Airway Bacterial Biodiversity in Exhaled Breath Condensates of Asthmatic Children—Does It Differ from the Healthy Ones?
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Sample Collection
2.3. Genetic Material Isolation and Amplification
2.3.1. DNA Extraction
2.3.2. Preparation of DNA Libraries for Sequencing
2.3.3. 16S rRNA Gene Sequencing
2.4. Data Analysis
3. Results
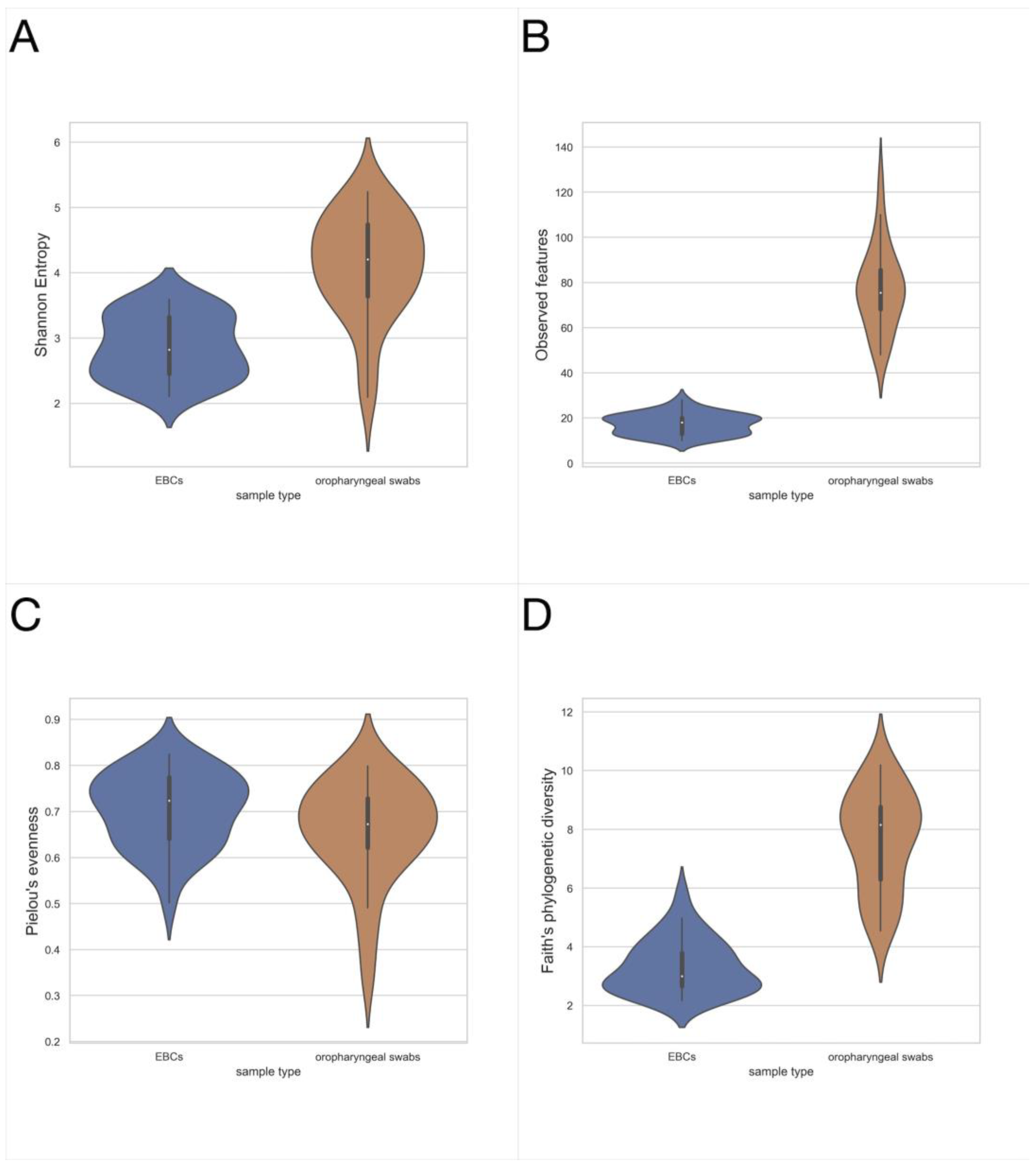
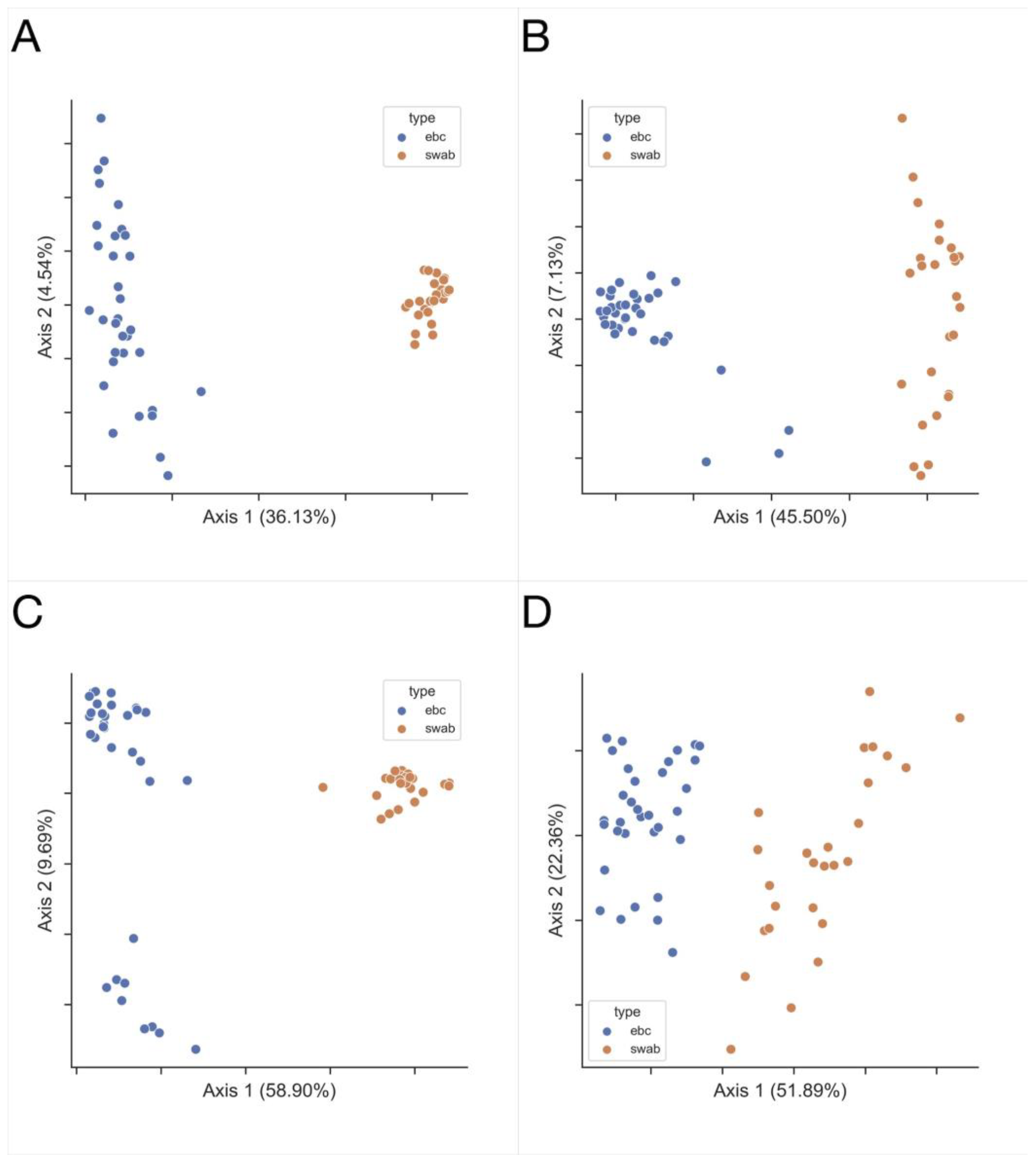
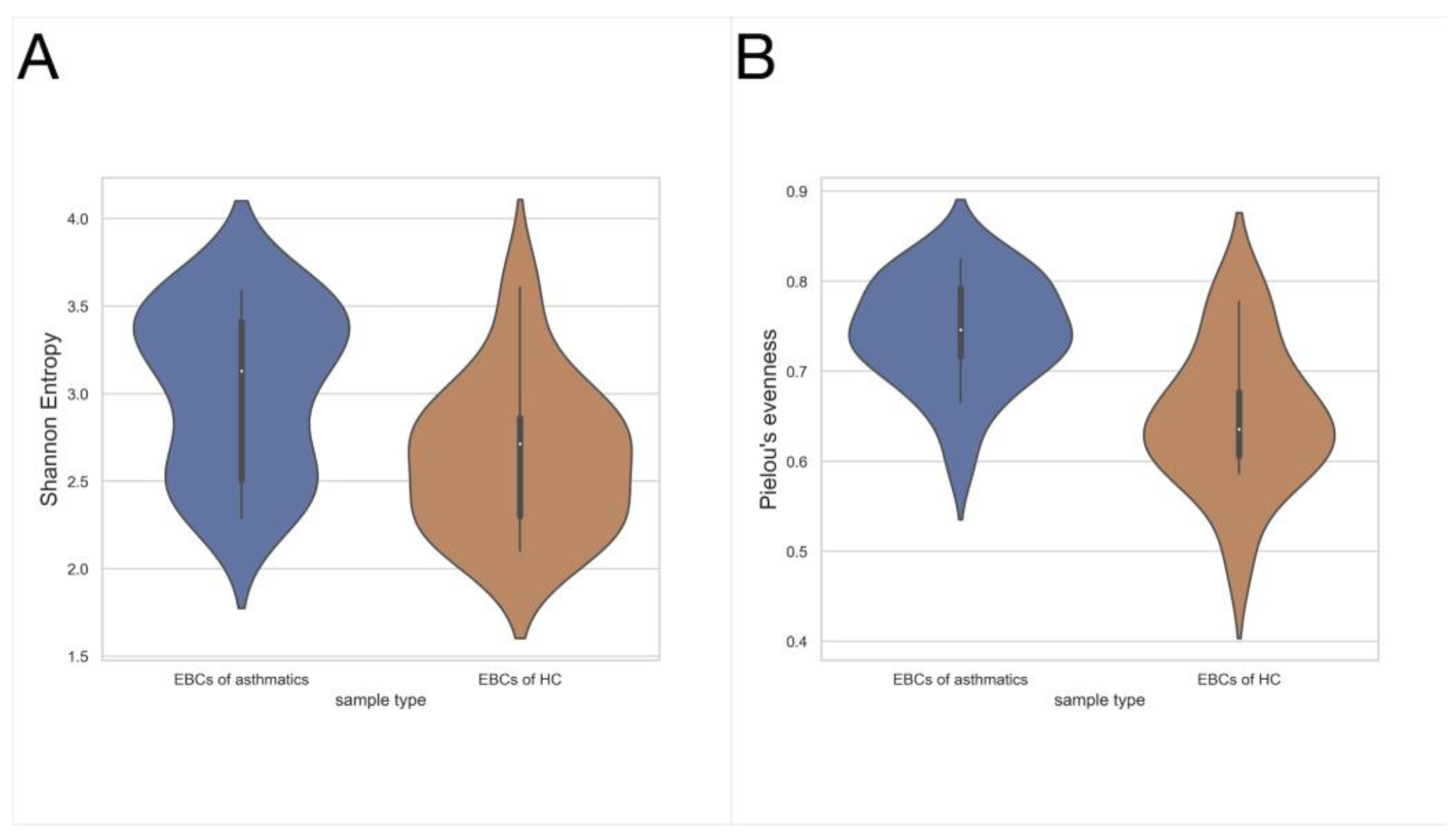
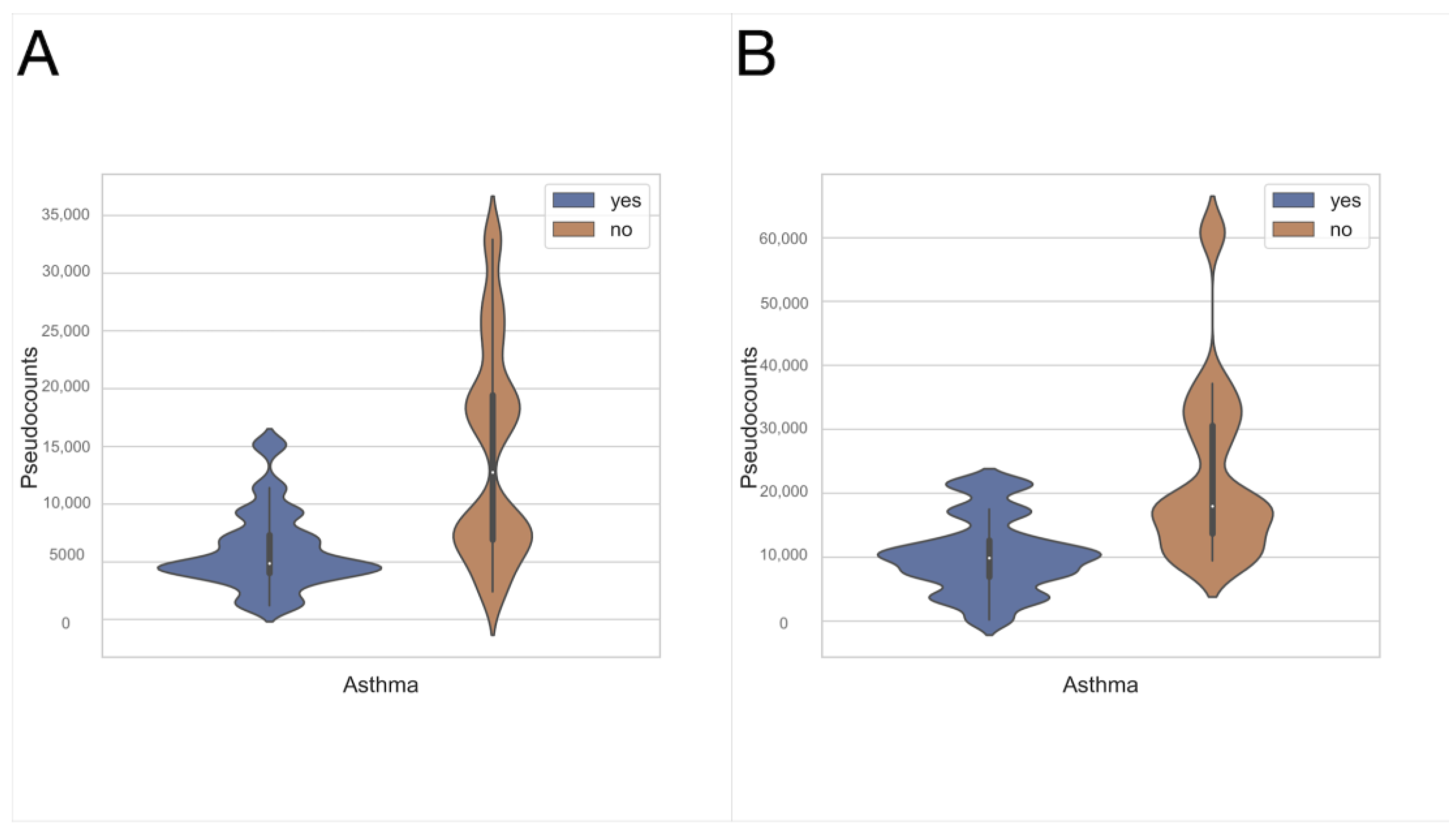
3.1. Exhaled Breath Condensate Samples Analysis
3.2. Oropharyngeal Swabs
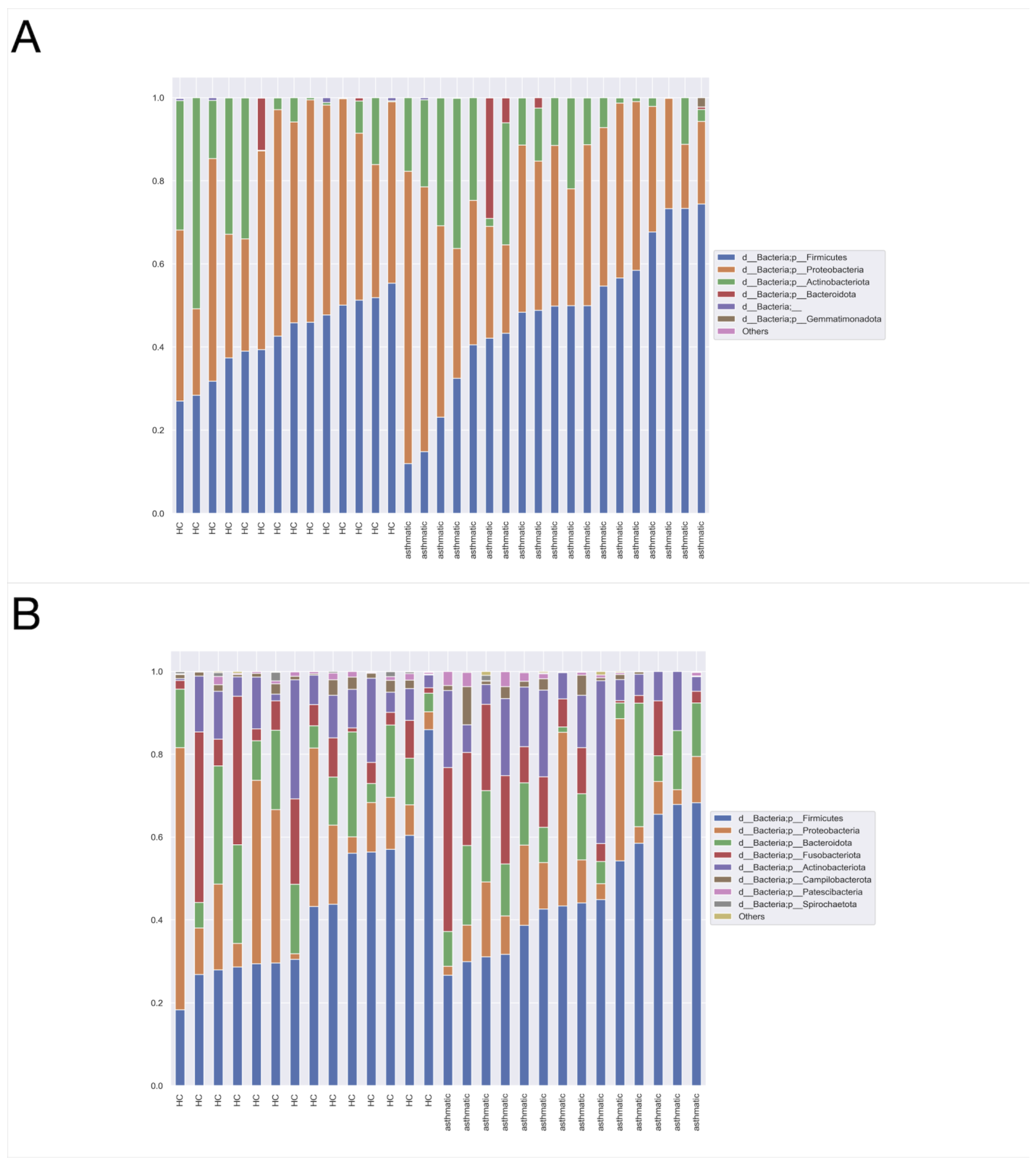
3.3. Dominant Taxa
3.4. Taxonomic Assignment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Patel, S.J.; Teach, S.J. Asthma. Pediatr. Rev. 2019, 40, 549–565. [Google Scholar] [CrossRef] [PubMed]
- GINA 2022 Main Report. Available online: https://ginasthma.org/gina-reports/ (accessed on 9 November 2022).
- Dharmage, S.C.; Perret, J.L.; Custovic, A. Epidemiology of Asthma in Children and Adults. Front. Pediatr. 2019, 7, 246. [Google Scholar] [CrossRef] [PubMed]
- Haahtela, T. A Biodiversity Hypothesis. Allergy Eur. J. Allergy Clin. Immunol. 2019, 74, 1445–1456. [Google Scholar] [CrossRef]
- Jartti, T.; Bønnelykke, K.; Elenius, V.; Feleszko, W. Role of Viruses in Asthma. Semin. Immunopathol. 2020, 42, 61–74. [Google Scholar] [CrossRef] [PubMed]
- von Mutius, E.; Smits, H.H. Primary Prevention of Asthma: From Risk and Protective Factors to Targeted Strategies for Prevention. Lancet 2020, 396, 854–866. [Google Scholar] [CrossRef]
- Beck, J.M.; Young, V.B.; Huffnagle, G.B. The Microbiome of the Lung. Transl. Res. 2012, 160, 258–266. [Google Scholar] [CrossRef]
- Rivas, M.N.; Crother, T.R.; Arditi, M. The Microbiome in Asthma. Curr. Opin. Pediatr. 2016, 28, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Horváth, I.; Barnes, P.J.; Loukides, S.; Sterk, P.J.; Högman, M.; Olin, A.-C.; Amann, A.; Antus, B.; Baraldi, E.; Bikov, A.; et al. A European Respiratory Society Technical Standard: Exhaled Biomarkers in Lung Disease. Eur. Respir. J. 2017, 49, 1600965. [Google Scholar] [CrossRef]
- An, S.Q.; Warris, A.; Turner, S. Microbiome characteristics of induced sputum compared to bronchial fluid and upper airway samples. Pediatr. Pulmonol. 2018, 53, 921–928. [Google Scholar] [CrossRef]
- Mutlu, G.M.; Garey, K.W.; Robbins, R.A.; Danziger, L.H.; Rubinstein, I. Collection and Analysis of Exhaled Breath Condensate in Humans. Am. J. Respir. Crit. Care Med. 2001, 164, 731–737. [Google Scholar] [CrossRef]
- Illumina MiSeq System. 16S Metagenomic Sequencing Library Preparation. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 22 December 2021).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Erratum in: Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2, High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7, improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Pielou, E.C. The Measurement of Diversity in Different Types of Biological Collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Robeson MS 2nd O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Ober, C.; Yao, T.C. The genetics of asthma and allergic disease: A 21st century perspective. Immunol. Rev. 2011, 242, 10–30. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Alamri, L.; Crandall, K.A.; Freishtat, R.J. Nasopharyngeal Microbiome Diversity Changes over Time in Children with Asthma. PLoS ONE 2017, 12, e0170543. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Authelet, K.J.; Hoptay, C.E.; Kwak, C.; Crandall, K.A.; Freishtat, R.J. Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. BMC 2018, 6, 179. [Google Scholar] [CrossRef]
- Depner, M.; Ege, M.J.; Cox, M.J.; Dwyer, S.; Walker, A.W.; Birzele, L.T.; Genuneit, J.; Horak, E.; Braun-Fahrländer, C.; Danielewicz, H.; et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J. Allergy Clin. Immunol. 2017, 139, 826–834.e13. [Google Scholar] [CrossRef]
- Boutin, S.; Depner, M.; Stahl, M.; Graeber, S.Y.; Dittrich, S.A.; Legatzki, A.; von Mutius, E.; Mall, M.; Dalpke, A.H. Comparison of Oropharyngeal Microbiota from Children with Asthma and Cystic Fibrosis. Mediat. Inflamm. 2017, 2017, 5047403. [Google Scholar] [CrossRef]
- Marri, P.R.; Stern, D.A.; Wright, A.L.; Billheimer, D.; Martinez, F.D. Asthma-Associated Differences in Microbial Composition of Induced Sputum. J. Allergy Clin. Immunol. 2013, 131, 346–352.e3. [Google Scholar] [CrossRef]
- Pang, Z.; Wang, G.; Gibson, P.; Guan, X.; Zhang, W.; Zheng, R.; Chen, F.; Wang, Z.; Wang, F. Airway Microbiome in Different Inflammatory Phenotypes of Asthma: A Cross-Sectional Study in Northeast China. Int. J. Med. Sci. 2019, 16, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; Daly, J.; Baines, K.J.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Hugenholtz, P.; Willner, D.; et al. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur. Respir. J. 2016, 47, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D.L.; Chen, Z.; Shankar, V.; Tyberg, M.; Vicencio, A.; Burk, R. Lower Airway Microbiota and Mycobiota in Children with Severe Asthma. J. Allergy Clin. Immunol. 2018, 141, 808–811.e7. [Google Scholar] [CrossRef]
- Cui, J.; Zhang, Y.; Zhao, H.; Sun, X.; Chen, Z.; Zhang, Q.; Yan, C.; Xue, G.; Li, S.; Feng, Y.; et al. The Relationship Between Lower Respiratory Tract Microbiome and Allergic Respiratory Tract Diseases in Children. Front. Microbiol. 2021, 12, 630345. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.F.M.; Pattaroni, C.; Cook, J.; Gregory, L.; Alonso, A.M.; Fleming, L.J.; Lloyd, C.M.; Bush, A.; Marsland, B.J.; Saglani, S. Lower airway microbiota associates with inflammatory phenotype in severe preschool wheeze. J. Allergy Clin. Immunol. 2019, 143, 1607–1610.e3. [Google Scholar] [CrossRef]
- Wu, L.; Shen, C.; Chen, Y.; Yang, X.; Luo, X.; Hang, C.; Yan, L.; Xu, X. Follow-up study of airway microbiota in children with persistent wheezing. Respir. Res. 2021, 22, 213. [Google Scholar] [CrossRef]
- Bisgaard, H.; Hermansen, M.N.; Buchvald, F.; Loland, L.; Halkjaer, L.B.; Bonnelykke, K.; Brasholt, M.; Heltberg, A.; Vissing, N.H.; Thorsen, S.V.; et al. Childhood Asthma after Bacterial Colonization of the Airway in Neonates. N. Engl. J. Med. 2007, 357, 1487–1495. [Google Scholar] [CrossRef]
- Huang, C.; Yu, Y.; Du, W.; Liu, Y.; Dai, R.; Tang, W.; Wang, P.; Zhang, C.; Shi, G. Fungal and Bacterial Microbiome Dysbiosis and Imbalance of Trans-Kingdom Network in Asthma. Clin. Transl. Allergy 2020, 10, 42. [Google Scholar] [CrossRef]
- Denner, D.R.; Sangwan, N.; Becker, J.B.; Hogarth, D.K.; Oldham, J.; Castillo, J.; Sperling, A.I.; Solway, J.; Naureckas, E.T.; Gilbert, J.A.; et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J. Allergy Clin. Immunol. 2016, 137, 1398–1405.e3. [Google Scholar] [CrossRef]
- Zhang, Q.; Cox, M.; Liang, Z.; Brinkmann, F.; Cardenas, P.A.; Duff, R.; Bhavsar, P.; Cookson, W.; Moffatt, M.; Chung, K.F. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS ONE 2016, 11, e0152724. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V.; Nariya, S.; Bhakta, N.R.; Beigelman, A.; Castro, M.; Dyer, A.-M.; Israel, E.; Kraft, M.; Martin, R.J.; et al. National Heart, Lung and Blood Institute’s “AsthmaNet”. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 2017, 140, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Christian, L.S.; Nariya, S.; Gonzalez, J.; Bhakta, N.R.; Ansel, K.M.; Beigelman, A.; Castro, M.; Dyer, A.-M.; Israel, E.; et al. Distinct associations of sputum and oral microbiota with atopic, immunologic, and clinical features in mild asthma. J. Allergy Clin. Immunol. 2020, 146, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, S.K.; Al-Asadi, J.N.; Al Yassen, A.Q. The Role of Caesarean Section in Childhood Asthma. Malays. Fam. Physician 2019, 14, 10–17. [Google Scholar]
- Castro-Rodriguez, J.A.; Forno, E.; Rodriguez-Martinez, C.E.; Celedón, J.C. Risk and Protective Factors for Childhood Asthma: What Is the Evidence? J. Allergy Clin. Immunol. Pract. 2016, 4, 1111–1122. [Google Scholar] [CrossRef]
- Słabuszewska-Jóźwiak, A.; Szymański, J.K.; Ciebiera, M.; Sarecka-Hujar, B.; Jakiel, G. Pediatrics Consequences of Caesarean Section—A Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2020, 17, 8031. [Google Scholar] [CrossRef]
- Fazlollahi, M.R.; Najmi, M.; Fallahnezhad, M.; Sabetkish, N.; Kazemnejad, A.; Bidad, K.; Shoormasti, R.S.; Mahloujirad, M. Paediatric asthma prevalence: The first national population-based survey in Iran. Clin. Respir. J. 2019, 13, 14–22. [Google Scholar] [CrossRef]
- Li, L.X.; Lin, S.Z.; Zhang, R.P.; Chen, S.W. Prevalence of pediatric asthma in the rural areas of China: A Meta analysis. Zhongguo Dang Dai Er Ke Za Zhi 2020, 22, 380–386. [Google Scholar]
- Boyle, R.J.; Shamji, M.H. Allergy prevention. Clin. Exp. Allergy 2021, 51, 4–5. [Google Scholar] [CrossRef]
- Kirjavainen, P.V.; Karvonen, A.M.; Adams, R.I.; Täubel, M.; Roponen, M.; Tuoresmäki, P.; Loss, G.; Jayaprakash, B.; Depner, M.; Ege, M.J.; et al. Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat. Med. 2019, 25, 1089–1095. [Google Scholar] [CrossRef]
- Bugova, G.; Janickova, M.; Uhliarova, B.; Babela, R.; Jesenak, M. The effect of passive smoking on bacterial colonisation of the upper airways and selected laboratory parameters in children. Acta Otorhinolaryngol. Ital. 2018, 38, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Brindisi, G.; Marazzato, M.; Brunetti, F.; De Castro, G.; Loffredo, L.; Carnevale, R.; Cinicola, B.; Palamara, A.T.; Conte, M.P.; Zicari, A.M. Allergic rhinitis, microbiota and passive smoke in children: A pilot study. Pediatr. Allergy Immunol. 2022, 33 (Suppl. 27), 22–26. [Google Scholar] [CrossRef] [PubMed]
- Northrup, T.F.; Stotts, A.L.; Suchting, R.; Matt, G.E.; Quintana, P.J.E.; Khan, A.M.; Green, C.; Klawans, M.R.; Johnson, M.; Benowitz, N.; et al. Thirdhand smoke associations with the gut microbiomes of infants admitted to a neonatal intensive care unit: An observational study. Environ. Res. 2021, 197, 111180. [Google Scholar] [CrossRef]
- Kelley, S.T.; Liu, W.; Quintana, P.J.E.; Hoh, E.; Dodder, N.G.; Mahabee-Gittens, E.M.; Padilla, S.; Ogden, S.; Frenzel, S.; Sisk-Hackworth, L.; et al. Altered microbiomes in thirdhand smoke-exposed children and their home environments. Pediatr. Res. 2021, 90, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidi, E.M.; Lappas, A.S.; Tzortzi, A.S.; Behrakis, P.K. Exhaled Breath Condensate: Technical and Diagnostic Aspects. Sci. World J. 2015, 2015, 435160. [Google Scholar] [CrossRef]


| Asthmatic Group n = 19 | Healthy Control n = 19 | |
|---|---|---|
| Swab collection | 12 (63.16%) | 14 (73.68%) |
| EBC † collection | 19 (100%) | 14 (73.68%) |
| Sex (women/men) | 6 (31.58%)/13 (68.42%) | 12 (63.16%)/7 (36.84%) |
| Mean age (years) | 10.3 | 12 |
| Place of living (town/village) | 11 (57.89%)/8 (42.11%) | 10 (52.63%)/9 (47.37%) |
| Overweight (>85 centile of BMI) | 3 (15.79%) | 3 (15.79%) |
| Underweight (<3 centile of BMI) | 1 (5.26%) | 1 (5.26%) |
| Asthma medications daily intake/Inhaled corticosteroids (ICS) treatment on a daily basis | 14 (73.68%)/10 (52.63%) | none |
| Natural birth/cesarean section (cc) | 13 (68.42%)/6 (31.58%) | 7 (36.84%)/12 (63.16%) |
| Inhalant allergies | 17 (89.47%) | none |
| Family history of allergic diseases | 14 (73.68%) | none |
| Active smokers within household | 7 (36.84%) | 4 (21.05%) |
| Taxon | Sample Presence [%] | Mean Relative Abundance [%] |
|---|---|---|
| d__Bacteria;p__Firmicutes;c__Bacilli †,‡,§ | 100.00 | 35.44 |
| d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria | 100.00 | 20.92 |
| d__Bacteria;p__Proteobacteria;c__Alphaproteobacteria | 100.00 | 17.86 |
| d__Bacteria;p__Actinobacteriota;c__Actinobacteria | 100.00 | 13.72 |
| Taxon | Sample Presence [%] | Mean Relative Abundance [%] |
|---|---|---|
| d__Bacteria;p__Firmicutes;c__Bacilli †,‡,§ | 100.00 | 33.63 |
| d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria | 100.00 | 16.63 |
| d__Bacteria;p__Bacteroidota;c__Bacteroidia | 100.00 | 13.35 |
| d__Bacteria;p__Fusobacteriota;c__Fusobacteriia | 96.43 | 11.71 |
| d__Bacteria;p__Actinobacteriota;c__Actinobacteria | 100.00 | 10.62 |
| d__Bacteria;p__Firmicutes;c__Negativicutes | 96.43 | 7.54 |
| d__Bacteria;p__Firmicutes;c__Clostridia | 92.86 | 3.71 |
| d__Bacteria;p__Campilobacterota;c__Campylobacteria | 92.86 | 1.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bar, K.; Żebrowska, P.; Łaczmański, Ł.; Sozańska, B. Airway Bacterial Biodiversity in Exhaled Breath Condensates of Asthmatic Children—Does It Differ from the Healthy Ones? J. Clin. Med. 2022, 11, 6774. https://doi.org/10.3390/jcm11226774
Bar K, Żebrowska P, Łaczmański Ł, Sozańska B. Airway Bacterial Biodiversity in Exhaled Breath Condensates of Asthmatic Children—Does It Differ from the Healthy Ones? Journal of Clinical Medicine. 2022; 11(22):6774. https://doi.org/10.3390/jcm11226774
Chicago/Turabian StyleBar, Kamil, Paulina Żebrowska, Łukasz Łaczmański, and Barbara Sozańska. 2022. "Airway Bacterial Biodiversity in Exhaled Breath Condensates of Asthmatic Children—Does It Differ from the Healthy Ones?" Journal of Clinical Medicine 11, no. 22: 6774. https://doi.org/10.3390/jcm11226774
APA StyleBar, K., Żebrowska, P., Łaczmański, Ł., & Sozańska, B. (2022). Airway Bacterial Biodiversity in Exhaled Breath Condensates of Asthmatic Children—Does It Differ from the Healthy Ones? Journal of Clinical Medicine, 11(22), 6774. https://doi.org/10.3390/jcm11226774






