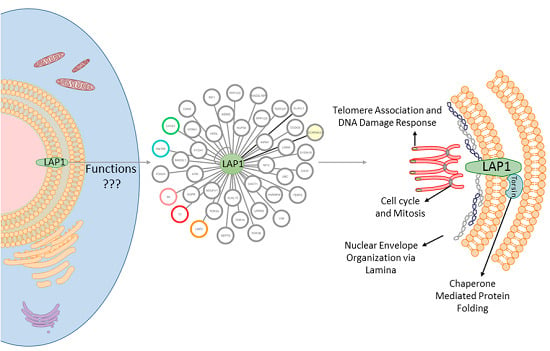
Lamina Associated Polypeptide 1 (LAP1) Interactome and Its Functional Features
Abstract
:1. Introduction
2. Results and Discussion
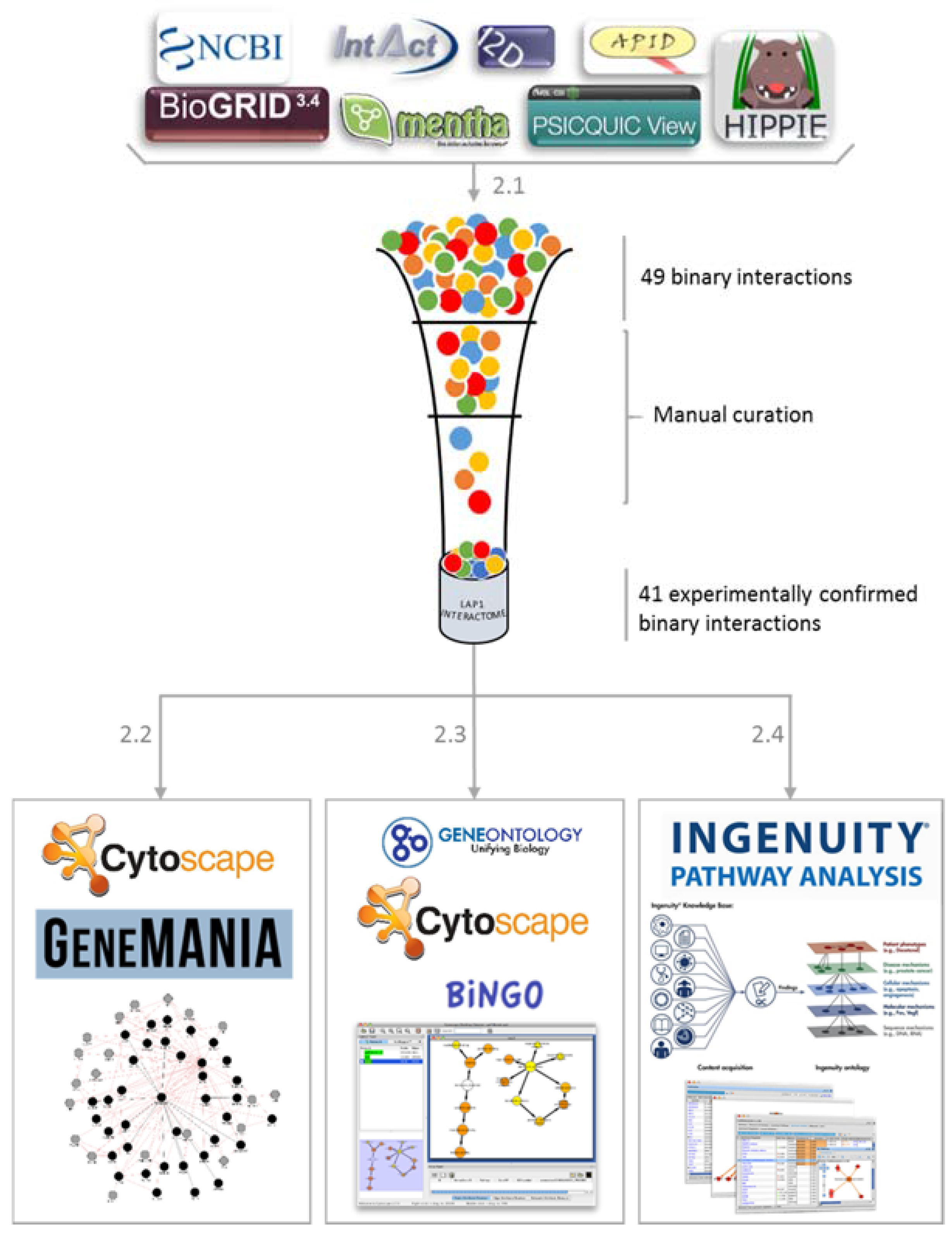
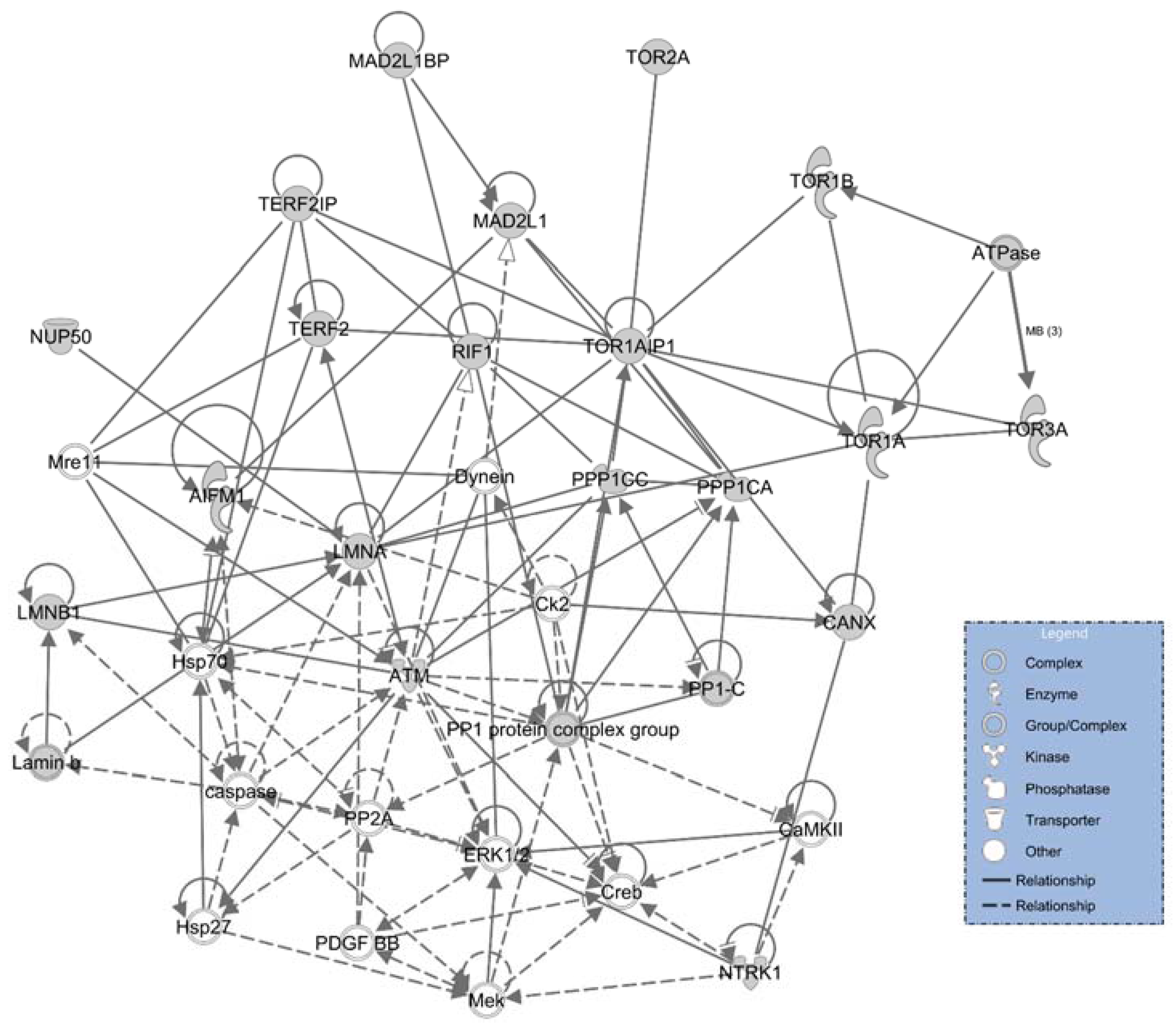
2.1. LAP1 Interactor’s Network
| Gene | Protein | Uniprot Accession Number | Species | Interaction Detection Method | References |
|---|---|---|---|---|---|
| 1C | Non-structural protein 1 | P04544 | TOR1AIP1 Hs—1C HRSVA | Affinity Capture-MS | [28] |
| AIFM1 | Apoptosis-inducing factor 1, mitochondrial | O95831 | Homo sapiens | Affinity Capture-MS | [31] |
| ATM | Serine-protein kinase ATM | Q13315 | Homo sapiens | Affinity Capture-MS | [32] |
| Atp1b4 | Protein ATP1B4 | Q99ME6 | Mus musculus | Two-hybrid | [33] |
| Q9R193 | Rattus norvegicus | Affinity Capture-WB | |||
| CA10 | Carbonic anhydrase-related protein 10 | Q9NS85 | Homo sapiens | Affinity Capture-MS | [34] |
| CANX | Calnexin | P35564 | Tor1aip1 Mm—CANX Hs | Affinity Capture-MS | [35] |
| CCDC8 | Coiled-coil domain-containing protein 8 | Q9H0W5 | Homo sapiens | Affinity Capture-MS | [36] |
| EGFR | Epidermal growth factor receptor | P00533 | Homo sapiens | Affinity Capture-MS | [37] |
| ELAVL1 * | ELAV-like protein 1 | Q15717 | Homo sapiens | Affinity Capture-RNA | [25] |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M | P52272 | Homo sapiens | Co-fractionation | [38] |
| HPDL | 4-hydroxyphenylpyruvate dioxygenase-like protein | Q96IR7 | Homo sapiens | Co-fractionation | [38] |
| KLHL15 | Kelch-like protein 15 | Q96M94 | Homo sapiens | Affinity Capture-MS | [31] |
| LMNA | Prelamin-A/C | P48678 | Mus musculus | Affinity Capture-MS | [39] |
| P02545 | Homo sapiens | Two-hybrid | [40] | ||
| Homo sapiens | Reconstituted Complex | [9] | |||
| Homo sapiens | Proximity Label-MS | [41] | |||
| Lmnb1 | Lamin-B1 | P70615 | Rattus norvegicus | Affinity Capture-WB | [42] |
| LMP2 | Latent membrane protein 2 | Q1HVJ2 | TOR1AIP1 Hs—LMP2 HHV-4 | Affinity Capture-MS | [43] |
| LRRK2 | Leucine-rich repeat serine/threonine-protein kinase 2 | Q5S007 | Homo sapiens | Affinity Capture-MS | [44] |
| Mad2l1 | MAD2 mitotic arrest deficient-like 1 | Q9Z1B5 | TOR1AIP1 Hs—Mad2l1 Mm | Affinity Capture-MS | [35] |
| MAD2L1BP | MAD2 mitotic arrest deficient-like 1 | Q15013 | Homo sapiens | Affinity Capture-MS | [31] |
| MDM2 | E3 ubiquitin-protein ligase Mdm2 | Q00987 | Homo sapiens | Affinity Capture-MS | [45] |
| NDUFV1 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | P49821 | Homo sapiens | Co-fractionation | [38] |
| NIT2 | Omega-amidase NIT2 | Q9NQR4 | Homo sapiens | Co-fractionation | [38] |
| NTRK1 | High affinity nerve growth factor receptor | P04629 | Homo sapiens | Affinity Capture-MS | [46] |
| NUP50 | Nuclear pore complex protein Nup50 | Q9UKX7 | Homo sapiens | Co-fractionation | [38] |
| OXCT1 | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | P55809 | Homo sapiens | Co-fractionation | [38] |
| PPP1CA | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | P62136 | Homo sapiens | Affinity Capture-WB | [47] |
| Two-hybrid | [48] | ||||
| PPP1CC | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | P36873 | Homo sapiens | Affinity Capture-WB | [47] |
| Two-hybrid | [49] | ||||
| PTCH1 | Protein patched homolog 1 | Q13635 | Homo sapiens | Affinity Capture-MS | [34] |
| RIF1 | Telomere-associated protein RIF1 | Q5UIP0 | Homo sapiens | Co-fractionation | [38] |
| SCARNA22 ** | Small Cajal body-specific RNA 22 | Gene ID: 677770 | Homo sapiens | Affinity Capture-RNA | [26] |
| S100A16 | Protein S100-A16 | Q96FQ6 | Homo sapiens | Co-fractionation | [38] |
| SEPT9 | Septin-9 | Q9UHD8 | Homo sapiens | Co-fractionation | [38] |
| Tat | Protein Tat | P04608 | TOR1AIP1 Hs—tat HIV-1 | Affinity Capture-MS | [27] |
| TERF2 | Telomeric repeat-binding factor 2 | Q15554 | Homo sapiens | Two-hybrid | [50] |
| TERF2IP | Telomeric repeat-binding factor 2-interacting protein 1 | Q9NYB0 | Homo sapiens | Two-hybrid | [50] |
| TOR1A | Torsin-1A | O14656 | Homo sapiens | Affinity Capture-MS/WB | [11,15,16,51,52] |
| Reconstituted Complex | [52] | ||||
| [16,52] | |||||
| Affinity Capture-MS | [53] | ||||
| TOR1AIP1 | Lamina-associated polypeptide 1B | Q5JTV8 | Tor1aip1 Mm—TOR1AIP1 Hs | Affinity Capture-MS | [35] |
| TOR1B | Torsin-1B | O14657 | Homo sapiens | Affinity Capture-WB | [53] |
| TOR2A | Torsin-2A | Q5JU69 | Homo sapiens | Affinity Capture-WB | [53] |
| TOR3A | Torsin-3A | Q9H497 | Homo sapiens | Affinity Capture-WB | [53] |
| UBC | Polyubiquitin-C | P0CG48 | Homo sapiens | Affinity Capture-MS | [54,55,56,57,58,59] |
| VIM | Vimentin | P08670 | Homo sapiens | Affinity Capture-WB | [60] |

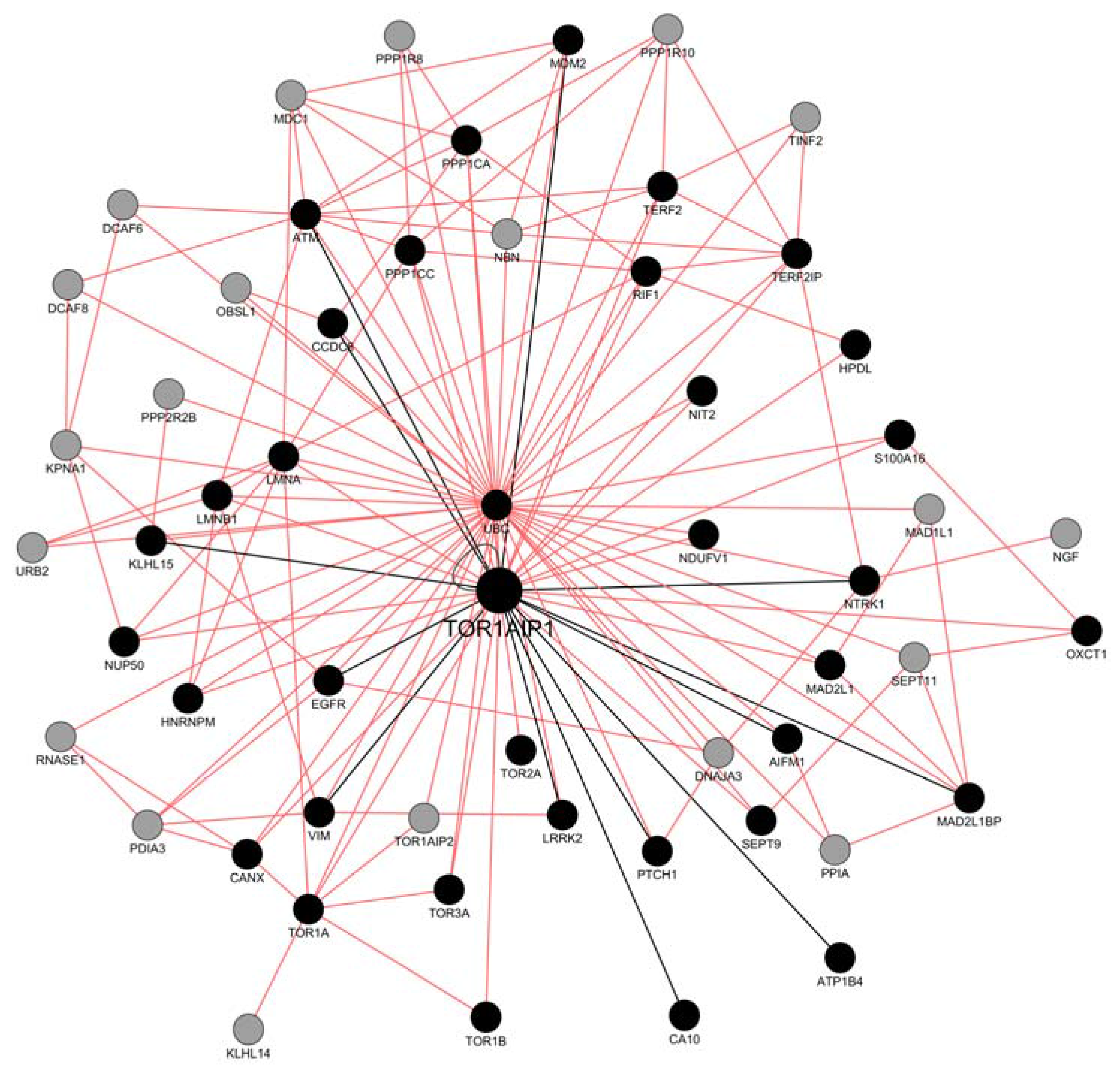
2.2. Network Construction with GeneMANIA
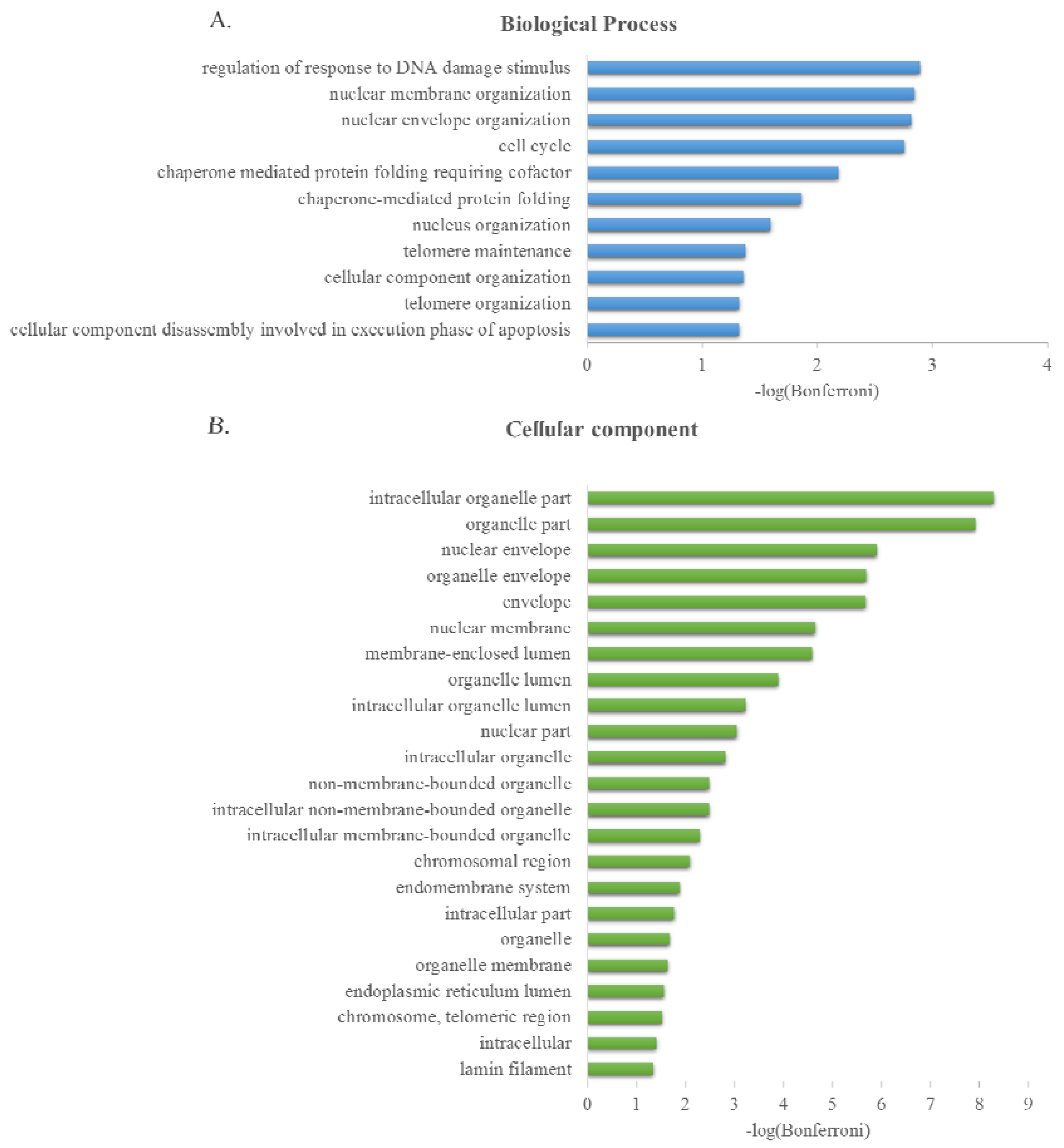
2.3. GO Term Enrichment Analysis
2.4. Ingenuity Pathways Analysis (IPA) Physiological and Functional Analysis
| Name | p-Value | Overlap % | |
|---|---|---|---|
| Telomerase Signalling | 2.99 × 10−5 | 4.00% | 4/99 |
| Telomere Extension by Telomerase | 3.28 × 10−4 | 13.30% | 2/15 |
| HER-2 Signalling in Breast Cancer | 3.47 × 10−4 | 3.90% | 3/76 |
| Glioma Signalling | 6.68 × 10−4 | 3.20% | 3/95 |
| Huntington’s Disease Signalling | 7.49 × 10−4 | 1.70% | 4/229 |
| Name | p-Value | # Molecules |
|---|---|---|
| Skeletal and Muscular System Development and Function | 3.61 × 10−3–1.82 × 10−6 | 7 |
| Tissue Development | 3.61 × 10−3–1.82 ×10−6 | 13 |
| Nervous System Development and Function | 3.61 × 10−3–3.93 × 10−6 | 13 |
| Organ Morphology | 3.61 × 10−3–3.93 × 10−6 | 9 |
| Tissue Morphology | 3.61 × 10−3–3.93 × 10−6 | 16 |
| ID | Associated Network Functions | Score |
|---|---|---|
| 1 | Cell Morphology, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair | 47 |
| 2 | Cancer, Organismal Injury and Abnormalities, Respiratory Disease | 32 |
| 3 | RNA Post-Transcriptional Modification, Protein Synthesis, Gene Expression | 3 |
| 4 | Developmental Disorder, Neurological Disease, Behaviour | 2 |
| Name | p-Value | Overlap % |
|---|---|---|
| Hypoxia-Inducible Factor Signalling | 2.72 ×10−4 | 4.3% 3/70 |
| Mitochondrial Dysfunction | 3.90 × 10−3 | 1.7% 3/176 |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 3.97 × 10−3 | 3.8% 2/52 |
| Cell Cycle: G1/S Checkpoint Regulation | 6.32 × 10−3 | 3.0% 2/66 |
| TR/RXR Activation | 1.03 × 10−2 | 2.4% 2/85 |
| Name | p-Value | # Molecules |
|---|---|---|
| Cell Morphology | 3.61 × 10−3–3.18 × 10−11 | 18 |
| Cellular Assembly and Organization | 3.61 × 10−3–3.18 × 10−11 | 20 |
| DNA Replication, Recombination, and Repair | 3.61 × 10−3–1.34 × 10−10 | 15 |
| Cell Cycle | 3.61 × 10−3–9.17 × 10−9 | 17 |
| Cell Death and Survival | 3.61 × 10−3–4.21 × 10−7 | 20 |
3. Experimental Section
3.1. Collection of Associated Interactions of LAP1
3.2. Gene Ontology Term Enrichment Analysis
3.3. Software Platforms and Plugins
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dauer, W.T.; Worman, H.J. The Nuclear Envelope as a Signaling Node in Development and Disease. Dev. Cell 2009, 17, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Margalit, A.; Goldman, R.D.; Shumaker, D.K.; Wilson, K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005, 6, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Burke, B.; Stewart, C.L. Functional architecture of the cell’s nucleus in development, aging, and disease. Curr. Top. Dev. Biol. 2014, 109, 1–52. [Google Scholar] [PubMed]
- Schirmer, E.C.; Florens, L.; Guan, T.; Yates, J.R.; Gerace, L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003, 301, 1380–1382. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.; Gerace, L. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J. Cell Biol. 1988, 107, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Crimaudo, C.; Gerace, L. cDNA cloning and characterization of lamina-associated polypeptide 1C (LAP1C), an integral protein of the inner nuclear membrane. J. Biol. Chem. 1995, 270, 8822–8828. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Kondoh, J.; Hayashi, D.; Ban, T.; Takagi, M.; Kamei, Y.; Tsuji, L.; Kim, J.; Yoneda, Y. Molecular cloning of one isotype of human lamina-associated polypeptide 1s and a topological analysis using its deletion mutants. Biochem. Biophys. Res. Commun. 2002, 294, 770–778. [Google Scholar] [CrossRef]
- Santos, M.; Domingues, S.C.; Costa, P.; Muller, T.; Galozzi, S.; Marcus, K.; da Cruz E Silva, E.F.; da Cruz E Silva, O.A.; Rebelo, S. Identification of a Novel Human LAP1 Isoform That Is Regulated by Protein Phosphorylation. PLoS ONE 2014, 9, e113732. [Google Scholar] [CrossRef] [PubMed]
- Foisner, R.; Gerace, L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 1993, 73, 1267–1279. [Google Scholar] [CrossRef]
- Gerace, L.; Huber, M.D. Nuclear lamina at the crossroads of the cytoplasm and nucleus. J. Struct. Biol. 2012, 177, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, R.E.; Dauer, W.T. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol. 2005, 168, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Ozelius, L.J.; Hewett, J.W.; Page, C.E.; Bressman, S.B.; Kramer, P.L.; Shalish, C.; de Leon, D.; Brin, M.F.; Raymond, D.; Corey, D.P.; et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 1997, 17, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Alegre, P. Aberrant Cellular Behavior of Mutant TorsinA Implicates Nuclear Envelope Dysfunction in DYT1 Dystonia. J. Neurosci. 2004, 24, 2593–2601. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, R.E.; Dauer, W.T. Mislocalization to the nuclear envelope: An effect of the dystonia-causing torsinA mutation. Proc. Natl. Acad. Sci. USA 2004, 101, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Naismith, T.V.; Dalal, S.; Hanson, P.I. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J. Biol. Chem. 2009, 284, 27866–27874. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Brown, R.S.; Chase, A.R.; Eisele, M.R.; Schlieker, C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc. Natl. Acad. Sci. USA 2013, 110, E1545–E1554. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Méndez-lópez, I.; Wang, Y.; Hays, A.P.; Tanji, K.; Lefkowitch, J.H.; Schulze, P.C.; Worman, H.J.; Dauer, W.T. Lamina-associated Polypeptide-1 Interacts with the Muscular Dystrophy Protein Emerin and is Essential for Skeletal Muscle Maintenance. Dev. Cell 2014, 26, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Bione, S.; Maestrini, E.; Rivella, S.; Mancini, M.; Regis, S.; Romeo, G.; Toniolo, D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994, 8, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Rebelo, S.; van Kleeff, P.J.M.; Kim, C.E.; Dauer, W.T.; Fardilha, M.; da Cruz E Silva, O.A.; da Cruz E Silva, E.F. The nuclear envelope protein, LAP1B, is a novel protein phosphatase 1 substrate. PLoS ONE 2013, 8, e76788. [Google Scholar] [CrossRef] [PubMed]
- Kayman-Kurekci, G.; Talim, B.; Korkusuz, P.; Sayar, N.; Sarioglu, T.; Oncel, I.; Sharafi, P.; Gundesli, H.; Balci-Hayta, B.; Purali, N.; et al. Mutation in TOR1AIP1 encoding LAP1B in a form of muscular dystrophy: A novel gene related to nuclear envelopathies. Neuromuscul. Disord. 2014, 24, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Dorboz, I.; Coutelier, M.; Bertrand, A.T.; Caberg, J.-H.; Elmaleh-Bergès, M.; Lainé, J.; Stevanin, G.; Bonne, G.; Boespflug-Tanguy, O.; Servais, L. Severe dystonia, cerebellar atrophy, and cardiomyopathy likely caused by a missense mutation in TOR1AIP1. Orphanet J. Rare Dis. 2014, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, S.; Edgar, F.; Odete, A.B. Mutation Research/Reviews in Mutation Research Genetic mutations strengthen functional association of LAP1 with DYT1 dystonia and muscular dystrophy. Mutat. Res. Mutat. Res. 2015, 2–7. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-Y.; Dauer, W.T.; Worman, H.J. Lamina-associated polypeptide 1: Protein interactions and tissue-selective functions. Semin. Cell Dev. Biol. 2014, 29, 164–168. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abdelmohsen, K.; Srikantan, S.; Yang, X.; Lal, A.; Kim, H.H.; Kuwano, Y.; Galban, S.; Becker, K.G.; Kamara, D.; de Cabo, R.; et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009, 28, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Su, M.Y.; Maggi, L.B.; Lu, L.; Mullins, C.; Crosby, S.; Huang, G.; Chng, W.J.; Vij, R.; Tomasson, M.H. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Invest. 2012, 122, 2793–2806. [Google Scholar] [CrossRef] [PubMed]
- Gautier, V.W.; Gu, L.; O’Donoghue, N.; Pennington, S.; Sheehy, N.; Hall, W.W. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology 2009, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Tran, K.C.; Teng, M.N.; Heesom, K.J.; Matthews, D.A.; Barr, J.N.; Hiscox, J.A. The Interactome of the Human Respiratory Syncytial Virus NS1 Protein Highlights Multiple Effects on Host Cell Biology. J. Virol. 2012, 86, 7777–7789. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.T.; Zimmerman, J.S.; Rowe, D.T. Epstein-Barr virus latent membrane protein 2B (LMP2B) co-localizes with LMP2A in perinuclear regions in transiently transfected cells. J. Gen. Virol. 2002, 83, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Etingov, I.; Pante, N. Effect of Viral Infection on the Nuclear Envelope and Nuclear Pore Complex; Elsevier: Amsterdam, The Netherlands, 2012; Volume 299. [Google Scholar]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Paulo, J.A.; Gygi, M.P.; Rad, R.; Kolippakkam, D.; Szpyt, J.; Zarraga, G.; Tam, S.; et al. High-Throughput Proteomic Mapping of Human Interaction Networks via Affinity-Purification Mass Spectrometry (Pre-Publication). Available online: http://thebiogrid.org/166968/publication/high-throughput-proteomic-mapping-of-human-interaction-networks-via-affinity-purification-mass-spectrometry.html (accessed on 23 October 2015).
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Pestov, N.B.; Ahmad, N.; Korneenko, T.V.; Zhao, H.; Radkov, R.; Schaer, D.; Roy, S.; Bibert, S.; Geering, K.; Modyanov, N.N. Evolution of Na,K-ATPase beta m-subunit into a coregulator of transcription in placental mammals. Proc. Natl. Acad. Sci. USA 2007, 104, 11215–11220. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [PubMed]
- Hutchins, J.R.A.; Toyoda, Y.; Hegemann, B.; Poser, I.; Hériché, J.-K.; Sykora, M.M.; Augsburg, M.; Hudecz, O.; Buschhorn, B.A.; Bulkescher, J.; et al. Systematic Analysis of Human Protein Complexes Identifies Chromosome Segregation Proteins. Science 2010, 328, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Hanson, D.; Stevens, A.; Murray, P.G.; Black, G.C.M.; Clayton, P.E. Identifying biological pathways that underlie primordial short stature using network analysis. J. Mol. Endocrinol. 2014, 52, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Taylor, P.; Moran, M.F. Proteomic Analysis of the Epidermal Growth Factor Receptor (EGFR) Interactome and Post-translational Modifications Associated with Receptor Endocytosis in Response to EGF and Stress. Mol. Cell. Proteomics 2014, 13, 1644–1658. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Hart, G.T.; Nepusz, T.; Yang, H.; Turinsky, A.L.; Li, Z.; Wang, P.I.; Boutz, D.R.; Fong, V.; Phanse, S.; et al. Census of Human Soluble Protein Complexes. Cell 2012, 150, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Kubben, N.; Voncken, J.W.; Demmers, J.; Calis, C.; van Almen, G.; Pinto, Y.M.; Misteli, T. Identification of differential protein interactors of lamin A and progerin. Nucleus 2010, 1, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Zhong, N.; Radu, G.; Ju, W.; Brown, W.T. Novel progerin-interactive partner proteins hnRNP E1, EGF, Mel 18, and UBC9 interact with lamin A/C. Biochem. Biophys. Res. Commun. 2005, 338, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Maison, C.; Pyrpasopoulou, A.; Theodoropoulos, P.A.; Georgatos, S.D. The inner nuclear membrane protein LAP1 forms a native complex with B-type lamins and partitions with spindle-associated mitotic vesicles. EMBO J. 1997, 16, 4839–4850. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Deo, R.C.; Padi, M.; Adelmant, G.; Calderwood, M.A.; Rolland, T.; Grace, M.; Dricot, A.; Askenazi, M.; Tavares, M.; et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 2012, 487, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Kim, J.W.; Lee, B.D.; Kang, H.C.; Xu, J.-C.; Jia, H.; Stankowski, J.; Kim, M.-S.; Zhong, J.; Kumar, M.; et al. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson’s disease. Cell 2014, 157, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.; Scherl, A.; Way, L.; Blackburn, E.A.; Walkinshaw, M.D.; Ball, K.L.; Hupp, T.R. A systems wide mass spectrometric based linear motif screen to identify dominant in-vivo interacting proteins for the ubiquitin ligase MDM2. Cell. Signal. 2014, 26, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Emdal, K.B.; Pedersen, A.-K.; Bekker-Jensen, D.B.; Tsafou, K.P.; Horn, H.; Lindner, S.; Schulte, J.H.; Eggert, A.; Jensen, L.J.; Francavilla, C.; et al. Temporal proteomics of NGF-TrkA signaling identifies an inhibitory role for the E3 ligase Cbl-b in neuroblastoma cell differentiation. Sci. Signal. 2015, 8, ra40. [Google Scholar] [CrossRef] [PubMed]
- Santos, M. Characterization of novel LAP1 complexes and their relevance in DYT1 dystonia. Ph.D. Thesis, University of Aveiro, Aveiro, Portugal, 2014. [Google Scholar]
- Esteves, S.L.; Domingues, S.C.; da Cruz e Silva, O.A.; Fardilha, M.; da Cruz e Silva, E.F. Protein phosphatase 1alpha interacting proteins in the human brain. Omics 2012, 16, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Esteves, S.L.C.; Korrodi-Gregório, L.; Cotrim, C.Z.; van Kleeff, P.J.M.; Domingues, S.C.; Da Cruz E Silva, O.A.B.; Fardilha, M.; Da Cruz E Silva, E.F. Protein phosphatase 1γ isoforms linked interactions in the brain. J. Mol. Neurosci. 2013, 50, 179–197. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.-H.; Kim, H.; He, Q.; Baek, H.J.; Yang, D.; Chen, L.-Y.; Liang, J.; Chae, H.K.; Safari, A.; Liu, D.; et al. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol. Cell. Proteomics 2011, 10, M110.001628. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Millen, L.; Mendoza, J.L.; Thomas, P.J. A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J. Biol. Chem. 2010, 285, 37271–37280. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.S.H.; Zhao, C.; Chase, A.R.; Wang, J.; Schlieker, C. The mechanism of Torsin ATPase activation. Proc. Natl. Acad. Sci. USA 2014, 111, E4822–E4831. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.E.; Perez, A.; Perkins, G.; Ellisman, M.H.; Dauer, W.T. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. USA 2010, 107, 9861–9866. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, J.M.R.; Sylvestersen, K.B.; Bekker-Jensen, S.; Szklarczyk, D.; Poulsen, J.W.; Horn, H.; Jensen, L.J.; Mailand, N.; Nielsen, M.L. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 2011, 10, M110.003590. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Nielsen, M.L.; Cox, J.; Mann, M.; Choudhary, C. A Proteome-wide, Quantitative Survey of In Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol. Cell. Proteomics 2011, 10, M111.013284. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, M.J.; Elia, A.E.H.; Xu, Q.; Thoma, C.R.; Izhar, L.; Leng, Y.; Guo, A.; Chen, Y.N.; Rush, J.; Hsu, P.W.C.; et al. Global identification of modular cullin-RING ligase substrates. Cell 2011, 147, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Povlsen, L.K.; Beli, P.; Wagner, S.A.; Poulsen, S.L.; Sylvestersen, K.B.; Poulsen, J.W.; Nielsen, M.L.; Bekker-Jensen, S.; Mailand, N.; Choudhary, C. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell Biol. 2012, 14, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Stes, E.; Laga, M.; Walton, A.; Samyn, N.; Timmerman, E.; de Smet, I.; Goormachtig, S.; Gevaert, K. A COFRADIC protocol to study protein ubiquitination. J. Proteome Res. 2014, 13, 3107–3113. [Google Scholar] [CrossRef] [PubMed]
- Hewett, J.W.; Zeng, J.; Niland, B.P.; Bragg, D.C.; Breakefield, X.O. Dystonia-causing mutant torsinA inhibits cell adhesion and neurite extension through interference with cytoskeletal dynamics. Neurobiol. Dis. 2006, 22, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, E.C.; Foisner, R. Proteins that associate with lamins: Many faces, many functions. Exp. Cell Res. 2007, 313, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, B.; Tomchick, D.R.; Machius, M.; Rizo, J.; Yu, H.; Luo, X. p31comet blocks Mad2 activation through structural mimicry. Cell 2007, 131, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.C.H.; Kulkarni, K.; Zhang, Z.; Kong, E.H.; Barford, D. Structure of the mitotic checkpoint complex. Nature 2012, 484, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Costa, P.; Martins, F.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A.B.; Rebelo, S. LAP1 is a crucial protein for the maintenance of the nuclear envelope structure and cell cycle progression. Mol. Cell. Biochem. 2015, 399, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Welcker, M.; Hizli, A.A.; Posakony, J.J.; Aebersold, R.; Clurman, B.E. Identification of CDK2 substrates in human cell lysates. Genome Biol. 2008, 9, R149. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef] [PubMed]
- Blethrow, J.D.; Glavy, J.S.; Morgan, D.O.; Shokat, K.M. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. USA 2008, 105, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shao, Y.; Voorhees, J.J.; Fisher, G.J. Ultraviolet Irradiation-Induces Epidermal Growth Factor Receptor (EGFR) Nuclear Translocation in Human Keratinocytes. J. Cell. Biochem. 2009, 107, 873–880. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.; Takai, H.; Buonomo, S.B.C.; Eisenhaber, F.; de Lange, T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004, 18, 2108–2019. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef] [PubMed]
- Gene, T.; Consortium, O.; Gene, T.; Go, O. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Available online: http://geneontology.org/ (accessed on 23 October 2015).
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Rebelo, S.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A.B. DYT1 dystonia-associated mutant affects cytoskeletal dynamics. Microsc. Microanal. 2015, 21, 26–27. [Google Scholar] [CrossRef]
- Stark, C. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; Del-Toro, N.; et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Calderone, A.; Castagnoli, L.; Cesareni, G. Mentha: A Resource for Browsing Integrated Protein-Interaction Networks. Nat. Methods 2013, 10, 690–691. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.R.; Jurisica, I. Online Predicted Human Interaction Database. Bioinformatics 2005, 21, 2076–2082. [Google Scholar] [CrossRef] [PubMed]
- Prieto, C.; De Las Rivas, J. APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 2006, 34, W298–W302. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.H.; Fontaine, J.F.; Vinayagam, A.; Porras, P.; Wanker, E.E.; Andrade-Navarro, M.A. Hippie: Integrating protein interaction networks with experiment based quality scores. PLoS ONE 2012, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez gene: Gene-centered information at NCBI. Nucleic Acids Res. 2007, 35, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Del-Toro, N.; Dumousseau, M.; Orchard, S.; Jimenez, R.C.; Galeota, E.; Launay, G.; Goll, J.; Breuer, K.; Ono, K.; Salwinski, L.; et al. A new reference implementation of the PSICQUIC web service. Nucleic Acids Res. 2013, 41, W601–W606. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Cytoscape Consortium. Available online: http://www.cytoscape.org/ (accessed on 23 October 2015).
- GeneMANIA Cytoscape plugin. Available online: http://apps.cytoscape.org/apps/genemania (accessed on 23 October 2015).
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Maere, S. BiNGO Cytoscape plugin. Available online: http://apps.cytoscape.org/apps/bingo (accessed on 23 October 2015).
- QIAGEN Ingenuity Pathway Analysis. Available online: www.qiagen.com/ingenuity (accessed on 23 October 2015).
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano, J.B.; Da Cruz e Silva, O.A.B.; Rebelo, S. Lamina Associated Polypeptide 1 (LAP1) Interactome and Its Functional Features. Membranes 2016, 6, 8. https://doi.org/10.3390/membranes6010008
Serrano JB, Da Cruz e Silva OAB, Rebelo S. Lamina Associated Polypeptide 1 (LAP1) Interactome and Its Functional Features. Membranes. 2016; 6(1):8. https://doi.org/10.3390/membranes6010008
Chicago/Turabian StyleSerrano, Joana B., Odete A. B. Da Cruz e Silva, and Sandra Rebelo. 2016. "Lamina Associated Polypeptide 1 (LAP1) Interactome and Its Functional Features" Membranes 6, no. 1: 8. https://doi.org/10.3390/membranes6010008
APA StyleSerrano, J. B., Da Cruz e Silva, O. A. B., & Rebelo, S. (2016). Lamina Associated Polypeptide 1 (LAP1) Interactome and Its Functional Features. Membranes, 6(1), 8. https://doi.org/10.3390/membranes6010008