Microbiome of Acute Otitis Externa
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Clinical Variables
2.2. Sample Collection and DNA Extraction
2.3. Library Construction and Sequencing
2.4. Bioinformatics Pipeline and Statistical Analysis
3. Results
3.1. Demographic Data
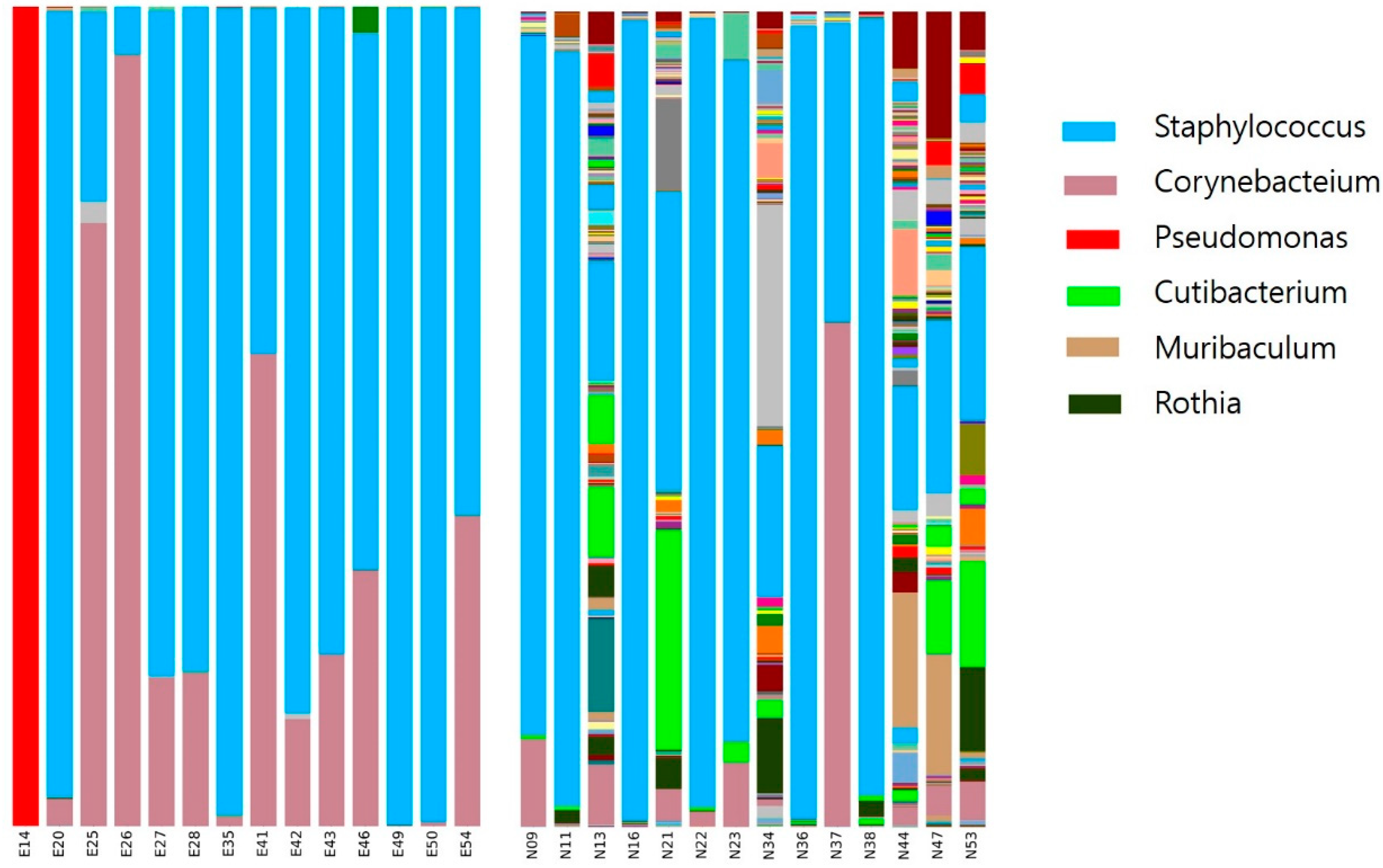
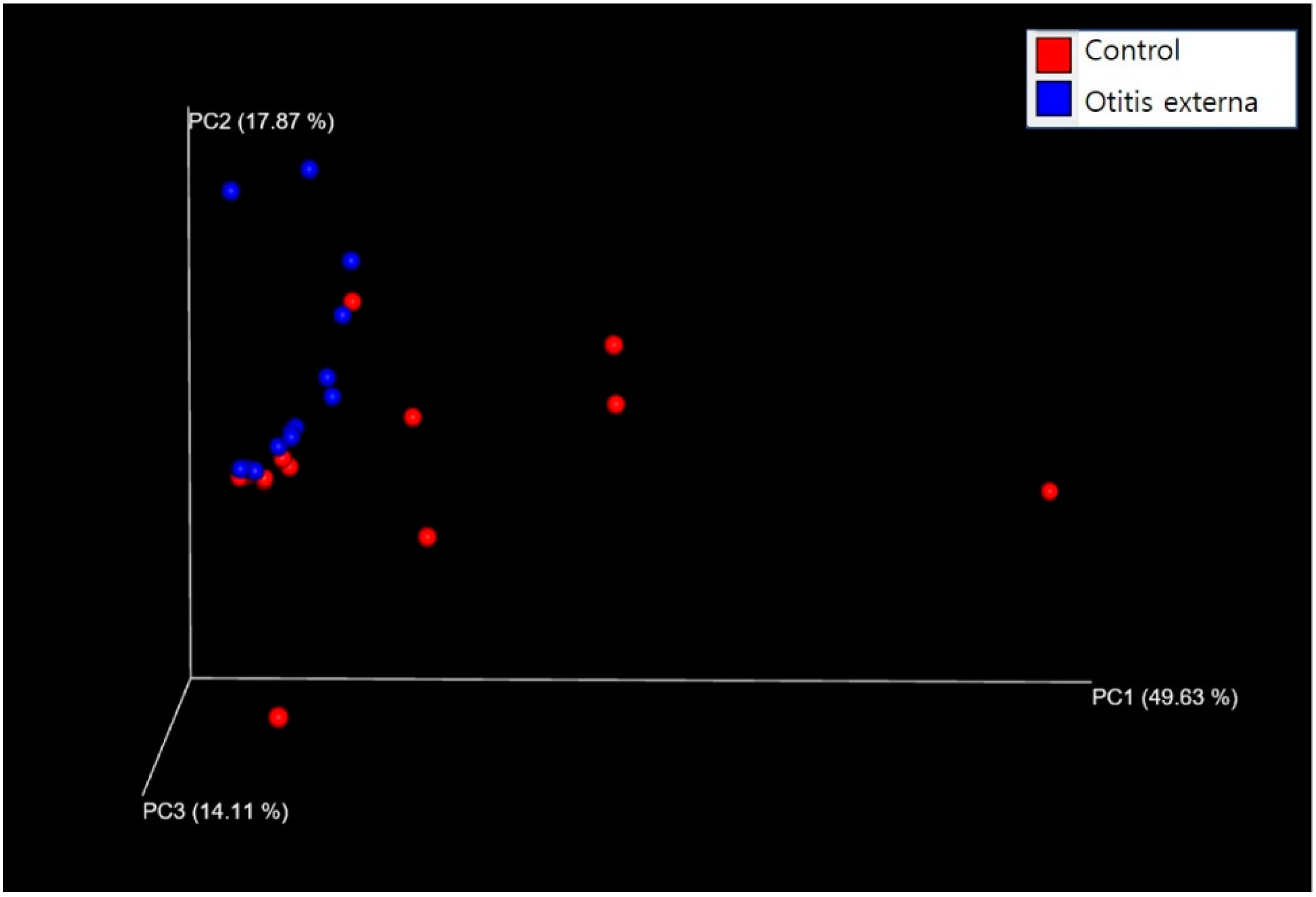
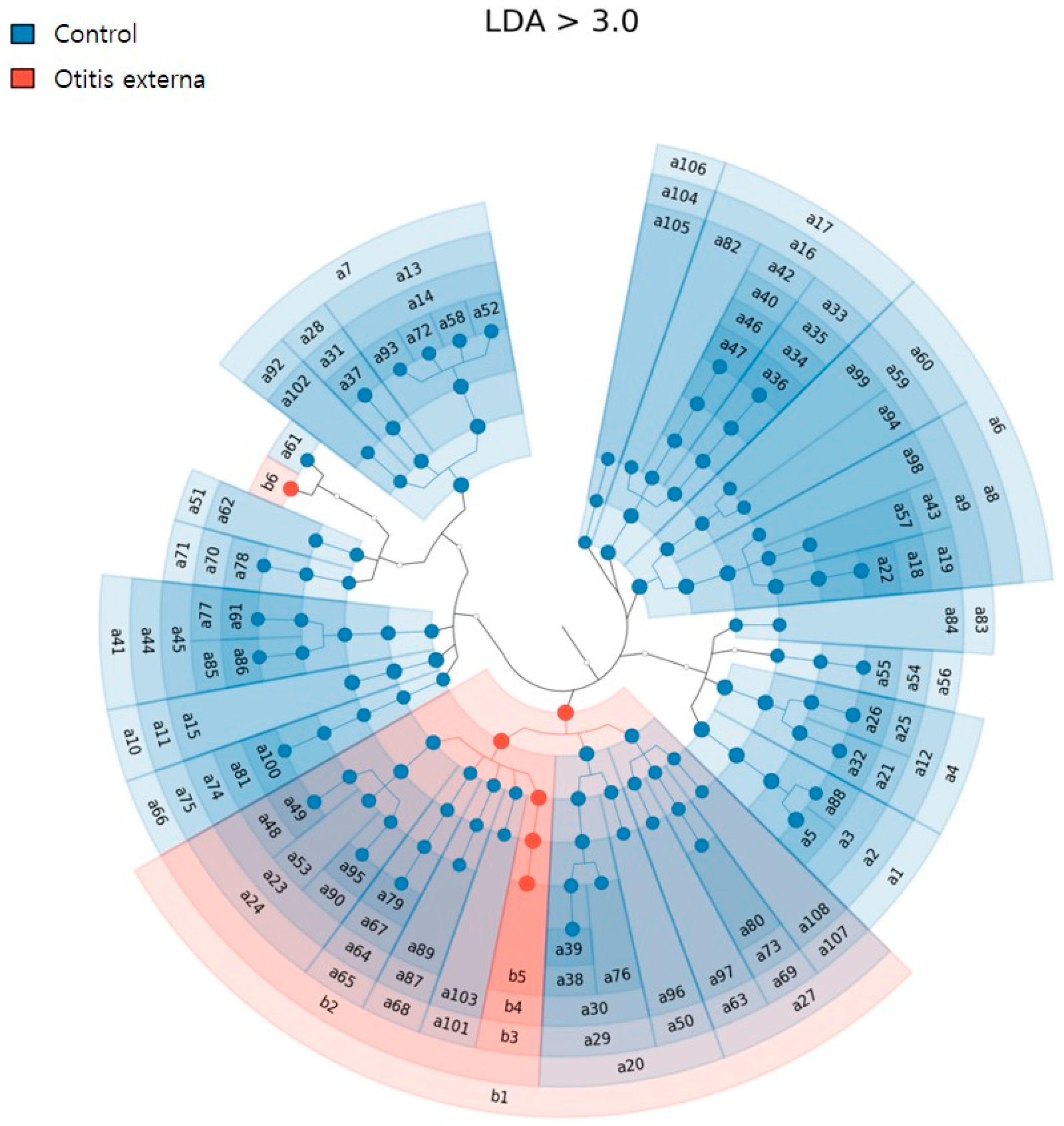
3.2. Bacterial Diversity and Community Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Norton, J.E.; Novy, L.E. Studies on the self disinfecting power of the skin. Am. J. Public Health Nations Health 1931, 21, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Roland, P.S.; Stroman, D.W. Microbiology of acute otitis externa. Laryngoscope 2002, 112, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.B.; Brook, I.; Bianki, D.; Thompson, D.H. Microbiology of otitis externa. Otolaryngol. Head Neck Surg. 1997, 116, 23–25. [Google Scholar] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Sun, J.; Chang, E.B. Exploring gut microbes in human health and disease: Pushing the envelope. Genes Dis. 2014, 1, 132–139. [Google Scholar] [CrossRef]
- Hrestak, D.; Matijašić, M.; Čipčić Paljetak, H.; Ledić Drvar, D.; Ljubojević Hadžavdić, S.; Perić, M. Skin Microbiota in Atopic Dermatitis. Int. J. Mol. Sci. 2022, 23, 3503. [Google Scholar] [CrossRef]
- Chung, K.F. Airway microbial dysbiosis in asthmatic patients: A target for prevention and treatment? J. Allergy Clin. Immunol. 2017, 139, 1071–1081. [Google Scholar] [CrossRef]
- Hyun, D.W.; Min, H.J.; Kim, M.S.; Whon, T.W.; Shin, N.R.; Kim, P.S.; Kim, H.S.; Lee, J.Y.; Kang, W.; Choi, A.M.K.; et al. Dysbiosis of Inferior Turbinate Microbiota Is Associated with High Total IgE Levels in Patients with Allergic Rhinitis. Infect. Immun. 2018, 86, e00934-17. [Google Scholar] [CrossRef]
- Kobayashi, T.; Glatz, M.; Horiuchi, K.; Kawasaki, H.; Akiyama, H.; Kaplan, D.H.; Kong, H.H.; Amagai, M.; Nagao, K. Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity 2015, 42, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]
- Roy Sarkar, S.; Banerjee, S. Gut microbiota in neurodegenerative disorders. J. Neuroimmunol. 2019, 328, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, S.; Devalia, H.; Smith, C.; Hubbard, R.; Dean, A. Otitis externa in UK general practice: A survey using the UK General Practice Research Database. Br. J. Gen. Pract. 2001, 51, 533–538. [Google Scholar] [PubMed]
- Kasai, T.; Fukui, Y.; Aoki, K.; Ishii, Y.; Tateda, K. Changes in the ear canal microbiota of dogs with otitis externa. J. Appl. Microbiol. 2021, 130, 1084–1091. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Main, T.; Lim, D. The human external auditory canal, secretory system--an ultrastructural study. Laryngoscope 1976, 86, 1164–1176. [Google Scholar] [CrossRef]
- Sjövall, A.; Aho, V.T.E.; Hyyrynen, T.; Kinnari, T.J.; Auvinen, P.; Silvola, J.; Aarnisalo, A.; Laulajainen-Hongisto, A. Microbiome of the Healthy External Auditory Canal. Otol. Neurotol. 2021, 42, e609–e614. [Google Scholar] [CrossRef]
- Stroman, D.W.; Roland, P.S.; Dohar, J.; Burt, W. Microbiology of normal external auditory canal. Laryngoscope 2001, 111, 2054–2059. [Google Scholar] [CrossRef]
- Burton, M.; Krumbeck, J.A.; Wu, G.; Tang, S.; Prem, A.; Gupta, A.K.; Dawson, T.L., Jr. The adult microbiome of healthy and otitis patients: Definition of the core healthy and diseased ear microbiomes. PLoS ONE 2022, 24, e0262806. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Hilty, M.; Qi, W.; Brugger, S.D.; Frei, L.; Agyeman, P.K.A.; Frey, P.; Aebi, S.; Mühlemann, K. Nasopharyngeal Microbiota in Infants with Acute Otitis Media. J. Infect. Dis. 2012, 205, 1048–1055. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.R.; Hauser, L.J.; Frank, D.N. The sinonasal bacterial microbiome in health and disease. Curr. Opin. Otolaryngol. Head Neck Surg. 2016, 24, 20–25. [Google Scholar] [CrossRef]
- Fyhrquist, N.; Muirhead, G.; Prast-Nielsen, S.; Jeanmougin, M.; Olah, P.; Skoog, T.; Jules-Clement, G.; Feld, M.; Barrientos-Somarribas, M.; Sinkko, H.; et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat. Commun. 2019, 10, 4703. [Google Scholar] [CrossRef]
- Larry, S.; McDaniel. Otitis Media and Ear Infections. In Bacteria, Encyclopedia of Infection and Immunity; Elsevier: Amsterdam, The Netherlands, 2022; Volume 3, pp. 259–262. [Google Scholar]
- Kloos, W. Natural populations of the genus Staphylococcus. Ann. Rev. Microbiol. 1980, 34, 559–592. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Hata, T.R.; Tong, Y.; Cheng, J.Y.; Shafiq, F.; Butcher, A.M.; Salem, S.S.; Brinton, S.L.; Rudman Spergel, A.K.; Johnson, K.; et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat. Med. 2021, 27, 700–709. [Google Scholar] [CrossRef]
- Shokry, E.; Filho, N.R.A. Insights into cerumen and application in diagnostics: Past, present and future prospective. Biochem. Med. 2017, 27, 030503. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.K.; Han, S.J.; Hong, S.J.; Hong, S.M. Microbiome of Acute Otitis Externa. J. Clin. Med. 2022, 11, 7074. https://doi.org/10.3390/jcm11237074
Kim SK, Han SJ, Hong SJ, Hong SM. Microbiome of Acute Otitis Externa. Journal of Clinical Medicine. 2022; 11(23):7074. https://doi.org/10.3390/jcm11237074
Chicago/Turabian StyleKim, Sung Kyun, Sung Jun Han, Seok Jin Hong, and Seok Min Hong. 2022. "Microbiome of Acute Otitis Externa" Journal of Clinical Medicine 11, no. 23: 7074. https://doi.org/10.3390/jcm11237074
APA StyleKim, S. K., Han, S. J., Hong, S. J., & Hong, S. M. (2022). Microbiome of Acute Otitis Externa. Journal of Clinical Medicine, 11(23), 7074. https://doi.org/10.3390/jcm11237074





