1. Introduction
Deep learning models are susceptible to noise in training data, as they learn by using stochastic gradient functions. This causes variance errors and may cause overfitting, resulting in low generalisations for validating data. A machine learning technique known as ensemble learning reduces predictive variance by combining the predictions of integrated models. Ensembles are often more accurate than individual classifiers that produce them [
1,
2,
3,
4,
5].
On the other hand, a deep convolutional neural networks (CNNs) drive process is a difficult optimising process that often does not converge. As a result, CNN’s latest drive weights may not show a consistent or optimal performance as the final model weights. To overcome this problem, the average performance of training weights is calculated as many points in the training cycle [
6,
7,
8]. In general, it could be called the average weight prediction based on the method developed by Polyak-Ruppert [
9,
10].
Additionally, every CNN is very sensitive to the volume of training data. The model will learn better if you have high-volume data. A special case of cross-validation is called leave-one-out cross-validation (LOOCV), and it is used to evaluate the efficiency of machine learning models with a small dataset. This is a lengthy and costly process, even though it provides a reliable and impartial estimate of model performance. While very simple in application, there are some limitations in using, as there is no need for its application if a large dataset or mathematically costly method is used. During the application of the LOOCV process, each machine learning model is adjusted at a higher number of times, representing a more robust assessment since each data can participate as the entire test dataset [
11,
12].
In recent years, deep transfer learning with an ensemble of multiple CNNs has been widely used in medical-image processing [
13,
14,
15,
16,
17]. The trained deep ensemble learning represents a single hypothesis. Empirically, ensembles yield better results when significant diversity among the models, even on a small dataset. Therefore, many ensemble methods seek to promote diversity among the combined models. An ensemble indicates different techniques, including simple averaging, weighted-averaging, majority voting (MVOT), bagging, boosting, CNN blocks, randomizing, and stacking using multi-model predictions on the same dataset [
18,
19,
20,
21].
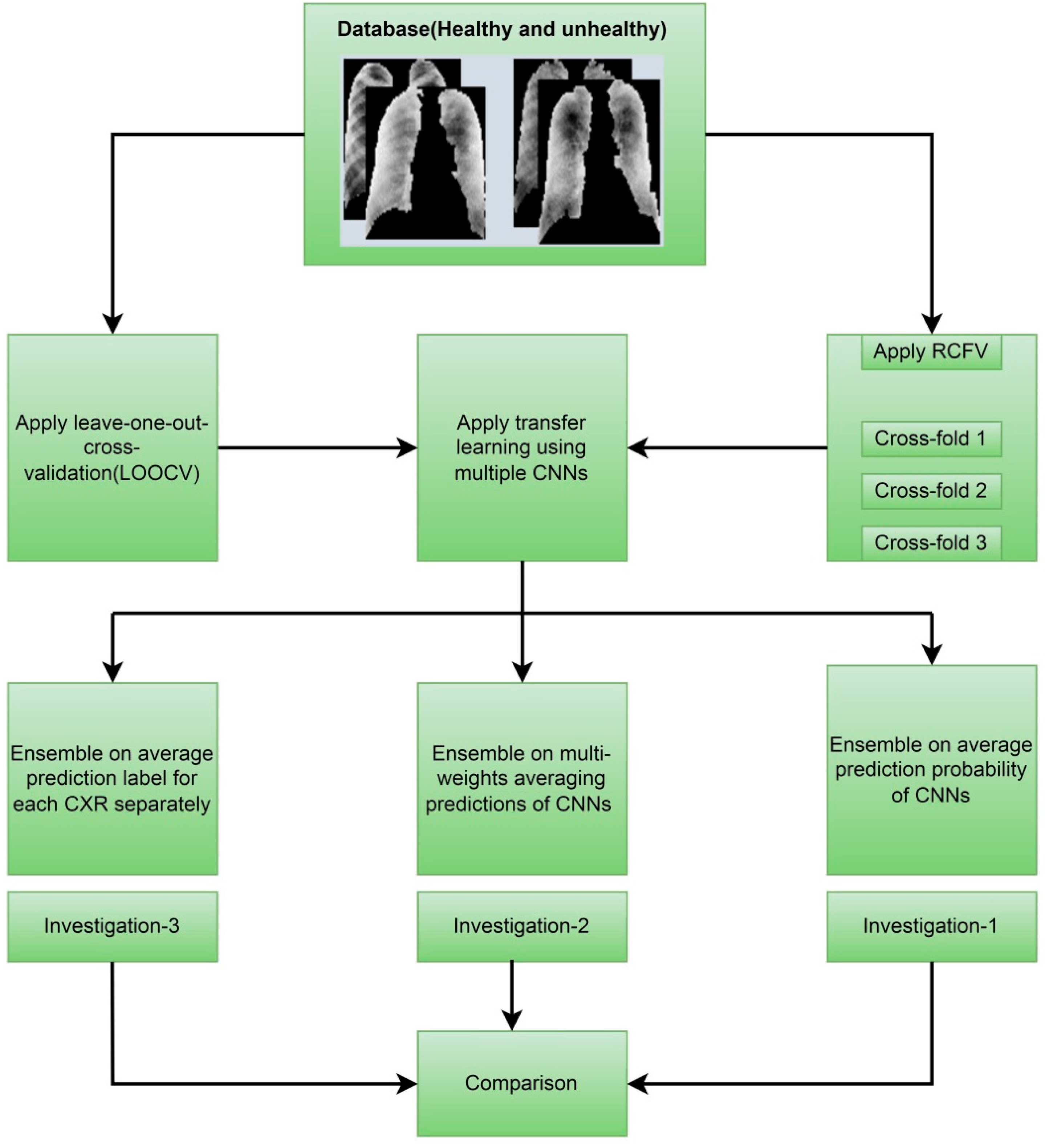
This paper proposed simple averaging, weighted-averaging, and MVOT techniques to detect pneumoconiosis in coal workers’ chest X-ray radiographs (CXRs). The summary of our list of contributions is as follows:
We have used databases of posterior-anterior (PA) CXRs collected from various hospitals by the Commonwealth Scientific and Industrial Research Organisation (CSIRO), Sydney, Australia. To overcome the problems associated with small datasets, we assessed proposed ensemble techniques, simple averaging, weighted-averaging, and MVOT using randomised cross-fold-validation (RCFV) and leave-one-out cross-validations (LOOCV) of the original dataset independently.
In all techniques, transfer learning has been implemented using multiple CNN are namely CheXNet [
22], DenseNet-121 [
23], Inception-V3 [
24], Xception [
25], and ResNet50 [
26]. We proposed ensemble techniques in three investigations: investigation-1 uses simple averaging on RCFV data, investigation-2 uses weighted averaging on RCFV data, and investigation-3 uses MVOT on LOOCV data.
Finally, we compared the investigation’s outcomes using five formulas of statistical measurements [
27], sensitivity, specificity, accuracy, precision, and F1-Score, with state-of-the-art approaches from the literature for the same dataset and highlighted the efficient CNN model in our dataset.
The following
Figure 1 depicts the overall contributions, providing an improved understanding of what we have performed in this study.
Section 2 presents background studies and findings for pneumoconiosis classification on the same dataset using various classical, traditional machine, and deep learning methods. The orientation of the dataset and the detailed methodologies within each investigation are presented separately in
Section 3.
Section 4 provides the outcomes of investigation-1, investigation-2, and investigation-3.
Section 5 summarised the outcomes of the investigation and compared them with state-of-the-art approaches from the background study for the same dataset. The assumptions and limitations are also highlighted there. Finally,
Section 6 provides the conclusion of this research study.
2. Background Study
The abnormality on a chest X-ray of the lung is signified by the increase or decrease in density areas. The chest X-ray lung abnormalities with increased density are also known as pulmonary opacities. Pulmonary opacities have three major patterns: consolidation, interstitial, and atelectasis. Among these, the interstitial patterns of pulmonary opacities are mainly responsible for pneumoconiosis disease [
28]. According to the International Labour Organization’s (ILO) classification, two abnormalities are observed for all types of pneumoconiosis—parenchymal and pleural. Parenchymal abnormalities are indicated by small opacity shape (round or irregular) and size (1.5 mm < diameter (round) < 10 mm and 1.5 mm < widths (irregular) < 10 mm) and large opacities of a round shape and size less than or equal to 50 mm. Pleural abnormalities are mainly indicated by angle obliteration and the diffusion of thickness in the CXR’s wall [
29].
There is no national approach to health screening of coal miners in Australia. In NSW, a chest X-ray is recommended every six years for mine-site workers but it is not mandatory. Medical screening has also failed to detect this potentially fatal disease [
30]. For these reasons, it is desirable to develop an established computer-based automatic system further to provide the quantitative evaluation of pneumoconiosis and serve as an initial screening process and a second opinion for medical doctors.
Past research on the automatic classification of pneumoconiosis classical, traditional machine, and deep learning methods were used. The texture features were mostly classified in classical methods using computer- and ILO-based standard classification [
31,
32,
33,
34,
35,
36,
37,
38,
39,
40,
41]. The profusion of small round opacities and ILO extent properties indicated normal and abnormal lungs. The backpropagation neural networks have been applied to find the shape and size of round opacities from the region of interest (ROI) portions of an image [
42,
43,
44,
45]. X-ray abnormalities were categorised and compared with the results of the standard ILO measurement of the size and shape of the round opacities.
In traditional machine learning, different methods for handcrafted feature extraction, or selection were used. The handcrafted features, such as texture features [
46,
47], from the left –right lung zones [
48,
49,
50,
51] were extracted. After the selection of important features, they were input into different machine learning classifiers, such as support vector machine (SVM) [
49,
52,
53,
54,
55,
56,
57,
58,
59,
60], decision trees (DT) [
55,
56], random trees (RT) [
57,
58,
59,
60], artificial neural networks (ANNs) [
61,
62,
63], K-nearest neighbours (KNN) [
64], self-organizing map (SOM) [
64], backpropagation (BP), radial basis function (RBF) neural networks (NN) [
57,
58,
59,
60,
64,
65], and ensemble classifier [
49,
52,
56].
In recent years, deep learning approaches have achieved state-of-the-art results due to their high dimensional feature representation of data [
66,
67]. Many deep convolutional neural networks performed better than humans, especially in medical image processing [
68]. Such examples include identifying indicators for cancer in blood [
69] and skin [
70,
71], malaria in blood cell [
72], tuberculosis (TB) from chest X-rays [
14,
16,
73], and more specifically pneumoconiosis in chest X-rays [
27,
74,
75,
76,
77,
78,
79,
80].
We have conducted different classical, traditional, and deep learning approaches in our previous published works on the same dataset used in this study. We used the ILO Standard Classification System in classical approaches, and the performance is presented in
Table 1.
We first extracted handcrafted features using different statistical image analysis methods in traditional machine learning approaches. Then, we input these features into different machine learning classifiers, such as support vector machine (SVM), MLP, NN, K-nearest neighbours (KNN), isolation forest, random forest, and ridge [
78]. We show these classifier results in
Table 1.
In deep learning approaches, first, we implemented, with and without transfer learning, convolutional neural networks (CNN) to detect pneumoconiosis. Deep transfer learning was implemented using seven pre-trained CNNs, VGG16 [
81], VGG19, Inception [
24], Xception [
25], ResNet50 [
26], DenseNet-121 [
23], and CheXNet [
22]. Then, we performed a performance comparison between them. The comparison was examined using different effects of dropout rates and different augmentation methods used in DL models, with and without transfer learning, to detect pneumoconiosis. We developed a cascade learning model, which outperforms others and achieved an overall classification accuracy of 90.24%, a specificity of 88.46%, and a sensitivity of 93.33% for detecting pneumoconiosis using generated synthesised images from real segmented CXR databases. We have also summarised deep CNNs results in
Table 1. The previous studies showed that the deep transfer learning performance of Inception-V3, Xception, ResNet50, DenseNet, and CheXNet was satisfactory compared to classical and traditional approaches.
3. Datasets and Methods
The first part of this section discusses our dataset and how it was processed using cross-validation to perform ensemble techniques. In contrast, the rest of the section describes the techniques used in three investigations.
3.1. Materials
Out of a collaboration between the University of Newcastle and the Commonwealth Scientific and Industrial Research Organisation (CSIRO) data61 Sydney NSW, Australia, chest X-ray image datasets were built with associated diagnostic labels for this study. CSIRO data61 collected the data from Coal Services Health NSW, St Vincent’s Hospital, Sydney, and Wesley Medical Imaging, Queensland. The publicly available NIOSH teaching chest X-ray dataset and ILO Standard Radiographs (International Labour Organization, (ILO) Genève, Switzerland) were also used to develop parts of the small dataset DL model. All radiographs used in this study are posterior-anterior (PA) radiographs. Seventy-one PA chest radiographs with small parenchymal opacities consistent with pneumoconiosis and 82 PA chest X-rays belonging to normal individuals were used. All data were collected from coal mine workers, including males and females. We conducted ensemble learning using randomised cross-fold-validation and leave-one-out cross-validation. The details are in the following subsections:
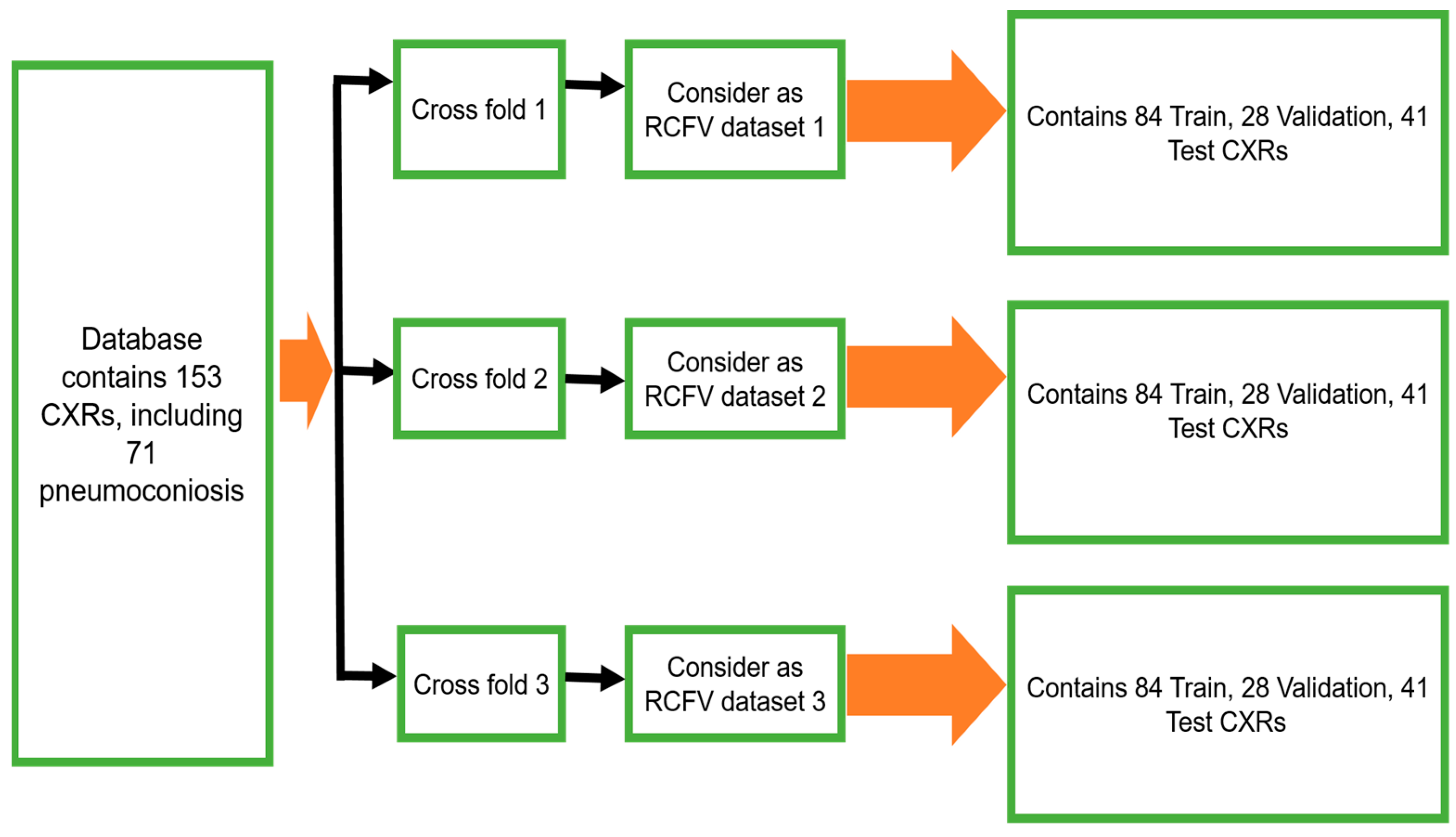
3.1.1. Randomised Cross-Fold-Validation
To maintain the balance of training data, 112 X-rays (56 normal and 56 pneumoconiosis) were used for training and 41 X-rays (26 normal and 15 pneumoconiosis) were used for testing. Twenty-five percent of training data were kept as a validation dataset for selecting the best model weights based on validation performance. We continued the randomised selection three times and then organised our total dataset into three different folds, namely, as randomised cross-fold-validation (RCFV) dataset 1, dataset 2, and dataset 3, as shown in
Figure 2. Therefore, we defined this cross-validation simply as RCFV.
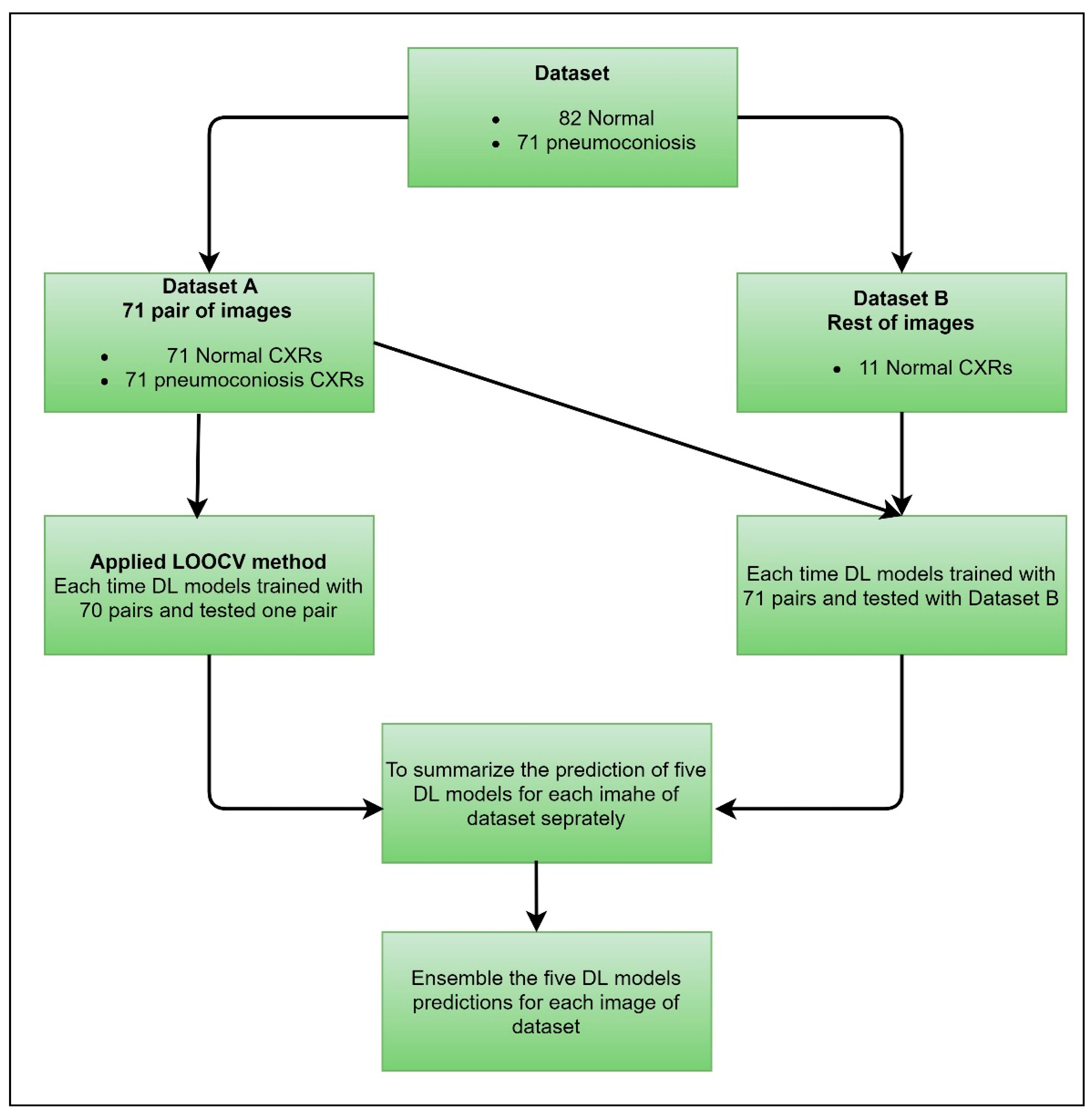
3.1.2. Leave-One-Out Cross-Validation
We proposed a case of cross-validation (LOOCV), which is used to assess the effectiveness of machine learning models with the same dataset. We organised our dataset into two groups, dataset A and dataset B, clearly mentioned in
Figure 3. Dataset A contained 71 pairs of images, including an equal number of normal and abnormal (pneumoconiosis) CXRs. Therefore, the remaining 11 normal images were in dataset B. As a result, no correlation exists between the pairs of images.
3.2. Methods
The proposed ensemble techniques, simple averaging, weighted averaging, and MVOT, were independently conducted using RCFV and LOOCV datasets. In all techniques, transfer learning was analysed by the same CNNs, namely CheXNet, DenseNet-121, Inception-V3, Xception, and ResNet50. We organized our proposed method into three investigations, as stated below.
3.2.1. Investigation-1: An Ensemble Learning Using Simple Averaging through RCFV Datasets
The deep learning model shows each test element’s probability value within the range [0, 1] during the forecast. Those fractional probabilities are converted to predict class labels using a threshold value condition. An ensemble is mapped using several CNNs model prediction probabilities as a combined decision instead of individually. Therefore, each value of the testing data was predicted by multiple models at once. After that, their average predictive probability interval between [0, 1] indicates the ensemble’s performance.
In this investigation, we implemented deep transfer learning throughout the ensemble using simple averaging of the probability of detection of pneumoconiosis predicted by five CNN models: CheXNet, DenseNet, Inception-V3, Xception, and ResNet50. Afterwards, we calculated the average prediction probabilities on the same RCFV testing datasets 1 to 3, as demonstrated in
Figure 2.
CNNs employ a stochastic learning algorithm to optimise training randomly. The optimisation is based on selecting the loss function while the model has been designed. The purpose of the loss function is to determine whether the model is operating properly or incorrectly. The cost function within the CNNs determines the difference in losses between true and predicted values. We applied the regularisation technique to reduce the complexity of a neural network model during training and, thus, prevented overfitting. There are very popular and efficient regularisation techniques called L2. The regularisation term is weighted by the scalar lambda divided by 2 m and added to the regular loss function chosen for the current task. This leads to a new expression for the loss function, as shown in the following Equation (1):
where
λ denotes the regularization parameter, and its value may optimise the learning rate for improved predictions. L2 regularisation is also known as the weight decay as it forces the weights to decay towards zero (but not exactly zero).
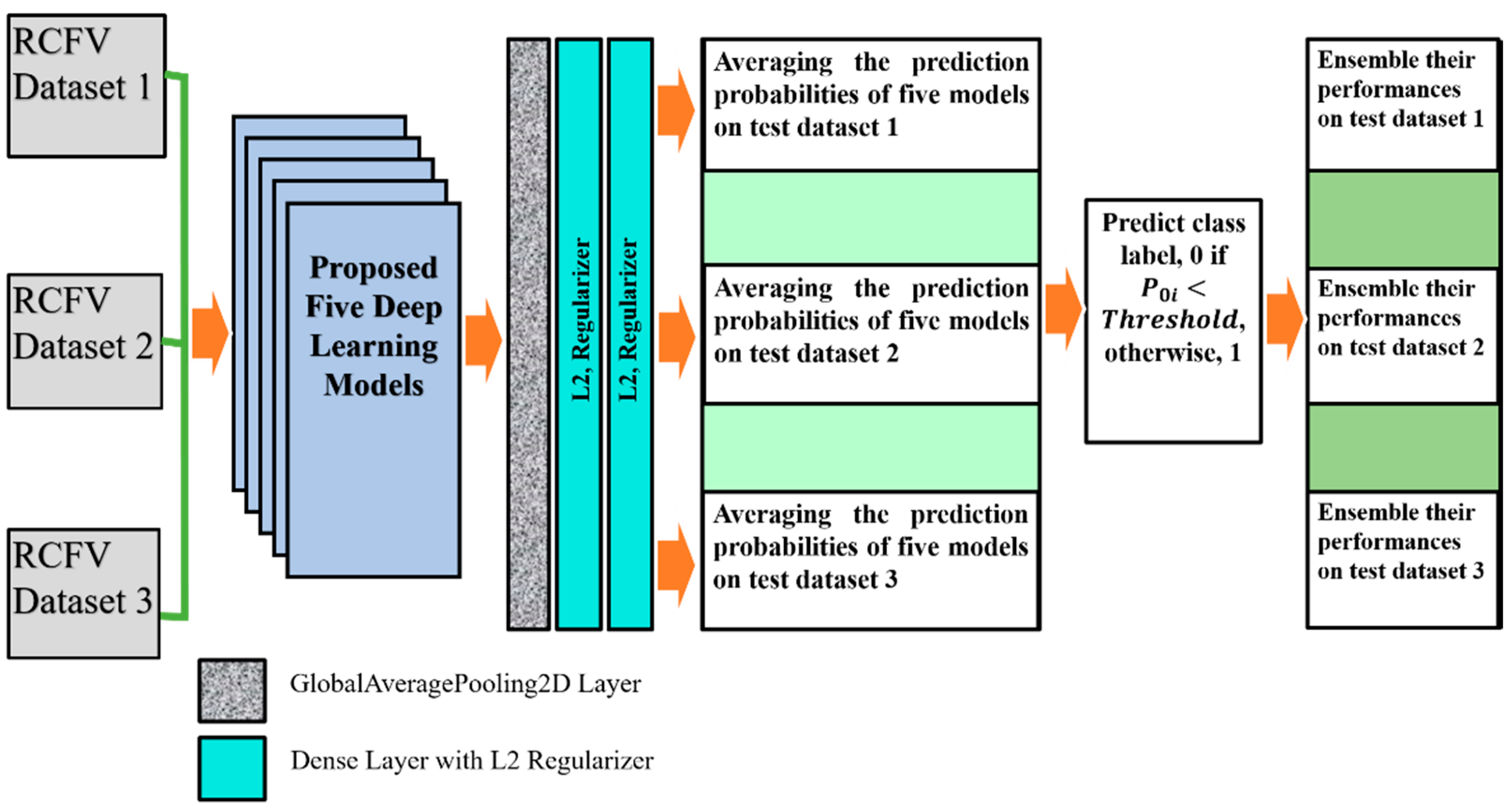
After taking the output of each of the five models, one GlobalAveragePooling2D layer was added. Three dense layers, with all their output nodes, were connected with all nodes of the next layer. Global Average Pooling is a transaction that computes the average performance of each entity map in the preceding layer. This relatively simple operation helps convert the data into a one-dimensional vector and avoids the overflow of features. There are no trainable parameters, similarly to the Max polling operation.
For L2 (0.001), two regularisers were used with the first-two dense layers for better optimisation with the proposed models. The last layer of the classifier used a sigmoid activation function and output probability scores for each class—normal and pneumoconiosis (see
Figure 4). We used 512 × 512 X-ray input forms for each proposed CNNs architecture, where the output of the prediction probability value ranged between [0, 1]. The regular loss-function, binary cross-entropy with an Adam optimiser of the learning rate, 0.0001, was also used during training.
We trained each DL model up to 50 epochs and used the last weights to find the prediction probability of normal and abnormal CXRs. For instance, in RCFV dataset-1, we applied five models independently and then calculated their prediction probabilities separately. Next, we calculated the average of their probability values for each unique test image using mathematical Equation (2). If the average value
, then its predicted label changes to 0; otherwise, it is 1, where
for normal images and
for pneumoconiosis images.
The ensemble performances of five models, CheXNet, DenaseNet, Inception-V3, Xception, and ResNet50, were computed using confusion matrix values, true positives, false negatives, true negatives, and false positives. The ensemble performance for RCFV datasets 2 and 3 was calculated according to the same process used for dataset 1. The details of the proposed workflow are demonstrated in
Figure 4. The last three columns illustrate the direction of the average probability forecasts, the forecast labels, and the ensemble performance of the five models across three different cross-datasets.
3.2.2. Investigation-2: An Ensemble Learning Using Weighted Averaging through RCFV Datasets
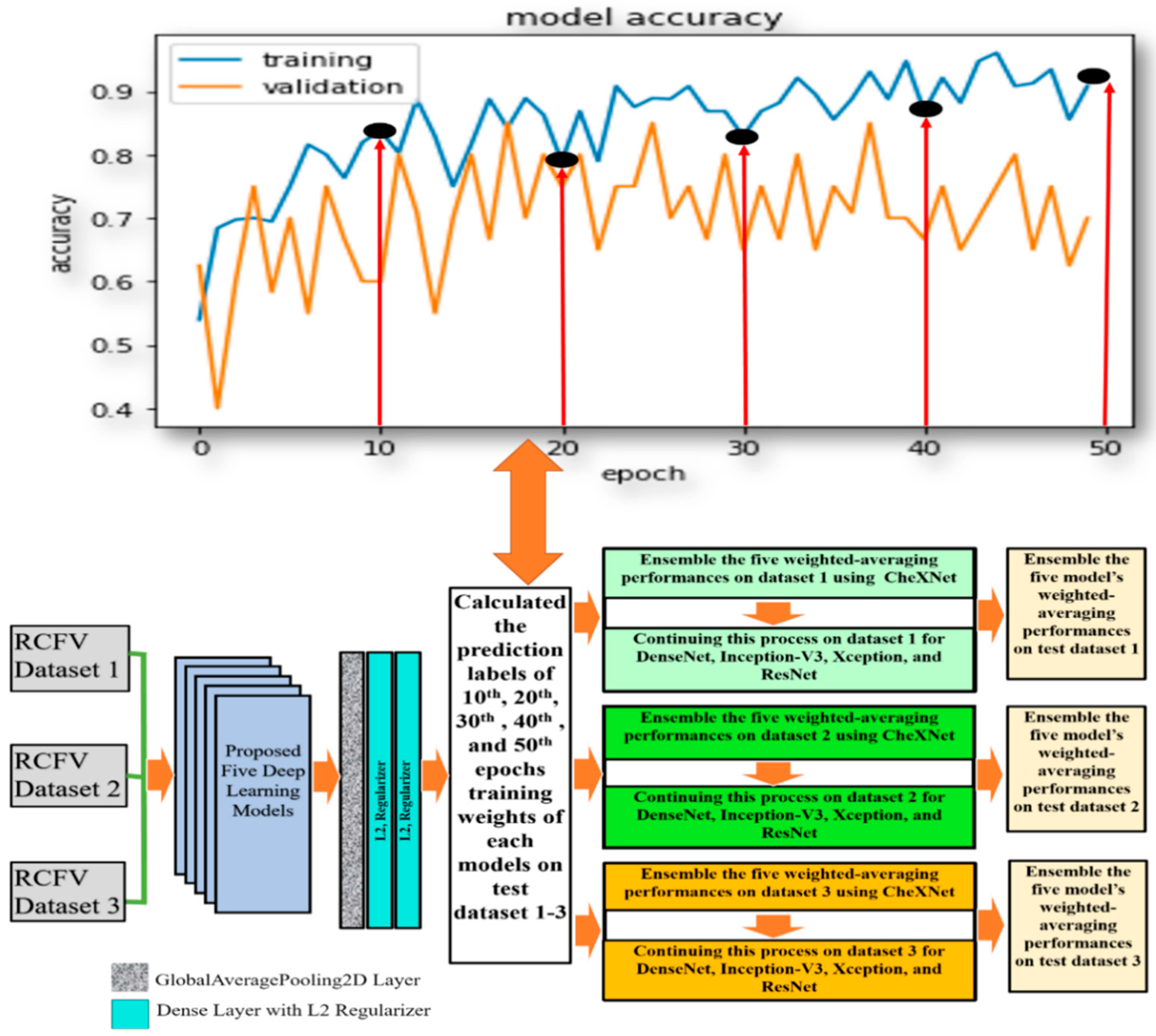
With the method used in the previous investigation, we investigated multi-model ensemble learning using the latest drive weights of each model in detecting pneumoconiosis diseases from CXR. We replicate the investigation in this section using the same five models, CheXNet, DenseNet, Inception-V3, Xception, and ResNet50, used in investigation-1. To find the optimal solution for pneumoconiosis detection, we carried out ensemble learning using the combination of the weighted average and majority voting techniques. Here, we focused on its different training epochs in calculating the weighted average ensemble for a single model. We kept the same training process and dataset, as described in investigation-1. In calculating a weighted average on a single model, we used the specified weights from the epochs (10th, 20th, 30th, 40th, and 50th set) for each proposed model, as defined in the central white box in
Figure 5.
For instance, in dataset 1, we trained the CheXNet model independently on training data and then computed the five sets of prediction labels of its 10th, 20th, 30th, 40th, and 50th epochs’ weights with test data. The weighted average ensemble prediction labels of the CheXNet were found using the majority voting (MVOT) decision on these five sets of predictions. As a result, if, and only if, the weight of the majority says that a BL image is BL, then the ensemble decision is BL; otherwise, it is normal.
Likewise, we continued this process for dataset 1 for DenseNet-121, Inception-V3, Xception, and ResNet50 models, as described in the second last column in
Figure 5. Finally, every single model’s weighted average ensemble return was used to calculate the multi-model ensemble for dataset-1. To accomplish this, MVOT was also applied to the five independent sets of five weighted average prediction labels in the models, as described in the last column of
Figure 5.
Similarly, we conducted this process for the testing datasets 2 and 3 and compared weighted-averaging ensemble performances of a single and integrated model using true positive, false negative, true negative, and false positive values from the predicted confusion matrix.
3.2.3. Investigation-3: An Ensemble Learning Using MVOT through the LOOCV Dataset
In this investigation, we implemented LOOCV to select a robust DL model from CheXNet, DenseNet, InceptionV3, Xception, and ResNet50 by using our organised dataset, as discussed in
Section 3.1.2, representing the best competence for detecting detection pneumoconiosis from CXRs.
Figure 6 shows how we handled training and testing using DL applications for each cross-data application. For dataset A, every DL model has trained on 70 pairs of images and tested one pair. We continued the process 71 times automatically. We trained the same model with dataset A for dataset B and then tested the performances on dataset B. Next, we independently calculated each DL model performance for each image using a combination of datasets A and B. Finally, each model’s predictions ensemble return is used to calculate the multi-model ensemble for all data in LOOCV using a simple MVOT technique. Therefore, if the majority of models predict as “normal”, then its ensemble prediction is defined as a “normal” or, otherwise, “abnormal” lung.
Finally, we compared the MVOT-based ensemble performances of a single and integrated model using true positive, false negative, true negative, and false positive values from the predicted confusion matrix.
5. Discussion
From investigation-1 to investigation-3, we applied different methodologies to improve pneumoconiosis detections in CXRs. In
Table 14, we summarised the best statistical combination derived from the investigated ensemble techniques. Here, the lower the standard deviation (SD), the closer the values are to the mean of the set of investigations. The higher the SD, the wider the range of investigations. All techniques were processed to find the optimal solution for detecting pneumoconiosis from X-ray images. Investigation-1 had the best combination of accuracy of 82.93%, a sensitivity of 88.00%, a specificity of 75.00%, a precision of 84.62%, and an F1-score 86.27%, as summarised in
Table 14, even though these are lower values than the individual performances of the CheXNet model without ensemble learning, as shown in
Table 2. When compared to an individual, the performance of the ensemble learning technique in the first investigation did not improve pneumoconiosis detection’s accuracy.
In investigation-2, we found that the detection performances slightly improved in the ensemble of multi-weighted averaging on a single model, CheXNet, as demonstrated in
Table 14, which has shown improved statistical combinations than the other methodological findings in investigation-1. In investigation-3, we first observed that the same CheXNet model independently improved the accuracy from 87.80 to 90.20%. In addition, the proposed ensemble learning obtained 91.50% peak performance for detecting pneumoconiosis in coal workers from CXRs with state-of-the-art methods. Investigation-3 had an excellent success rate of more than 90.00% for all five observations. Therefore, as ground truth, our proposed ensemble learning outperformed other state-of-the-art classical and traditional machine and deep learning methods, as summarised in
Table 1.
The University of Newcastle’s (Australia) high-performance computing (HPC) system was used for all investigations. Python 3.6 was used to run deep learning platform Keras 2.2.2 and machine learning platform Scikit-learn 0.19.1. In addition, we also looked at how long it took to train five different models, CheXNet, DenseNet, Inception-V3, Xception, and ResNet50, which took 19, 20, 16, 13, and 11 min, respectively, for 50 epochs. Furthermore, the model training and validation performance were monitored from the continuity of average
and standard deviation
of accuracies and losses on each epoch.
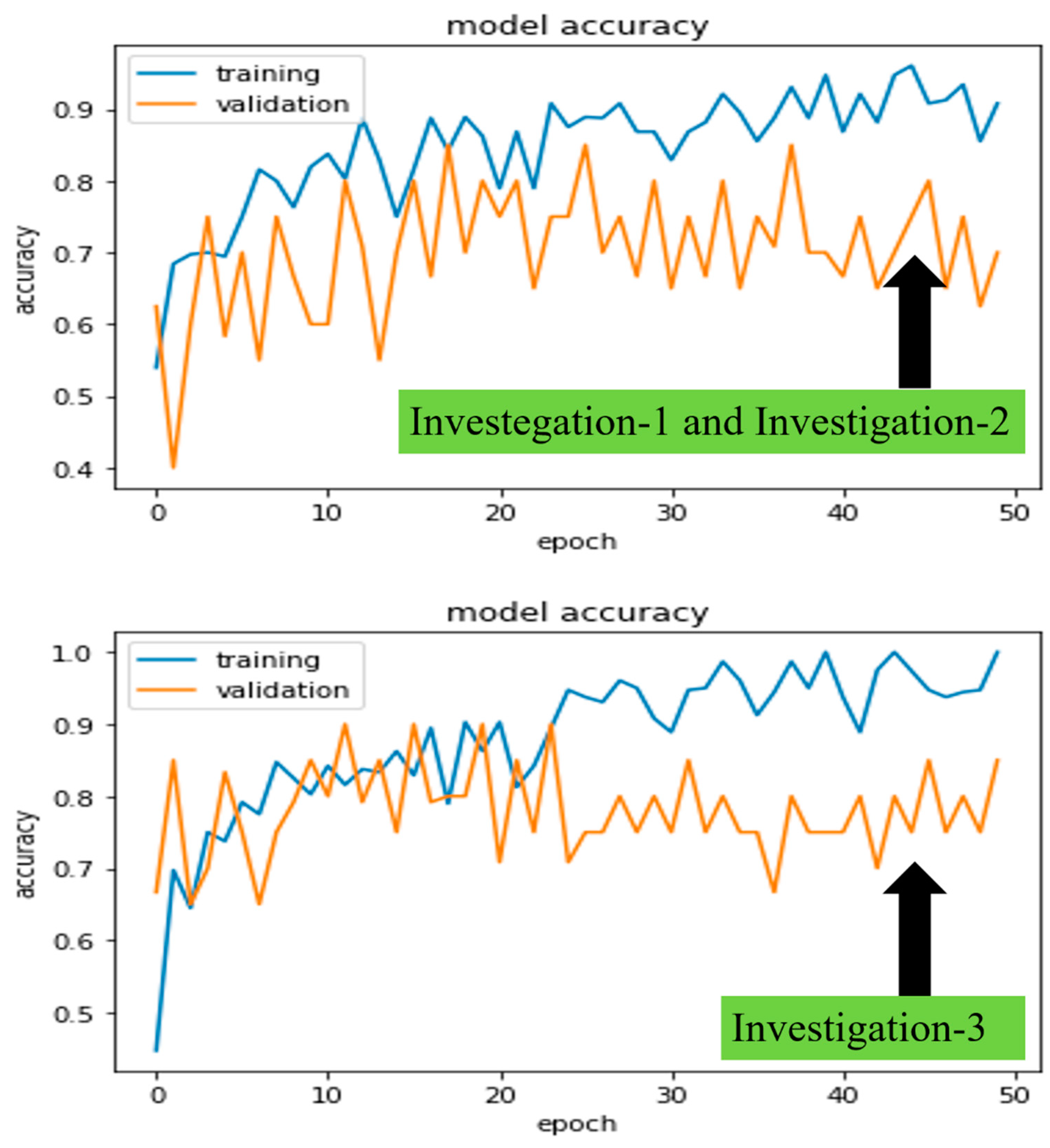
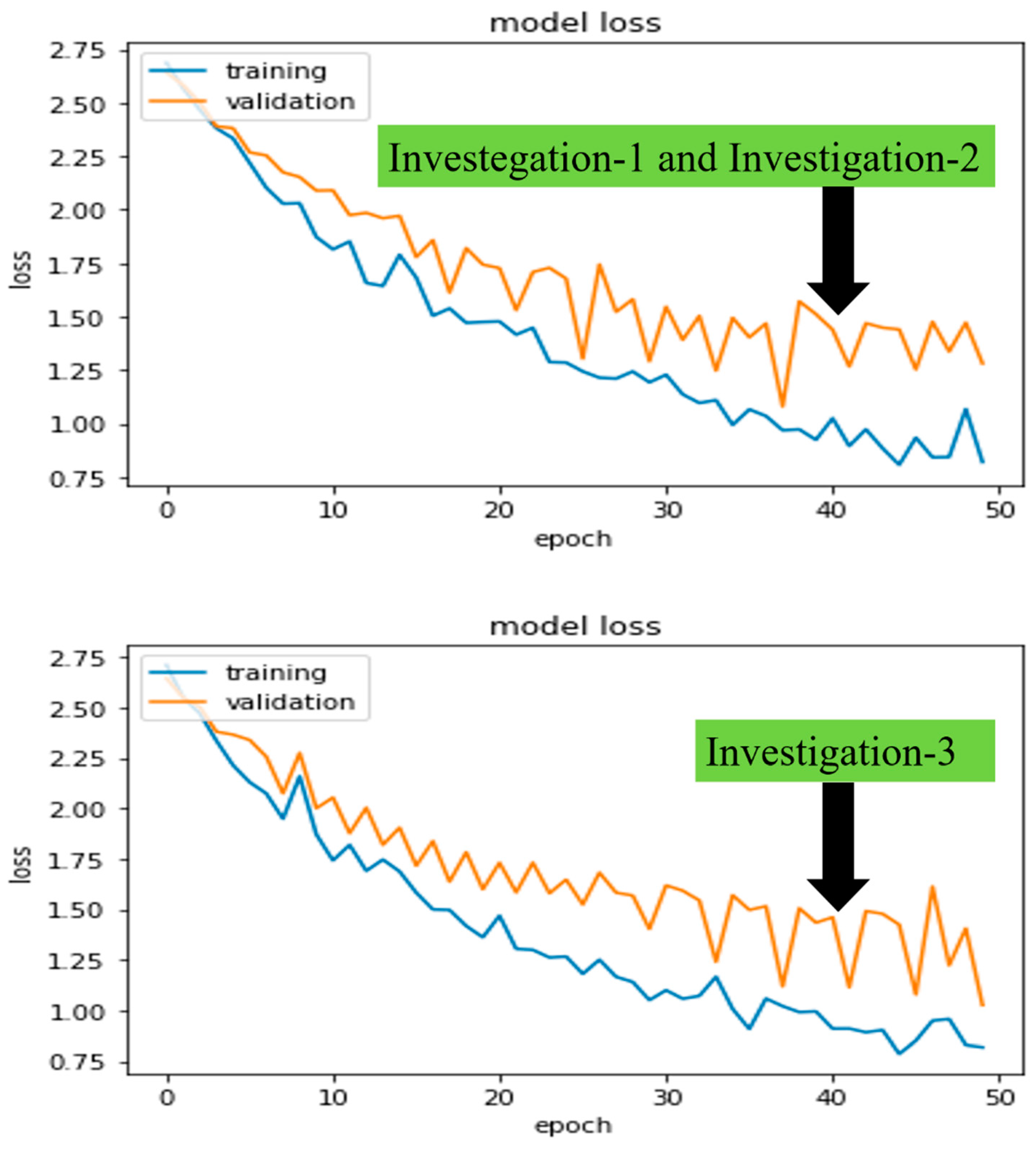
Figure 7 and
Figure 8 demonstrated the robust model, CheXNet training, and validation accuracies and losses for all proposed investigations. The investigated DL model was validated using Equations (3) and (4), where
indicates the
epoch’s
accuracy or loss values of a trained model. By comparing
and
, we were able to pick the best-trained model to perform the test. In the following paragraphs, we present these values for the same robust model, CheXNet.
In investigations 1 and 2, the and of training and validation accuracies were calculated as , , , and approximately. Similarly, losses were calculated as , , , and approximately.
Finally, in investigation 3, the and of training and validation accuracies were calculated as , , , and approximately. Similarly, losses were calculated as , , , and approximately.
The de-identified private CXRs database was gathered from the Coal Services Health NSW, St Vincent’s Hospital, Sydney, Wesley Medical Imaging, Queensland, and ILO standard, which are supposed to comprise 100% correct assumptions for this research study. However, our proposed ensemble technique achieved an accuracy of 91.50%, a true positive rate (sensitivity) of 90.14%, and a true negative rate (specificity) of 92.68%, which were, on average, 10% lower than our assumptions.
This research study has a few limitations as well. First and foremost, the CSIRO’s Sydney, Australia, office anonymised this private dataset, which cannot be accessed without their written consent [
77]. However, if the proposed dataset is large, the outperformed ensemble investigation-3 may be mathematically expensive and take longer to obtain a robust assessment than other investigations. Future studies will focus on testing the proposed model in a clinical setting and gathering input to improve the methodology further. Furthermore, we also recommend some form of variations in tool coupling to at least retain the best case.