Expression, Purification, and Monitoring of Conformational Changes of hCB2 TMH67H8 in Different Membrane-Mimetic Lipid Mixtures Using Circular Dichroism and NMR Techniques
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Materials
2.2. hCB2 TMH67H6 Expression and Purification
2.3. Reconstitution of TMH67H8 and NMR Experiments
2.4. Circular Dichroism Experiments
2.5. Molecular Dynamics of TMH67H8
3. Results
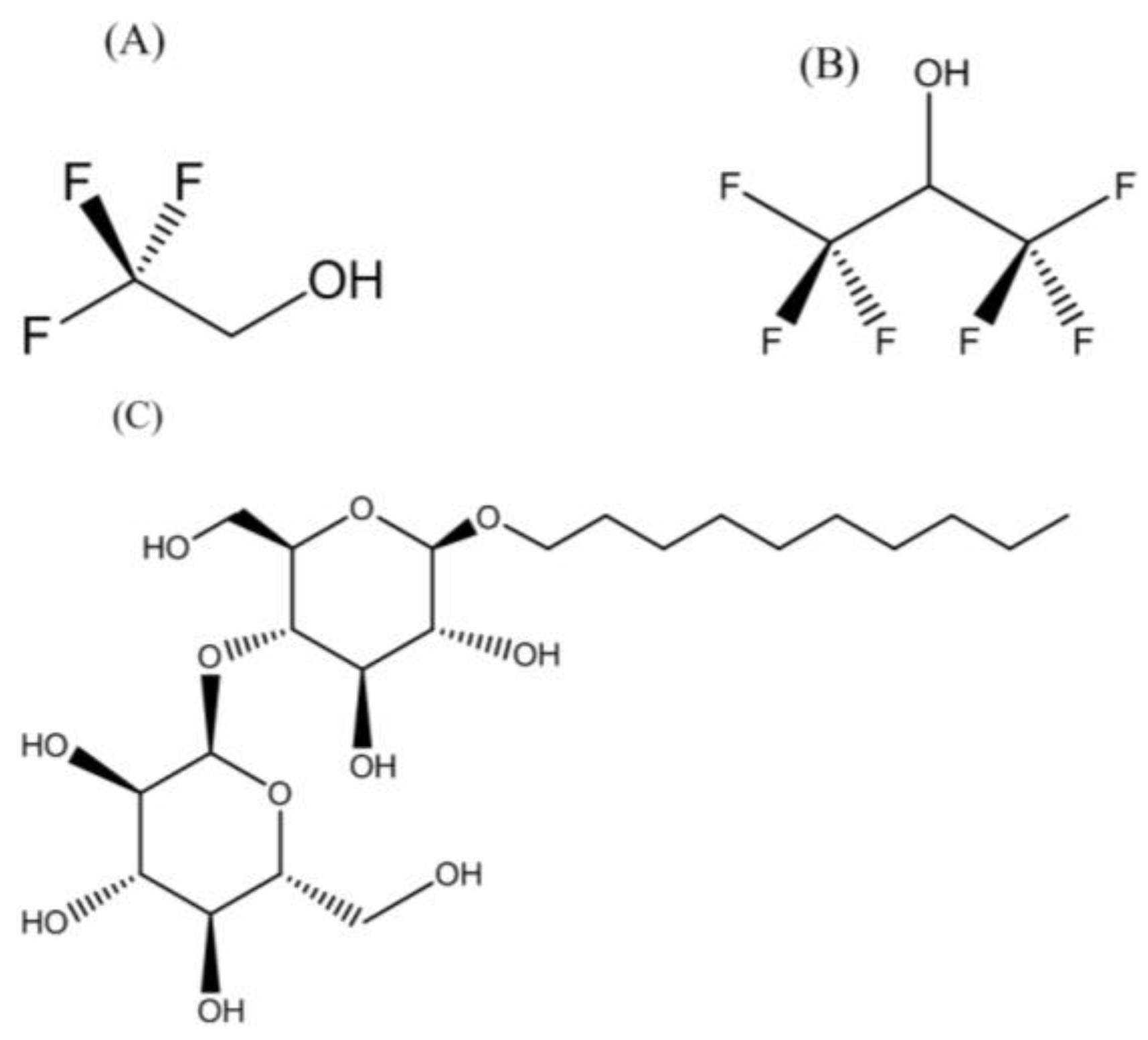
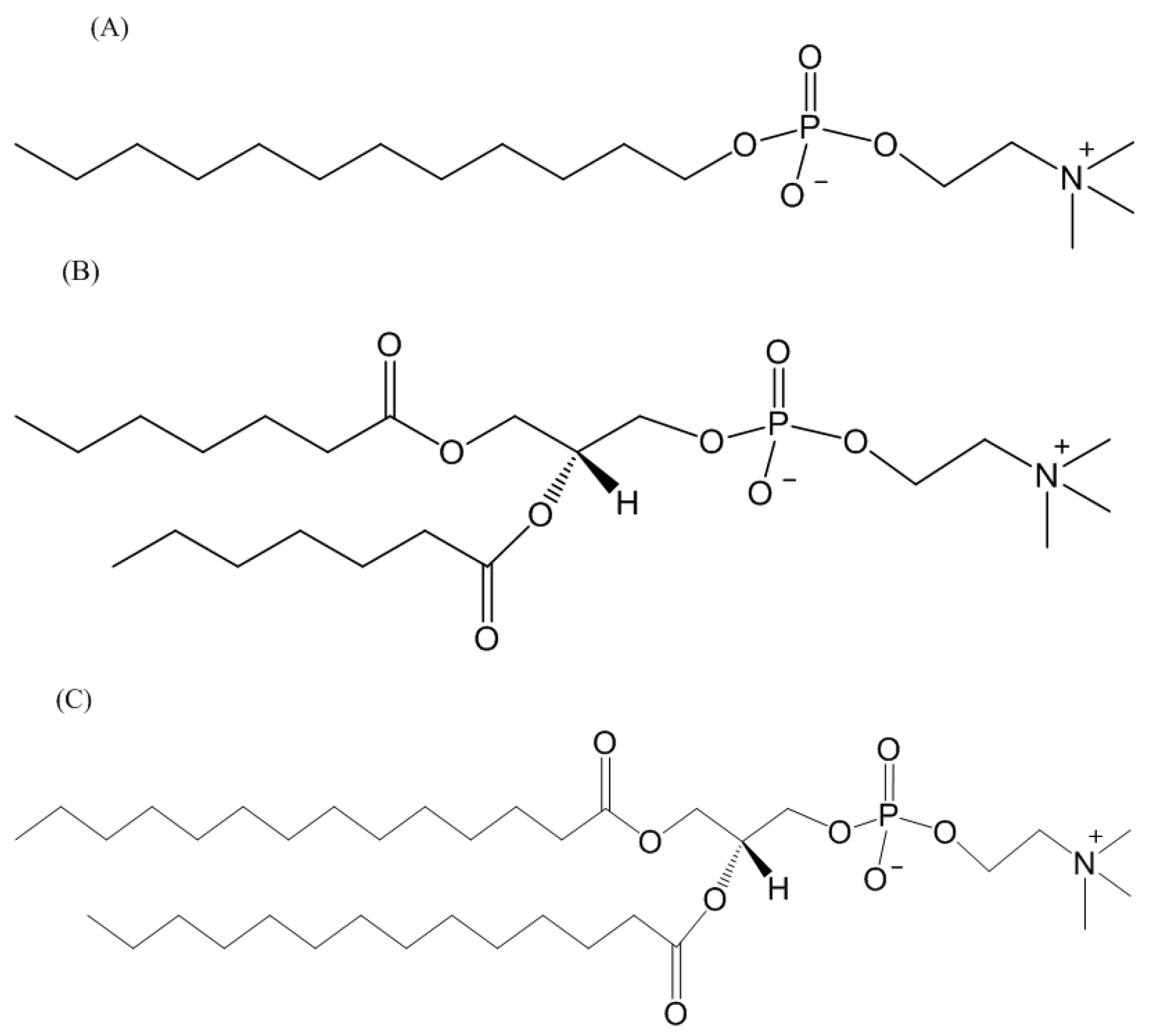
3.1. Membrane-Mimetic Chemical Structures
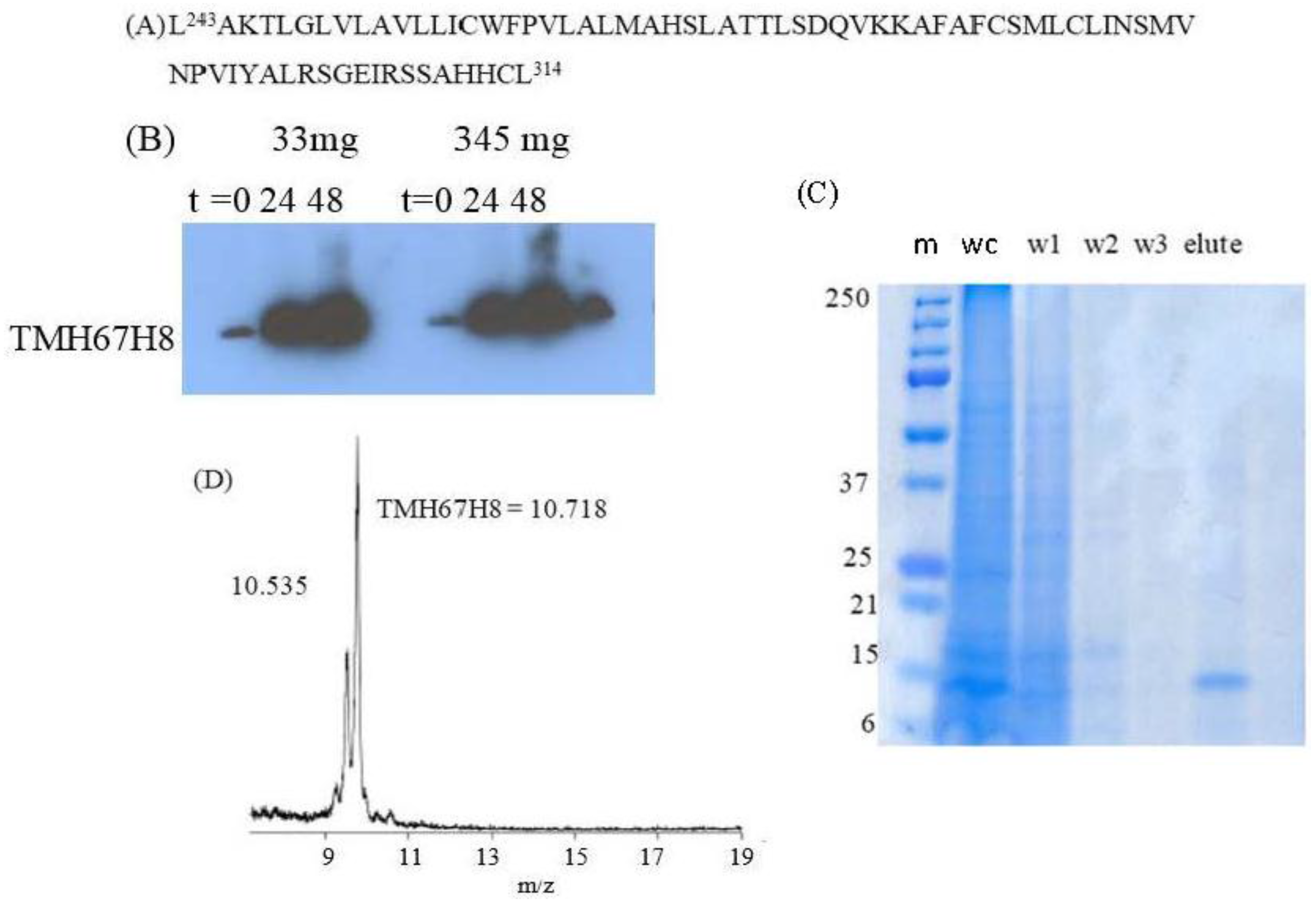
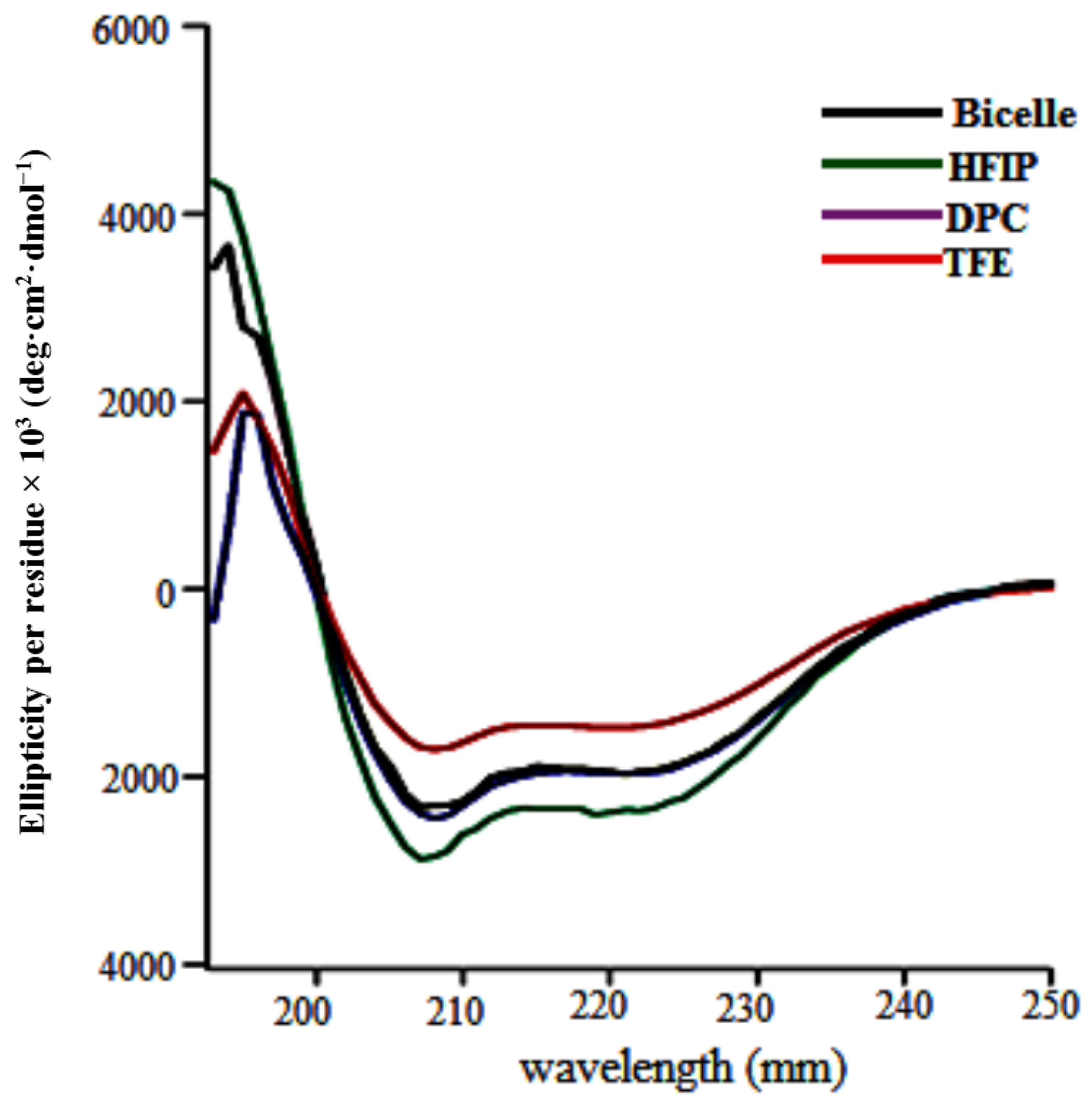
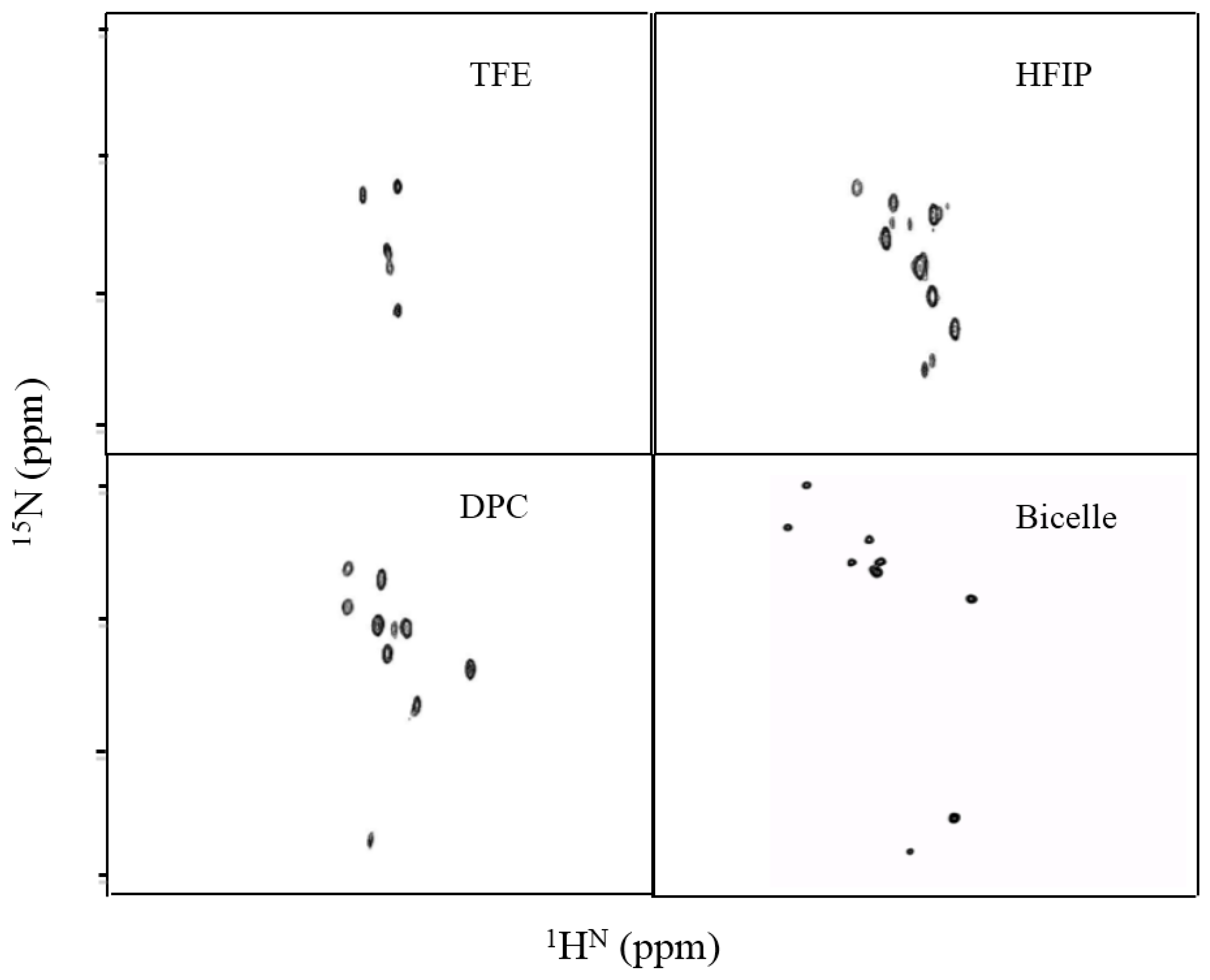
3.2. Insect Cells Expression, Western Blots Analysis, CD Measurements, and Solution NMR of TMH67H8
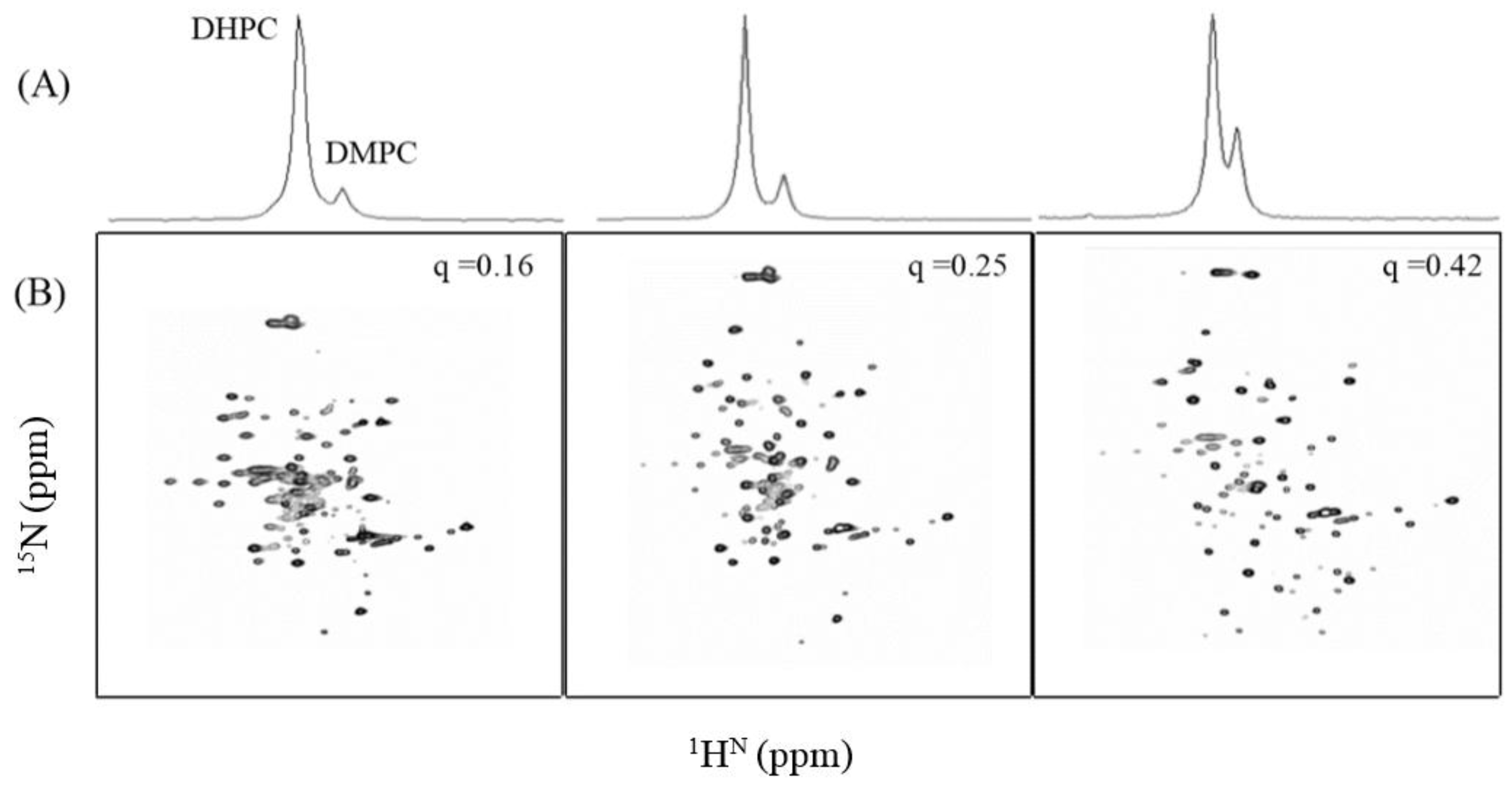
3.3. NMR Analysis of TMH67H8 as a Function of Bicelle q Ratio
3.4. Modeling TMH67H8 in a Membrane-Mimetic Environment
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DHPC | Dihexanoylphosphocholine |
| DMPC | Dimyristoylphosphocholine |
| DDM | n-Dodecyl-β-d-Maltopyranoside |
| DPC | n-Dodecylphosphocholine |
| DSS | 4,4-dimethyl-4-silapentane-1-sulfonic acid |
| GPCRs | G-protein coupled receptors |
| hCB2 | human cannabinoid receptor 2 |
| HSQC | heteronuclear single quantum coherence or correlation |
| Leu | Leucine |
| MALDI-TOF | Matrix-assisted laser desorption ionization time-of-flight |
| TFE | Trifluoroethanol |
| HFIP | 1,1,1,3,3,3-hexafluoro-2-propanol |
| RMSD | Root-mean-square-deviation |
| Sf9 | Spodoptera frugiperda |
| TMH | Transmembrane helix |
| TROSY | Transverse relaxation-optimized spectroscopy |
References
- Kenakin, T. New concepts in drug discovery: Collateral efficacy and permissive antagonism. Nat. Rev. Drug Discov. 2005, 4, 919–927. [Google Scholar] [CrossRef]
- Jaakola, V.P.; Lane, J.R.; Lin, J.Y.; Katritch, V.; Ijzerman, A.P.; Stevens, R.C. Ligand binding and subtype selectivity of the human A(2A) adenosine receptor: Identification and characterization of essential amino acid residues. J. Biol. Chem. 2010, 285, 13032–13044. [Google Scholar] [CrossRef]
- Katritch, V.; Cherezov, V.; Stevens, R.C. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol. Sci. 2011, 33, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Lagerstrom, M.C.; Schioth, H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Yoshiura, C.; Kofuku, Y.; Ueda, T.; Mase, Y.; Yokogawa, M.; Osawa, M.; Terashima, Y.; Matsushima, K.; Shimada, I. NMR analyses of the interaction between CCR5 and its ligand using functional reconstitution of CCR5 in lipid bilayers. J. Am. Chem. Soc. 2010, 132, 6768–6777. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Scheerer, P.; Hofmann, K.P.; Choe, H.W.; Ernst, O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 2008, 454, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Palczewski, K.; Kumasaka, T.; Hori, T.; Behnke, C.A.; Motoshima, H.; Fox, B.A.; le Trong, I.; Teller, D.C.; Okada, T.; Stenkamp, R.E.; et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 2000, 289, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Cherezov, V.; Rosenbaum, D.M.; Hanson, M.A.; Rasmussen, S.G.; Thian, F.S.; Kobilka, T.S.; Choi, H.J.; Kuhn, P.; Weis, W.I.; Kobilka, B.K.; et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 2007, 318, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Warne, T.; Moukhametzianov, R.; Baker, J.G.; Nehme, R.; Edwards, P.C.; Leslie, A.G.; Schertler, G.F.; Tate, C.G. The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor. Nature 2011, 469, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Chien, E.Y.; Liu, W.; Zhao, Q.; Katritch, V.; Han, G.W.; Hanson, M.A.; Shi, L.; Newman, A.H.; Javitch, J.A.; Cherezov, V.; et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 2010, 330, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Topiol, S.; Sabio, M. X-ray structure breakthroughs in the GPCR transmembrane region. Biochem. Pharmacol. 2009, 78, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Casagrande, F.; Chu, M.; Maier, K.; Kiefer, H.; Opella, S.J. Optimization of purification and refolding of the human chemokine receptor CXCR1 improves the stability of proteoliposomes for structure determination. Biochim. Biophys. Acta 2011, 1818, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.; Adeishvili, K.; Wuthrich, K. Transverse relaxation-optimized NMR spectroscopy with the outer membrane protein OmpX in dihexanoyl phosphatidylcholine micelles. Proc. Natl. Acad. Sci. USA 2001, 98, 2358–2363. [Google Scholar] [CrossRef] [PubMed]
- Tiburu, E.K.; Tyukhtenko, S.; Deshmukh, L.; Vinogradova, O.; Janero, D.R.; Makriyannis, A. Structural biology of human cannabinoid receptor-2 helix 6 in membrane-mimetic environments. Biochem. Biophys. Res. Commun. 2009, 384, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Carlotti, C.; Bontems, F.; Dufourc, E.J. Evidence for an α-helix → π-bulge helicity modulation for the neu/erbB-2 membrane-spanning segment. A 1H NMR and circular dichroism study. Biochemistry 2001, 40, 6534–6540. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, V.; Tillman, T.; Xu, Y.; Tang, P. NMR structure of the transmembrane domain of the n-acetylcholine receptor β2 subunit. Biochim. Biophys. Acta 2010, 1798, 1608–1614. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Abildgaard, F.; Bushweller, J.H.; Tamm, L.K. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat. Struct. Biol. 2001, 8, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Van Horn, W.D.; Kim, H.J.; Ellis, C.D.; Hadziselimovic, A.; Sulistijo, E.S.; Karra, M.D.; Tian, C.; Sonnichsen, F.D.; Sanders, C.R. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science 2009, 324, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Hiller, S.; Garces, R.G.; Malia, T.J.; Orekhov, V.Y.; Colombini, M.; Wagner, G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 2008, 321, 1206–1210. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Prytulla, S.; de Angelis, A.A.; Brown, J.M.; Kiefer, H.; Opella, S.J. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. J. Am. Chem. Soc. 2006, 128, 7402–7403. [Google Scholar] [CrossRef] [PubMed]
- Tiburu, E.K.; Tyukhtenko, S.; Zhou, H.; Janero, D.R.; Struppe, J.; Makriyannis, A. Human cannabinoid 1 GPCR C-terminal domain interacts with bilayer phospholipids to modulate the structure of its membrane environment. AAPS J. 2011, 13, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Sanders, C.R., Jr.; Landis, G.C. Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry 1995, 34, 4030–4040. [Google Scholar] [CrossRef] [PubMed]
- Bocharov, E.V.; Mineev, K.S.; Volynsky, P.E.; Ermolyuk, Y.S.; Tkach, E.N.; Sobol, A.G.; Chupin, V.V.; Kirpichnikov, M.P.; Efremov, R.G.; Arseniev, A.S. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J. Biol. Chem. 2008, 283, 6950–6956. [Google Scholar] [CrossRef] [PubMed]
- Lau, T.L.; Partridge, A.W.; Ginsberg, M.H.; Ulmer, T.S. Structure of the integrin β3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry 2008, 47, 4008–4016. [Google Scholar] [CrossRef] [PubMed]
- ALeitz, J.; Bayburt, T.H.; Barnakov, A.N.; Springer, B.A.; Sligar, S.G. Functional reconstitution of β2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques 2006, 40, 601–612. [Google Scholar]
- Gautier, A.; Mott, H.R.; Bostock, M.J.; Kirkpatrick, J.P.; Nietlispach, D. Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat. Struct. Mol. Biol. 2010, 17, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.S.; Arshava, B.; Neumoin, A.; Becker, J.M.; Guntert, P.; Zerbe, O.; Naider, F. Comparative NMR analysis of an 80-residue G protein-coupled receptor fragment in two membrane mimetic environments. Biochim. Biophys. Acta 2011, 1808, 2674–2684. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Wang, S.; Wang, C.; Zhu, T.; Li, F.; Sun, H. HFIP-induced structures and assemblies of the peptides from the transmembrane domain 4 of membrane protein Nramp1. Biopolymers 2006, 84, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Tyukhtenko, S.; Tiburu, E.K.; Deshmukh, L.; Vinogradova, O.; Janero, D.R.; Makriyannis, A. NMR solution structure of human cannabinoid receptor-1 helix 7/8 peptide: Candidate electrostatic interactions and microdomain formation. Biochem. Biophys. Res. Commun. 2009, 390, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, D.M.; Cherezov, V.; Hanson, M.A.; Rasmussen, S.G.; Thian, F.S.; Kobilka, T.S.; Choi, H.J.; Yao, X.J.; Weis, W.I.; Stevens, R.C.; et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 2007, 318, 1266–1273. [Google Scholar] [CrossRef]
- Pertwee, R.G.; Howlett, A.C.; Abood, M.E.; Alexander, S.P.; di Marzo, V.; Elphick, M.R.; Greasley, P.J.; Hansen, H.S.; Kunos, G.; Mackie, K.; et al. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: Beyond CB1 and CB2. Pharmacol. Rev. 2010, 62, 588–631. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, V.P.; Griffith, M.T.; Hanson, M.A.; Cherezov, V.; Chien, E.Y.; Lane, J.R.; Ijzerman, A.P.; Stevens, R.C. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 2008, 322, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; McIntosh, H.H.; Houston, D.B.; Howlett, A.C. The CB(1) cannabinoid receptor juxtamembrane C-terminal peptide confers activation to specific G proteins in brain. Mol. Pharmacol. 2000, 57, 162–170. [Google Scholar] [PubMed]
- Marcellino, D.; Ferre, S.; Casado, V.; Cortes, A.; le Foll, B.; Mazzola, C.; Drago, F.; Saur, O.; Stark, H.; Soriano, A.; et al. Identification of dopamine D1-D3 receptor heteromers. Indications for a role of synergistic D1-D3 receptor interactions in the striatum. J. Biol. Chem. 2008, 283, 26016–26025. [Google Scholar] [CrossRef] [PubMed]
- Marcellino, D.; Carriba, P.; Filip, M.; Borgkvist, A.; Frankowska, M.; Bellido, I.; Tanganelli, S.; Muller, C.E.; Fisone, G.; Lluis, C.; et al. Antagonistic cannabinoid CB1/dopamine D2 receptor interactions in striatal CB1/D2 heteromers. A combined neurochemical and behavioral analysis. Neuropharmacology 2008, 54, 815–823. [Google Scholar] [CrossRef]
- Wasilko, D.J.; Lee, S.E.; Stutzman-Engwall, K.J.; Reitz, B.A.; Emmons, T.L.; Mathis, K.J.; Bienkowski, M.J.; Tomasselli, A.G.; Fischer, H.D. The titerless infected-cells preservation and scale-up (TIPS) method for large-scale production of NO-sensitive human soluble guanylate cyclase (sGC) from insect cells infected with recombinant baculovirus. Protein Expr. Purif. 2009, 65, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Brocks, B.; Rode, H.J.; Klein, M.; Gerlach, E.; Dubel, S.; Little, M.; Pfizenmaier, K.; Moosmayer, D. A TNF receptor antagonistic scFv, which is not secreted in mammalian cells, is expressed as a soluble mono- and bivalent scFv derivative in insect cells. Immunotechnology 1997, 3, 173–184. [Google Scholar] [CrossRef]
- Cascio, M. Baculovirus expression of receptors and channels. Methods Neurosci. 1995, 25, 175–200. [Google Scholar]
- Booth, V.; Waring, A.J.; Walther, F.J.; Keough, K.M. NMR structures of the C-terminal segment of surfactant protein B in detergent micelles and hexafluoro-2-propanol. Biochemistry 2004, 43, 15187–15194. [Google Scholar] [CrossRef] [PubMed]
- Subasinghage, A.P.; O'Flynn, D.; Conlon, J.M.; Hewage, C.M. Conformational and membrane interaction studies of the antimicrobial peptide alyteserin-1c and its analogue [E4K]alyteserin-1c. Biochim. Biophys. Acta 2011, 1808, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Crescenzi, O.; Tomaselli, S.; Guerrini, R.; Salvadori, S.; D’Ursi, A.M.; Temussi, P.A.; Picone, D. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 2002, 269, 5642–5648. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.A.; Sherman, S.A.; Keiderling, T.A. β-hairpin stabilization in a 28-residue peptide derived from the β-subunit sequence of human chorionic gonadotropin hormone. Biopolymers 1999, 50, 413–423. [Google Scholar] [CrossRef]
- Arshava, B.; Taran, I.; Xie, H.; Becker, J.M.; Naider, F. High resolution NMR analysis of the seven transmembrane domains of a heptahelical receptor in organic-aqueous medium. Biopolymers 2002, 64, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Call, M.E.; Schnell, J.R.; Xu, C.; Lutz, R.A.; Chou, J.J.; Wucherpfennig, K.W. The structure of the ζζ transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 2006, 127, 355–368. [Google Scholar] [CrossRef]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiburu, E.K.; Zhuang, J.; Fleischer, H.N.A.; Arthur, P.K.; Awandare, G.A. Expression, Purification, and Monitoring of Conformational Changes of hCB2 TMH67H8 in Different Membrane-Mimetic Lipid Mixtures Using Circular Dichroism and NMR Techniques. Membranes 2017, 7, 10. https://doi.org/10.3390/membranes7010010
Tiburu EK, Zhuang J, Fleischer HNA, Arthur PK, Awandare GA. Expression, Purification, and Monitoring of Conformational Changes of hCB2 TMH67H8 in Different Membrane-Mimetic Lipid Mixtures Using Circular Dichroism and NMR Techniques. Membranes. 2017; 7(1):10. https://doi.org/10.3390/membranes7010010
Chicago/Turabian StyleTiburu, Elvis K., Jianqin Zhuang, Heidimarie N. A. Fleischer, Patrick K. Arthur, and Gordon A. Awandare. 2017. "Expression, Purification, and Monitoring of Conformational Changes of hCB2 TMH67H8 in Different Membrane-Mimetic Lipid Mixtures Using Circular Dichroism and NMR Techniques" Membranes 7, no. 1: 10. https://doi.org/10.3390/membranes7010010
APA StyleTiburu, E. K., Zhuang, J., Fleischer, H. N. A., Arthur, P. K., & Awandare, G. A. (2017). Expression, Purification, and Monitoring of Conformational Changes of hCB2 TMH67H8 in Different Membrane-Mimetic Lipid Mixtures Using Circular Dichroism and NMR Techniques. Membranes, 7(1), 10. https://doi.org/10.3390/membranes7010010







