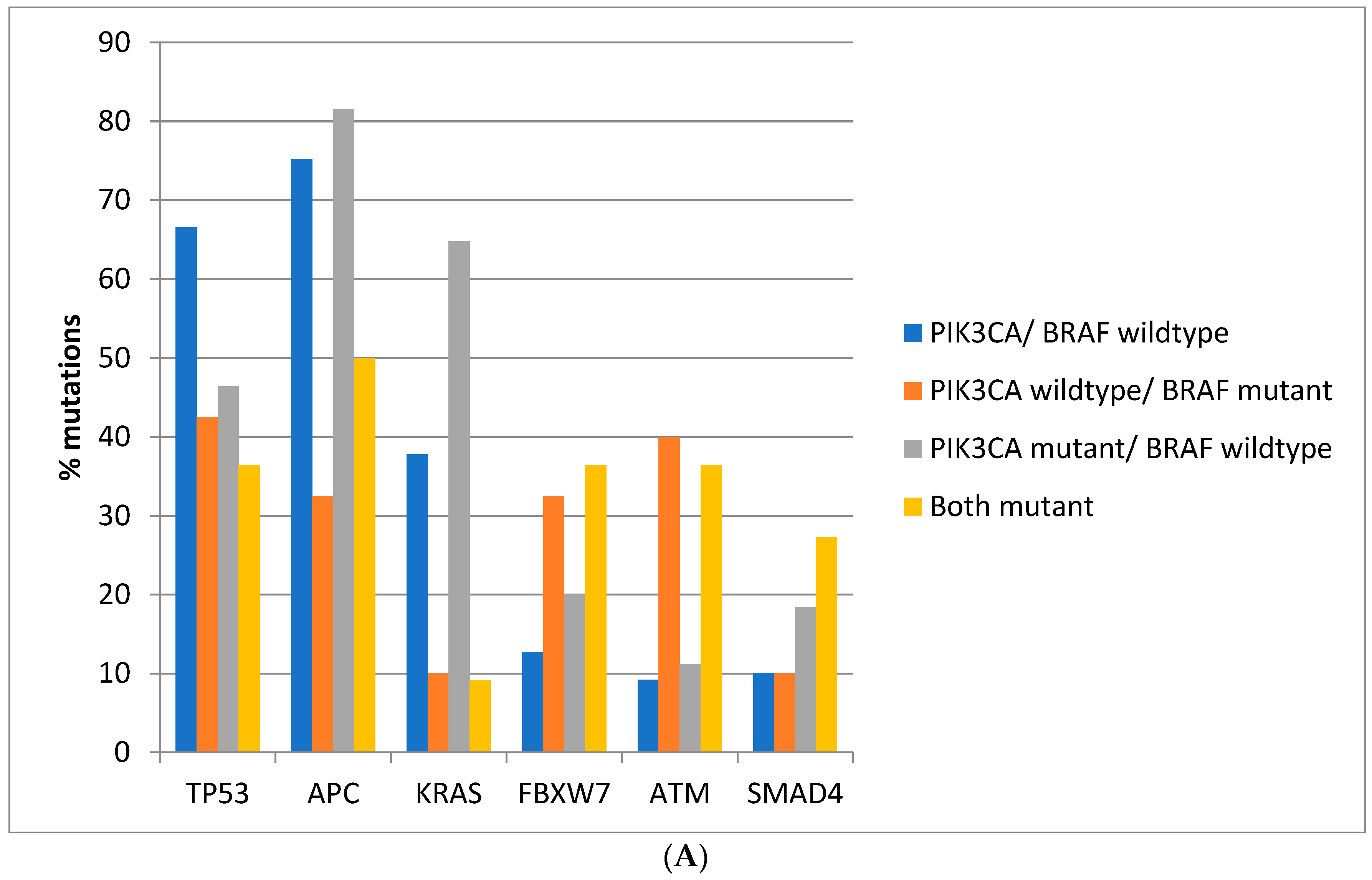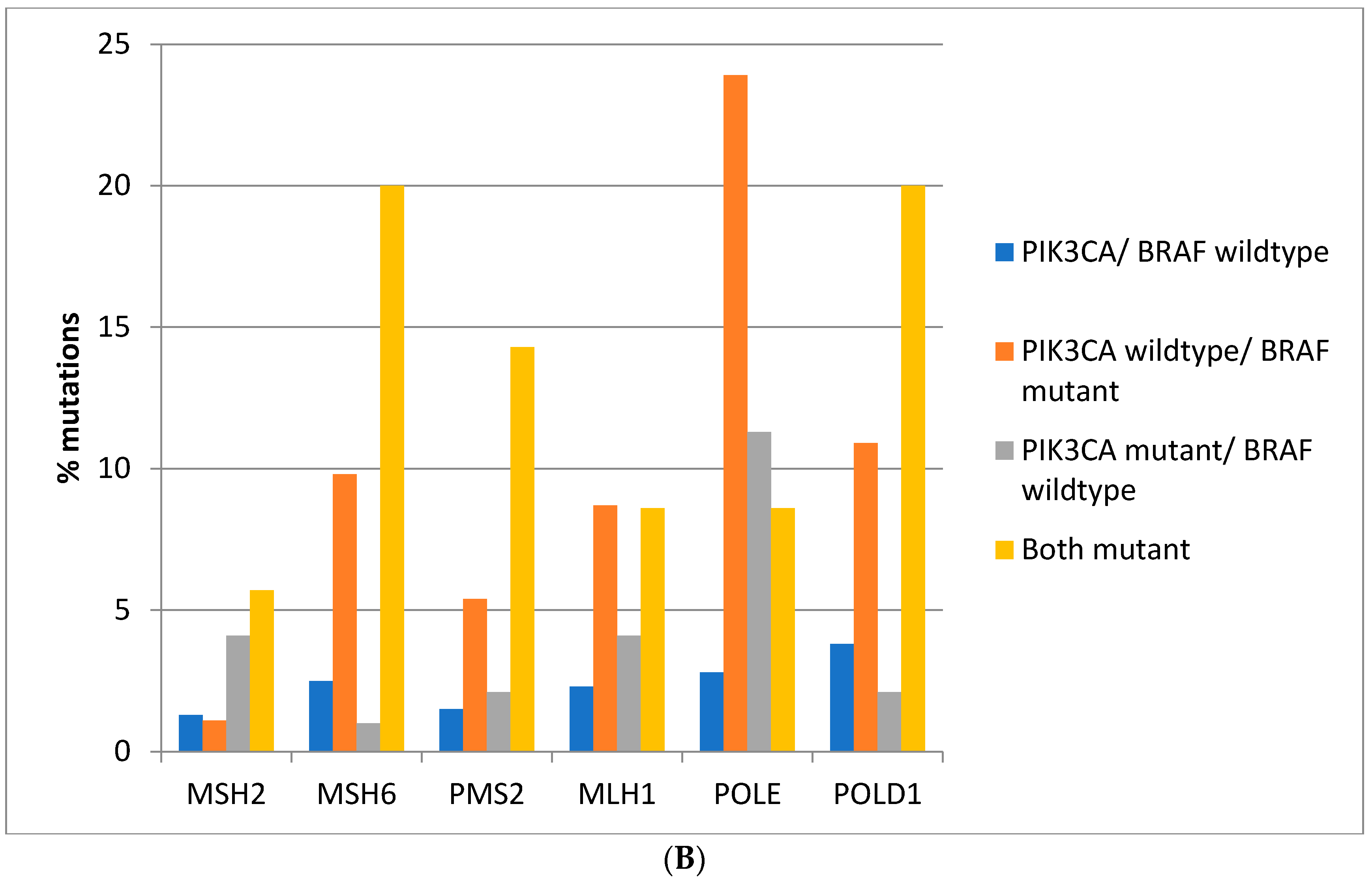1. Introduction
BRAF is a serine threonine kinase which is part of the RAF/MEK/ERK cascade transducing signals from growth factors for cell proliferation and apoptosis inhibition [
1]. Two other RAF homologues, ARAF and CRAF (also known as RAF1), exist in human cells encoded by distinct genes.
BRAF is encoded by a gene at human chromosome 7q34 while genes encoding for ARAF and CRAF are located at chromosomes Xp11.3 and 3p25.2, respectively. The three proteins can homodimerize or heterodimerize with another protein of the family [
2]. Wild type RAF proteins are activated by RAS to dimerize and phosphorylate kinases of the MEK family which in their turn activate ERK kinases. ERK kinases inhibit apoptosis and promote proliferation by activating transcription factors, the prototypic being the AP1 (Activating Protein 1) complex.
In contrast to other RAF homologues,
BRAF is commonly mutated in cancers. The most common canonical mutations at position V600 are currently effectively targeted with small molecule inhibitors in malignant melanoma and other cancers [
3]. Mutations in
BRAF occur in about 10% of colorectal cancers and have been examined for therapeutic targeting with the same small molecule inhibitors used in melanoma [
4]. A
BRAF inhibitor is currently approved in combination with the anti-EGFR monoclonal antibody cetuximab for second line treatment of metastatic colorectal cancers with
BRAF mutations [
5]. However, as shown in the registration trial, only one in five patients respond to therapy, despite all patients bearing the targeted
BRAF mutation [
6]. Development of resistance leading to progression in responding patients is also the rule [
7].
The other signal transduction cascade activated by growth factors and receptor tyrosine kinases is the PI3K/AKT/mTOR cascade. The cascade is triggered through the activation of kinase PI3K by diverse receptor tyrosine kinases, such as the Epidermal Growth Factor Receptor (EGFR) family receptors, the Fibroblast Growth Factor Receptor (FGFR) family receptors, the Platelet-derived Growth Factor Receptor (PGFR), and the Insulin-like Growth Factor Receptor (IGFR) [
8]. Production of the lipid phosphatidylinositol-3,4,5-triphosphate in the cell membrane by activated PI3K enables proximity of kinase PDK1, the mTORC2 complex and kinase AKT, resulting in phosphorylation and activation of the latter [
9]. AKT has several substrates involved in carcinogenesis and results in the promotion of cell proliferation and apoptosis inhibition. Through down-stream activation of the mTORC1 complex, AKT positively regulates protein translation and cell growth [
10,
11]. The most common mutation in the PI3K/AKT pathway is in the gene encoding for the alpha catalytic sub-unit of PI3K,
PIK3CA, occurring in about one in four colorectal cancers [
12]. Half of the activating mutations in
PIK3CA occurring in colorectal cancers concern hotspots of amino acid positions E545, E542 and Q546 of the helical domain or position H1047 of the kinase domain, while the other half is distributed across the gene [
12]. Mutations in
PIK3CA are not mutually exclusive with
BRAF mutations and occur in both
BRAF mutated and wild type colorectal cancers. Thus, these mutations may play a role in resistance of a subset of
BRAF mutated cancers to targeted therapies, through the extensive crosstalk of the KRAS/RAF/MEK/ERK and the PI3K/AKT/mTOR pathways [
13,
14]. For example, mTORC1 activation by PI3K/AKT provides a feedback inhibition of receptor tyrosine kinase signaling through Insulin Receptor Substrate proteins IRS1 and IRS2 and adaptor protein GRB10 (Growth Factor Receptor Bound protein 10) [
15,
16]. Other crosstalk points between the pathways are mediated by activation of mTORC1 and inhibition of TSC1 and TSC2 (Tuberous Sclerosis Complex 1 and 2) by MEK kinases and activation of PI3K by KRAS [
17].
This paper examines colorectal cancers with BRAF mutations with and without concomitant PIK3CA mutations using data from published genomic studies. A detailed understanding of the molecular processes associated with these distinct subsets of colorectal cancers and additional molecular aberrations that support BRAF mutations in the neoplastic process is needed for improving therapeutic results.
3. Results
In total, 22 of 534 profiled colorectal cancer patients in the TCGA cohort (4.1%) had mutations in both BRAF and PIK3CA oncogenes. This represents 35% of the total number of cases (62 cases) with BRAF mutations. A total of 15 of the 22 patients had V600E BRAF mutations (with additional non-classical BRAF mutations in 2 of the 15 patients). Three patients had mutations at codons 594 and 597 of BRAF that are also considered pathogenic (categorized as classes II and III as opposed to codon V600 mutations that are categorized as class I) and 4 patients had other BRAF mutations of unknown significance. In 20 of the 22 patients, PIK3CA mutations that are considered pathogenic were present, while in 2 patients PIK3CA mutations were evaluated as of unknown significance. Three cases had PIK3CA mutations at hotspot amino acid position E545, 1 patient at position E542, and 10 patients had PIK3CA mutations at hotspot position H1047. The six remaining cases with PIK3CA pathogenic mutations were mutations in amino acid positions other than E545, E542, and H1047.
In the DFCI cohort, 35 of 619 patients (5.7%) had mutations in both BRAF and PIK3CA oncogenes. This represented 27.6% of the total number of cases with BRAF mutations in the DFCI cohort. In total, 28 of the 35 patients had classic V600E mutations, while 6 patients had non-classic mutations also considered pathogenic (in 1 patient with two additional mutations of unknown significance) and 1 patient had a BRAF mutation of unknown significance. The most common PIK3CA mutation in BRAF mutated cases were at H1047 but the predominance of H1047 position mutations compared to other classic hotspot positions was less pronounced than in TCGA. Mutations in the hotspot amino acid positions E542 (4 patients), E545 (3 patients), Q546 (6 patients), and H1047 (6 patients) of PIK3CA were present in 19 of 35 BRAF/PIK3CA double mutant patients, and 12 additional patients had pathogenic non-hotspot PIK3CA mutations. Four patients had mutations of unknown significance.
Compared with colorectal cancer patients without
BRAF mutations,
BRAF mutant colorectal cancer patients in the TCGA cohort presented at a more advanced age (
Table 1). This was true independently of whether they had concomitant
PIK3CA mutations. However, no significant differences were observed in age at presentation of
BRAF mutant colorectal cancer patients in the DFCI cohort (
Table 2).
BRAF mutant patients and even more so
BRAF/
PIK3CA double mutant colorectal cancers in TCGA tended to be of earlier stage (stages I and II) than
BRAF wild type disease (
p = 0.003,
Table 1). In the double mutant group, 86.4% of patients presented with stage I or II cancers. This was also observed in the DFCI cohort, where
BRAF mutant colorectal cancers with or without concomitant
PIK3CA mutations presented less often as stage III or IV than colorectal cancer with both genes on a wild type configuration (
Table 2). At odds with TCGA, the group with mutant
PIK3CA and wild type
BRAF had similar prevalence of stage III and stage IV presentation, compared with the double mutant group in the DFCI cohort.
BRAF mutant cancers were located almost exclusively in the colon and rarely in the rectum. Rectal cancers constituted 25.9% and 22.2% of cases in the whole TCGA and DFCI cohorts, respectively.
BRAF mutant cancers were more commonly MSI high or POLE positive. Compared with
BRAF mutant/
PIK3CA wild type cancers, double mutant cancers were even more often MSI or POLE positive (
Table 3). Consistent with the high prevalence of MSI and POLE subtype,
BRAF/
PIK3CA double mutant colorectal cancers in TCGA had a high Tumor Mutation Burden (TMB) above 200 in 86.4% of cases (
Table 3).
BRAF mutant cancers with wild type
PIK3CA had TMB above 200 in 70% of cases. In the DFCI cohort, 77.1% of
BRAF/
PIK3CA double mutant colorectal cancers and 65.2% of
BRAF mutant cancers with wild type
PIK3CA have a TMB above 200 (
Table 4). In contrast, in the TCGA cohort, colorectal cancers with wild type
BRAF had TMB above 200 in 16.8% of cases when
PIK3CA was mutant and in 7.3% of cases when
PIK3CA was wild type (
Table 3). In the DFCI cohort, colorectal cancers with wild type
BRAF had TMB above 200 in 35% of cases when
PIK3CA was mutant and in 18.5% of cases when
PIK3CA was also wild type (
Table 4).
In the metastatic colon cancer study cohort published by MSKCC, with 1099 patients,
BRAF mutations were present in 10.8% of patients. Among these patients, about one third or 3.4% of the whole cohort had
BRAF and
PIK3CA double mutant cancers. Patients in this study, although all with metastatic disease, had genomic evaluation either from a metastatic site biopsy (n = 522) or from the primary tumor (n = 596, a few patients had biopsies from both sites).
BRAF mutated cancers evaluated from primary tumor biopsy showed MSI high in 51.9% of samples while in
BRAF mutated cancers evaluated from metastatic site biopsies showed MSI high in 11.5% of samples (Fisher’s exact test
p = 0.02,
Figure 1). These data suggest that, in MSI high
BRAF mutated colorectal cancers, metastatic disease develops from clones that have become MSS or had been MSS from the beginning.
BRAF mutant colorectal cancers show a higher prevalence of chromosomal stability as determined by low Aneuploidy Score (AS) and low Fraction of Genome Altered (FGA) score.
BRAF/
PIK3CA double mutant cancers had higher rates of chromosomal stability than
BRAF mutant cancers with wild type
PIK3CA (
Table 3). On the other hand,
BRAF wild type colorectal cancers show higher rates of chromosomal instability (CIN) as determined by higher AS and FGA scores. The most frequent amplified locus in colorectal cancer at chromosome 20q11.21 is observed exclusively in cancers with wild type
BRAF, with or without mutations in
PIK3CA (in about 10% of cases in TCGA, not shown). The CpG Island Methylator Phenotype (CIMP) is more prevalent in
BRAF mutant cancers than in
BRAF wild type cancers (
Table 4).
Regarding the prevalence of mutations in other commonly mutated oncogenes and tumor suppressors, there are significant differences between colorectal cancers depending on the presence of
BRAF and
PIK3CA mutations. Mutations in the tumor suppressor
TP53 are more prevalent in colorectal cancers of the TCGA cohort with
BRAF and
PIK3CA wild type (66.6%) compared with cancers with either or both
BRAF and
PIK3CA mutated, where mutations in
TP53 are encountered in 36.4–46.4% of cases (Fisher’s exact test
p < 0.0001,
Figure 2A). Similarly, in the DFCI cohort, the prevalence of
TP53 mutations was 57.7% in
BRAF and
PIK3CA wild type cancers and 39.2–43.5% in cancers with either or both
BRAF and
PIK3CA mutated (Fisher’s exact test
p < 0.0001,
Figure 2B). Mutations in the tumor suppressor
APC are more prevalent in
BRAF wild type colorectal cancers independently of the presence or absence of concomitant
PIK3CA mutations in both cohorts (Fisher’s exact test
p < 0.0001 for the comparisons in both cohorts,
Figure 2A,B). Oncogene
KRAS mutations are rather rare in
BRAF mutant colorectal cancers, occurring in 10% of such cancers in TCGA and in about 5% in the DFCI cohort, compared with 37.8% and 31.6% in cancers with
BRAF and
PIK3CA wild type and 64.8% and 43.3% of cancers with
BRAF wild type and
PIK3CA mutations (
Figure 2A,B). Tumor suppressor
FBXW7 and
ATM mutations are more common in
BRAF mutant cancers with or without
PIK3CA mutations than in
BRAF wild type colorectal cancers (Fisher’s exact test
p = 0.0004 for
FBXW7 and
p < 0.0001 for
ATM in TCGA and Fisher’s exact test
p = 0.0003 for
FBXW7 and
p < 0.0001 for
ATM in the DFCI cohort,
Figure 2).
SMAD4 mutations show a higher prevalence in
BRAF/
PIK3CA double mutant cancers of the TCGA cohort but not in cancers with
BRAF mutations without
PIK3CA mutations. In addition, no significant differences in
SMAD4 mutations’ prevalence were observed in the DFCI cohort.
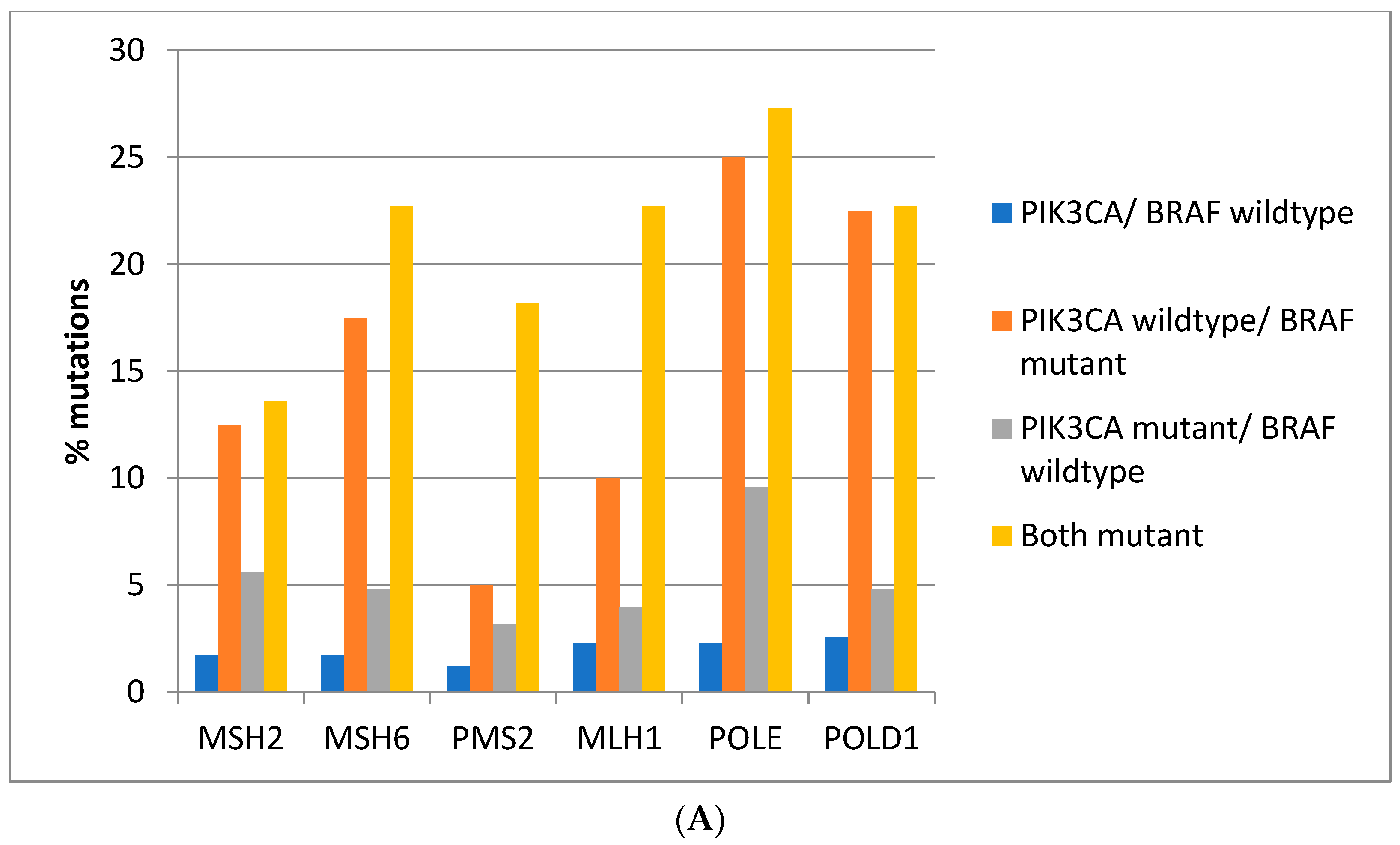
Mutations in the genes associated with MMR (
MSH2,
MSH6,
PMS2, and
MLH1) and those encoding for proof-reading polymerases epsilon (
POLE) and delta (
POLD1) show higher mutation rates in
BRAF/
PIK3CA double mutant colorectal cancers while cancers with
BRAF mutations and
PIK3CA wild type display similarly high mutation rates except for
PMS2 and
MLH1 which show significantly lower mutation rates (
Figure 3A). Consistently, in the DFCI cohort, the highest prevalence of MMR/proof-reading polymerases mutations is in double
BRAF and
PIK3CA mutant colorectal cancers with the exception of
POLE which shows the highest mutation prevalence in
BRAF mutant cancers without
PIK3CA mutations (
Figure 3B).
The majority of mutations observed in TCGA and DFCI cohorts in the three most frequently mutated cancer-related genes in colorectal cancer,
TP53,
KRAS, and
APC are oncogenic or likely oncogenic. In the colorectal cancer TCGA cohort, 99.1% of
TP53 mutations, 98.7% of
KRAS mutations, and 89.7% of
APC mutations are deemed oncogenic or likely oncogenic by the OncoKB database. Similarly, in the DFCI cohort, 98.6%, 97.2%, and 89.2% of mutations in
TP53,
KRAS, and
APC are oncogenic or likely oncogenic. The functional implications of other frequent mutations in colorectal cancer, occurring in more than 10% of cases, are shown in
Table 5. The oncogenicity or likely oncogenicity of these mutations varies from 15.3% for atypical cadherin gene
FAT1 to 84.1% for ubiquitin ligase gene
FBXW7, in the entire cohort, while the rest of the observed mutations are variants of unknown significance. Although the number of mutations in individual genes are low, when colorectal cancers are categorized according to
BRAF and
PIK3CA mutations, the prevalence of oncogenic or likely oncogenic mutations in these genes as a whole is higher in colorectal cancers with both
BRAF and
PIK3CA wild type (63.4%) than in double mutant cancers (47.3%) or cancers with
BRAF mutated and
PIK3CA wild type (41.7%) and with
PIK3CA mutant and
BRAF wild type (49.8%, χ
2 p < 0.001,
Table 5). However, in the DFCI cohort, although double
BRAF and
PIK3CA wild type colorectal cancers have a numerically higher prevalence of oncogenic/likely oncogenic mutations in commonly mutated cancer-associated genes (53.8%), the differences from the three other groups (
BRAF or
PIK3CA mutated or both mutated) were small and borderline statistically insignificant (χ
2 p = 0.051,
Table 6).
Mutations in genes associated with MMR and encoding for proof-reading polymerases
POLE and
POLD1 are designated as oncogenic or likely oncogenic in 20–25% of cases and show no significant differences in their oncogenic functional status in the four groups according to
BRAF and
PIK3CA mutations in either TCGA or DFCI cohorts (
Table 7 and
Table 8).
Survival of colorectal cancer patients of the TCGA was not different in the groups with
BRAF and
PIK3CA mutated or both genes mutated compared with cancers with both genes being wild type (Log Rank
p = 0.9,
Figure 4).
4. Discussion
BRAF is an oncogenic serine/threonine kinase which is frequently mutated in various cancers. In colorectal cancers, the prevalence of
BRAF mutations is 8–12% [
26]. The majority of
BRAF mutations result in the classical V600E position substitution. In a smaller number of cases, mutations resulting in substitutions on other amino acid positions of the protein, including at positions G469, G496, K601, L597, D594, G596, and G466, are present [
27]. While V600E substitutions enable the protein to signal autonomously as a monomer and activate the downstream ERK cascade, alternative substitutions require
BRAF homodimerization or heterodimerization with other RAF proteins. At odds with canonical V600E mutations, some alternative site mutations retain dependence to upstream KRAS signals for activation [
27].
KRAS mutations are more common than
BRAF mutations and are present in 30–40% of colorectal cancers [
18].
KRAS codon 12 or 13 mutations are mutually exclusive with
BRAF V600 mutations. For example, in a TCGA colorectal study with 534 analyzed cases only one sample had a concomitant
KRAS G12D substitution with a
BRAF V600E substitution. In the DFCI series, only one of the six
KRAS mutations occurring in samples with
BRAF mutations was a classic G13D substitution and the concomitant
BRAF mutation was a G469E substitution. In contrast to
KRAS mutations, mutations of the gene encoding for the catalytic sub-unit alpha of kinase PI3K,
PIK3CA, which are common in colorectal cancer are not mutually exclusive with
BRAF mutations. Among patients with
BRAF mutations, 35.5% of patients in TCGA and 27.5% of patients in the DFCI cohort had concomitant
PIK3CA mutations. This prevalence is higher than the overall prevalence of
PIK3CA mutations in the two series (27.5% in TCGA and 21.3% in DFCI).
In the current work, using data from TCGA and the DFCI cohorts, it is shown that double mutant
BRAF/
PIK3CA colorectal cancers and
BRAF mutant colorectal cancers without
PIK3CA mutations, which represent 4.1% and 7.5% of cases in TCGA and 5.7% and 14.9% of cases in the DFCI cohort, respectively, differ in their clinical and genomic characteristics from the groups of patients without
BRAF mutations.
BRAF mutations are the defining molecular alteration that links double mutant
BRAF/
PIK3CA colorectal cancers with MSI, a higher TMB, high CIMP, and low CIN, given that cancers with
BRAF mutant/
PIK3CA wild type cancers have similar rates of these characteristics without statistically significant differences compared to double mutants. Although not all
BRAF mutated colorectal cancers are MSI high, the association of
BRAF mutations with MSI and CIMP has been previously reported and MSI is commonly the result of MLH1 suppression through promoter methylation in cancers with
BRAF mutations [
28]. About 60% of sporadic MSI high colorectal cancers exhibit
BRAF mutations [
29]. However, the presence of
PIK3CA mutations in
BRAF mutated colorectal cancers leads to a numeric increase in the prevalence of MSI (from 59.5% to 68.2% in TCGA and from 47.1% to 62.9% in the DFCI cohort). In contrast, colorectal cancers with
PIK3CA mutations but without
BRAF mutations have statistically significant lower MSI, TMB, and CIMP rates and higher CIN rates compared with double mutants.
Despite different platforms used in the two studied cohorts, molecular alterations observed are consistent between them, with few differences. One such difference is in the mutation rate of tumor suppressor SMAD4, which showed a higher prevalence in double mutant cancers in TCGA but not in the DFCI cohort, where all four groups had similar
SMAD4 mutation rates. SMAD4 loss of function has been implicated in the serrated pathway of colorectal carcinogenesis where
BRAF mutations are also present [
30]. In a mouse model of colorectal carcinogenesis in vivo,
BRAF V600E mutations in intestinal stem cells promote differentiation and require inactivation of SMAD4 or of intestinal differentiation transcription factor CDX2 for efficient tumor formation [
31]. SMAD4 inactivation has been suggested as an important molecular event in the group of
BRAF mutated serrated carcinomas that are microsatellite stable [
30]. However, mutations of
SMAD4 occur only in a minority of
BRAF mutated colorectal cancers and accumulation of alternative lesions may be needed to advance
BRAF-associated carcinogenesis. Indeed, activation of the WNT/β-catenin pathway is required even in the presence of SMAD4 inactivation in
BRAF-associated colorectal cancers [
30]. Given that the prevalence of
APC mutations is shown to be lower in
BRAF mutated colorectal cancers, alternative modes of pathway activation are at play and may involve inhibition of kinase GSK3 through PI3K/AKT signaling [
32,
33]. Thus, concomitant
PIK3CA mutations could provide the required WNT/β-catenin pathway activation in
BRAF mutated cancers.
BRAF mutations are used as biomarkers of therapy guidance in metastatic colorectal cancer based on the results of the phase III randomized BEACON trial [
5]. This trial established that the combination of the
BRAF inhibitor encorafenib with EGFR monoclonal antibody cetuximab was superior to chemotherapy in pretreated patients with metastatic colorectal cancer bearing
BRAF V600E mutations. The study included a triplet arm with the MEK inhibitor binimetinib in addition to encorafenib and cetuximab which had a higher overall response rate than the doublet arm (26.8% versus 19.5% in the doublet arm). Despite that, median overall survival (OS) was 9.3 months in both targeted therapies arms [
6]. Median OS was 5.9 months in the control arm treated with chemotherapy. Thus, the encorafenib with cetuximab doublet is the preferred second line treatment for metastatic colorectal cancers with V600E
BRAF mutations. Besides showing that MEK inhibition with binimetinib has no benefit for OS, the BEACON results establish that pretreated patients with
BRAF mutations have short survivals even with the improved outcomes provided by the targeted treatment. It is worth noting that only 10% of the patients that participated in BEACON had MSI high cancers compared with a prevalence of MSI exceeding 50% in
BRAF mutant colorectal cancers of the TCGA and DFCI cohorts. Similarly, in a “real life” cohort of
BRAF mutant metastatic colorectal cancers treated with targeted therapies as used in the BEACON trial in several Italian centers, the prevalence of MSI or Mismatch Repair deficiency was 15% [
34]. This suggests that MSS cancers are enriched in the metastatic setting and that MSI-associated
BRAF mutated cancers progress less often to a metastatic stage. Consistently, data from a metastatic cohort, presented here, confirm that
BRAF mutations in biopsies from the patients’ primary tumor display MSI high in 37.8% of cases while biopsies from metastatic sites in similar
BRAF mutated cancer patients from the same cohort showed MSI high in 11.5% of cases. The prevalence of high TMB, in the range associated with putative responses to immune checkpoint inhibitors, is also lower in metastatic
BRAF mutated cancers, when evaluated from biopsy samples of metastatic sites compared with samples from biopsies of the primary tumor [
20]. In contrast, MSI high,
BRAF mutated cancers have in general a better prognosis [
35,
36]. In patients with stage III colorectal cancers who participated in the NCCTG N0147 trial, event-free survival (EFS) of the group with
BRAF mutations and MMR deficiency was similar to cancers with MMR proficiency and no
BRAF or
KRAS mutations, while EFS of proficient for MMR cancers with
BRAF mutations was inferior [
35]. OS is also inferior in stage III
BRAF mutant MSS colorectal cancers compared to MSS colorectal cancers with wild type
BRAF, but no difference dependent on
BRAF status is present in MSI high cancers [
37]. In addition, in metastatic colorectal cancer patients participating in four trials,
BRAF mutations were associated with worse PFS and OS in MMR proficient but not in MMR deficient cancers [
36].
BRAF mutant cancers have been divided in two subsets, based on unsupervised genomic clustering, that do not correlate with their MSI status or
PIK3CA mutation status [
38]. One of the sub-types called BM1 displays activation of KRAS/AKT and mTOR/protein translation pathways as well as features of epithelial to mesenchymal transition. The other sub-type BM2, which is more frequent, presents deregulation of cell cycle checkpoints as the main feature [
38]. Despite the lack of association of this clustering directly with the presence of concomitant mutations in
PIK3CA, the fact that the BM1 sub-type includes activation of the pathway among its defining characteristics suggests that this activation, independently of the specific nature of the underlying molecular lesion(s) producing it, is important in the pathogenesis of this
BRAF mutated sub-set. Moreover, in some cancers of the BM2 sub-type the presence of
PIK3CA mutations is not sufficient to activate the KRAS/AKT and mTOR/protein translation pathways. Despite the key function of PI3K/AKT/mTOR activity in a sub-set of
BRAF mutated colorectal cancers, few trials have attempted to target the pathway in these cancers or to systematically exploit therapeutically the sub-set with concomitant
BRAF and
PIK3CA mutations. A phase Ib trial of encorafenib, cetuximab with or without alpelisib in 54 metastatic
BRAF mutated colorectal cancer patients showed no significant difference in median PFS which was 3.7 months with the dual combination and 4.2 moths with the addition of alpelisib [
39]. A next generation sequence analysis which was performed in a subset of 21 patients (13 patients in the doublet arm and 8 patients in the triplet arm) showed that 3 of 7 patients (2 in the triplet arm and 1 patient in the doublet arm) with median PFS around or longer than 6 months had concomitant mutations in
PIK3CA while none of 14 patients with median PFS shorter than 6 months had such mutations [
39].
An additional key observation of the current report is that
BRAF mutated colorectal cancers with or without
PIK3CA mutations display lower prevalence of APC mutations than colorectal cancers with wild type
BRAF. Moreover,
BRAF mutated colorectal cancers with or without
PIK3CA mutations, as well as
PIK3CA mutated cancers without
BRAF mutations possess lower mutation rates of
TP53, suggesting that activation of either or both oncogenes decrease pressure for disabling of
TP53 in colorectal cancer cells. A negative correlation of
PIK3CA mutations and
TP53 mutations was also observed in a recently published series, which did not examine
BRAF mutations [
40]. In contrast to
APC and
TP53, the prevalence of mutations in tumor suppressors
FBXW7 and
ATM is higher in
BRAF mutated cancers. A high prevalence of
ATM mutations is observed in
BRAF mutated colorectal cancers in both TCGA and DFCI cohorts. In TCGA,
ATM mutations are present in 40% of cases with
PIK3CA wild type and in 36.4% of cases with
PIK3CA mutations. In the DFCI cohort,
ATM mutations are present in 28.3% of cases with
PIK3CA wild type and in 25.7% of cases with
PIK3CA mutations. The prevalence of
ATM mutations in the entire TCGA and DFCI cohorts is 13.1 % and 10.3%, respectively. Other series have shown an
ATM mutation prevalence of 15% in metastatic colorectal cancer but no increased prevalence in
BRAF mutated cases compared to
BRAF wild type metastatic colorectal cancers [
41]. This suggests that
ATM mutations in
BRAF mutated colorectal cancers are associated with a better prognosis sub-group such as MSI high. Indeed, 21 of 24
BRAF and
ATM mutated colorectal cancers (87.5%) in TCGA are MSI or POLE subtype while among
BRAF mutated, in
ATM wild type cancers only 55.3% belong to these subtypes (Fisher’s exact test
p = 0.01). In total, 20 of the 24
ATM mutations (83.3%) in
BRAF mutated colorectal cancers are categorized as likely oncogenic. In addition to
ATM, other genes related to DNA damage response (DDR) are mutated in smaller percentages of colorectal cancers and show a predilection for
BRAF mutated cancers. Mutations in DDR involved genes leading to homologous recombination defects sensitized to PARP inhibitors [
42]. In colorectal cancer, a phase II trial examining the PARP inhibitor olaparib as monotherapy in pretreated metastatic colorectal cancer patients showed no responses in either MSI or MSS disease [
43]. A phase I trial of olaparib in combination with irinotecan in unselected metastatic colorectal cancer patients showed also no responses and 9 of 25 patients had stable disease as the best response [
44]. The olaparib/irinotecan combination was used in a heavily pretreated metastatic colorectal cancer patient with an
ATM mutation who obtained stable disease for 3 months with a longer clinical and serologic markers improvement [
45]. Although PARP inhibitors in unselected colorectal cancer patients seem to have minimal activity, study of specific molecular subsets and of combinations with other targeted treatments deserve consideration. Combinations with
BRAF inhibitors in
BRAF mutated cancers with
ATM mutations or other homologous recombination defects could be a prime target.
In conclusion, the current study shows that the complex landscape of
BRAF mutated colorectal cancer with or without concomitant
PIK3CA mutations offers several leads for therapeutic targeting to improve outcomes of this subset of metastatic cancer patients associated with adverse survival. Newer
BRAF inhibitors in development which avoid the paradoxical activation of wild type
BRAF mediated by current inhibitors in use are also expected to advance therapeutics of these difficult to treat, resistant cancers [
46].