on the incidence of hemolytic anemia. Blood 2008, 111, 1820–1826. [Google Scholar] [CrossRef]
- De Back, T.R.; Kater, A.; Tonino, S.H. Autoimmune cytopenias in chronic lymphocytic leukemia: A concise review and treatment recommendations. Expert. Rev. Hematol. 2018, 11, 613–624. [Google Scholar] [CrossRef]
- Barcellini, W.; Fattizzo, B.; Zaninoni, A.; Radice, T.; Nichele, I.; Di Bona, E.; Lunghi, M.; Tassinari, C.; Alfinito, F.; Ferrari, A.; et al. Clinical heterogeneity and predictors of outcome in primary autoimmune hemolytic anemia: A GIMEMA study of 308 patients. Blood 2014, 124, 2930–2936. [Google Scholar] [CrossRef]
- Fattizzo, B.; Barcellini, W. Autoimmune Cytopenias in Chronic Lymphocytic Leukemia: Focus on Molecular Aspects. Front. Oncol. 2019, 9, 1435. [Google Scholar] [CrossRef] [PubMed]
- Jager, U.; Barcellini, W.; Broome, C.M.; Gertz, M.A.; Hill, A.; Hill, Q.A.; Jilma, B.; Kuter, D.J.; Michel, M.; Montillo, M.; et al. Diagnosis and treatment of autoimmune hemolytic anemia in adults: Recommendations from the First International Consensus Meeting. Blood Rev. 2020, 41, 100648. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, C.; Rajapakse, S.; Gooneratne, L. Rituximab in the treatment of autoimmune haemolytic anaemia. Br. J. Clin. Pharm. 2015, 79, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Birgens, H.; Frederiksen, H.; Hasselbalch, H.C.; Rasmussen, I.H.; Nielsen, O.J.; Kjeldsen, L.; Larsen, H.; Mourits-Andersen, T.; Plesner, T.; Rønnov-Jessen, D.; et al. A phase III randomized trial comparing glucocorticoid monotherapy versus glucocorticoid and rituximab in patients with autoimmune haemolytic anaemia. Br. J. Haematol. 2013, 163, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.; Terriou, L.; Roudot-Thoraval, F.; Hamidou, M.; Ebbo, M.; Le Guenno, G.; Galicier, L.; Audia, S.; Royer, B.; Morin, A.-S.; et al. A randomized and double-blind controlled trial evaluating the safety and efficacy of rituximab for warm auto-immune hemolytic anemia in adults (the RAIHA study). Am. J. Hematol. 2017, 92, 23–27. [Google Scholar] [CrossRef]
- D’Arena, G.; Laurenti, L.; Capalbo, S.; D’Arco, A.M.; De Filippi, R.; Marcacci, G.; Di Renzo, N.; Storti, S.; Califano, C.; Vigliotti, M.L.; et al. Rituximab therapy for chronic lymphocytic leukemia-associated autoimmune hemolytic anemia. Am. J. Hematol. 2006, 81, 598–602. [Google Scholar] [CrossRef]
- Nikitin, E.; Kislova, M.; Morozov, D.; Belyakova, V.; Suvorova, A.; Sveshnikova, J.; Vyscub, G.; Matveeva, I.; Shirokova, M.; Shipaeva, A.; et al. Ibrutinib in combination with rituximab is highly effective in treatment of chronic lymphocytic leukemia patients with steroid refractory and relapsed autoimmune cytopenias. Leukemia 2023, 1–10. [Google Scholar] [CrossRef]
- Rack, K.A.; Van Den Berg, E.; Haferlach, C.; Beverloo, H.B.; Costa, D.; Espinet, B.; Foot, N.; Jeffries, S.; Martin, K.; O’Connor, S.; et al. European recommendations and quality assurance for cytogenomic analysis of haematological neoplasms. Leukemia 2019, 33, 1851–1867. [Google Scholar] [CrossRef]
- Rack, K.A.; Berg, E.V.D.; Haferlach, C.; Beverloo, H.B.; Costa, D.; Espinet, B.; Foot, N.; Jeffries, S.; Martin, K.; O’connor, S.; et al. European recommendations and quality assurance for cytogenomic analysis of haematological neoplasms: Reponse to the comments from the Francophone Group of Hematological Cytogenetics (GFCH). Leukemia 2020, 34, 2262–2264. [Google Scholar] [CrossRef]
- Pellestor, F.; Gaillard, J.B.; Schneider, A.; Puechberty, J.; Gatinois, V. Chromoanagenesis, the mechanisms of a genomic chaos. Semin. Cell Dev. Biol. 2022, 123, 90–99. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Leibowitz, M.L.; Pellman, D. Chromothripsis and beyond: Rapid genome evolution from complex chromosomal rearrangements. Genes. Dev. 2013, 27, 2513–2530. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.M. Chromoplexy: A new category of complex rearrangements in the cancer genome. Cancer Cell 2013, 23, 567–569. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Granada, I.; Espinet, B.; Collado, R.; Ruiz-Xivillé, N.; Puiggros, A.; Uribe, M.; Arias, A.; Gómez, C.; Delgado, J.; et al. Balanced and unbalanced translocations in a multicentric series of 2843 patients with chronic lymphocytic leukemia. Genes Chromosom. Cancer 2022, 61, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Gozzetti, A.; Calabrese, S.; Crupi, R.; Zaja, F.; Tozzuoli, D.; Tassi, M.; Raspadori, D.; Lenoci, M.; Lauria, F. Trisomy 8 in chronic lymphocytic leukemia: A report of two cases. Cancer Genet. Cytogenet. 2007, 175, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, J.; Holzmann, K.; Miller, F.; Winkler, D.; Bühler, A.; Zenz, T.; Bullinger, L.; Kühn, M.W.M.; Gerhardinger, A.; Bloehdorn, J.; et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood 2012, 120, 4783–4794. [Google Scholar] [CrossRef]
- Gunnarsson, R.; Mansouri, L.; Isaksson, A.; Göransson, H.; Cahill, N.; Jansson, M.; Rasmussen, M.; Lundin, J.; Norin, S.; Buhl, A.M.; et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica 2011, 96, 1161–1169. [Google Scholar] [CrossRef]
- Urbankova, H.; Papajik, T.; Plachy, R.; Holzerova, M.; Balcarkova, J.; Divoka, M.; Prochazka, V.; Pikalova, Z.; Indrak, K.; Jarosova, M. Array-based karyotyping in chronic lymphocytic leukemia (CLL) detects new unbalanced abnormalities that escape conventional cytogenetics and CLL FISH panel. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc Czech Repub. 2014, 158, 56–64. [Google Scholar] [CrossRef]
- Tausch, E.; Schneider, C.; Robrecht, S.; Zhang, C.; Dolnik, A.; Bloehdorn, J.; Bahlo, J.; Al-Sawaf, O.; Ritgen, M.; Fink, A.-M.; et al. Prognostic and predictive impact of genetic markers in patients with CLL treated with obinutuzumab and venetoclax. Blood 2020, 135, 2402–2412. [Google Scholar] [CrossRef]
- Schweighofer, C.D.; Coombes, K.R.; Majewski, T.; Barron, L.L.; Lerner, S.; Sargent, R.L.; O’Brien, S.; Ferrajoli, A.; Wierda, W.G.; Czerniak, B.A.; et al. Genomic variation by whole-genome SNP mapping arrays predicts time-to-event outcome in patients with chronic lymphocytic leukemia: A comparison of CLL and HapMap genotypes. J. Mol. Diagn. 2013, 15, 196–209. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Wen, S.; O’Brien, S.; McLaughlin, P.; Wierda, W.G.; Ferrajoli, A.; Faderl, S.; Manning, J.; Lerner, S.; Mai, C.V.; et al. Assessment of chronic lymphocytic leukemia and small lymphocytic lymphoma by absolute lymphocyte counts in 2126 patients: 20 years of experience at the University of Texas M.D. Anderson Cancer Center. J. Clin. Oncol. 2007, 25, 4648–4656. [Google Scholar] [CrossRef]
- Jarosova, M.; Hruba, M.; Oltova, A.; Plevova, K.; Kruzova, L.; Kriegova, E.; Fillerova, R.; Koritakova, E.; Doubek, M.; Lysak, D.; et al. Chromosome 6q deletion correlates with poor prognosis and low relative expression of FOXO3 in chronic lymphocytic leukemia patients. Am. J. Hematol. 2017, 92, E604–E607. [Google Scholar] [CrossRef] [PubMed]
- Houldsworth, J.; Guttapalli, A.; Thodima, V.; Yan, X.J.; Mendiratta, G.; Zielonka, T.; Nanjangud, G.; Chen, W.; Patil, S.; Mato, A.; et al. Genomic imbalance defines three prognostic groups for risk stratification of patients with chronic lymphocytic leukemia. Leuk. Lymphoma 2014, 55, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Dalsass, A.; Mestichelli, F.; Ruggieri, M.; Gaspari, P.; Pezzoni, V.; Vagnoni, D.; Angelini, M.; Angelini, S.; Bigazzi, C.; Falcioni, S.; et al. 6q deletion detected by fluorescence in situ hybridization using bacterial artificial chromosome in chronic lymphocytic leukemia. Eur. J. Haematol. 2013, 91, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsson, R.; Isaksson, A.; Mansouri, M.; Göransson, H.; Jansson, M.; Cahill, N.; Rasmussen, M.; Staaf, J.; Lundin, J.; Norin, S.; et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: A high-resolution genomic screening of newly diagnosed patients. Leukemia 2010, 24, 211–215. [Google Scholar] [CrossRef]
- Zavacka, K.; Plevova, K. Chromothripsis in Chronic Lymphocytic Leukemia: A Driving Force of Genome Instability. Front. Oncol. 2021, 11, 771664. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).