Amyloid β 1-42 Can Form Ion Channels as Small as Gramicidin in Model Lipid Membranes
Abstract
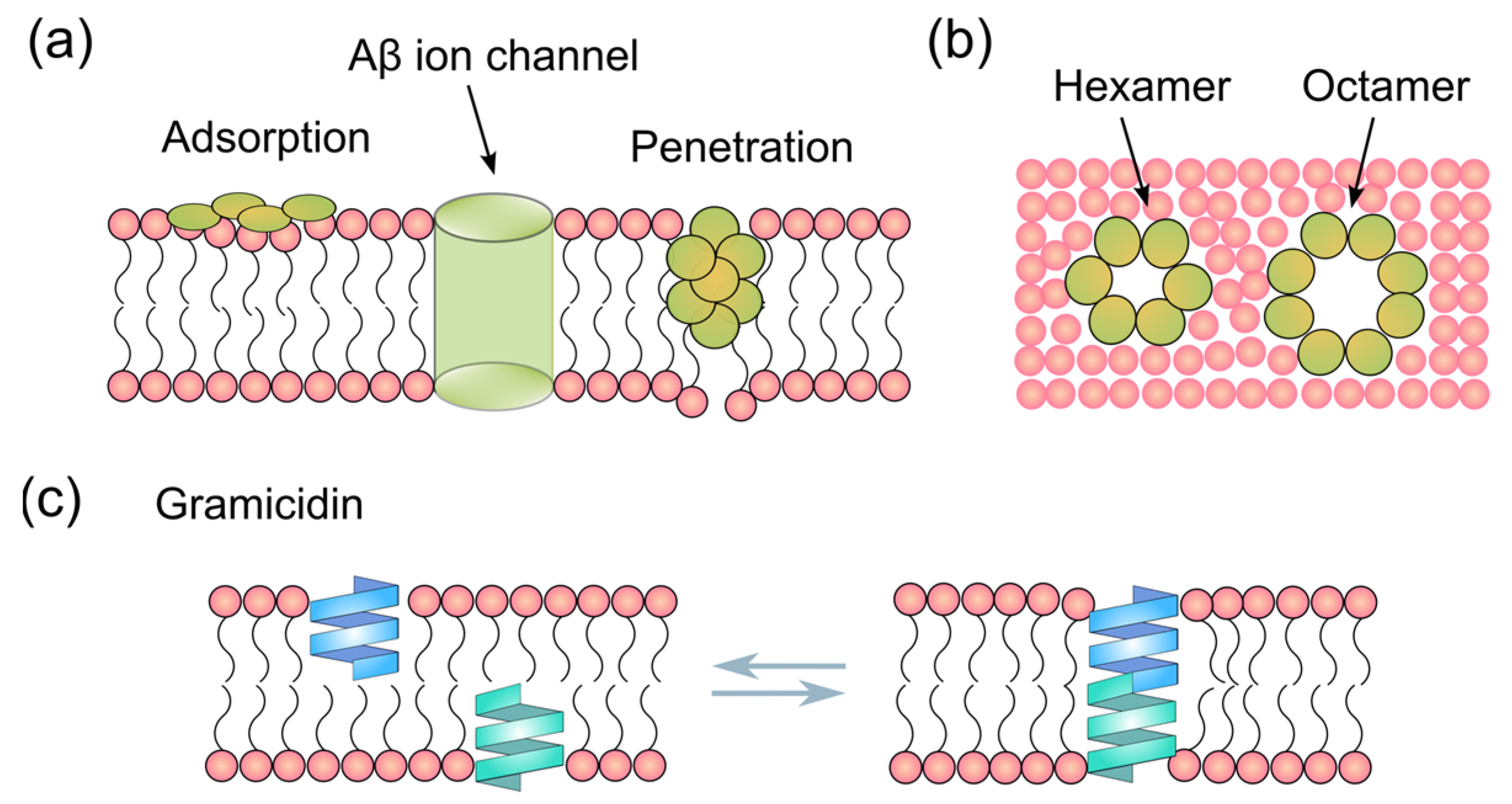
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Sample Preparation
2.2.1. Lipid Preparation for Artificial Planar Bilayers
2.2.2. Amyloid β Oligomer Preparation
2.2.3. Gramicidin Preparation
2.3. Black Lipid Membrane
Suspended Membrane Preparation
2.4. Atomic Force Microscopy (AFM)
Amyloid Oligomers and Fibrils for AFM Imaging
2.5. Data Collection and Statistical Analysis
3. Results and Discussion
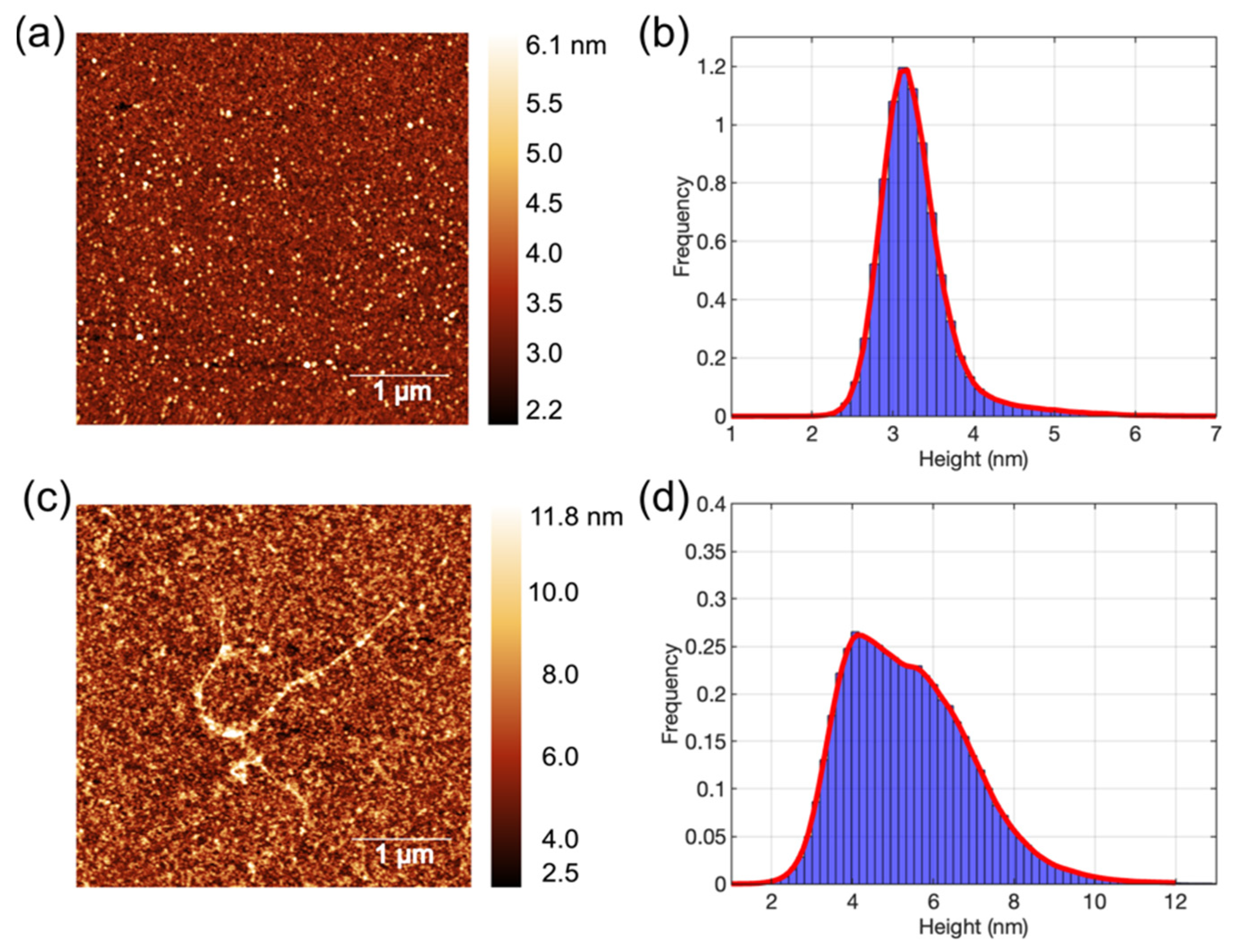
3.1. AFM Images of Aβ1-42 Oligomers and Fibrils
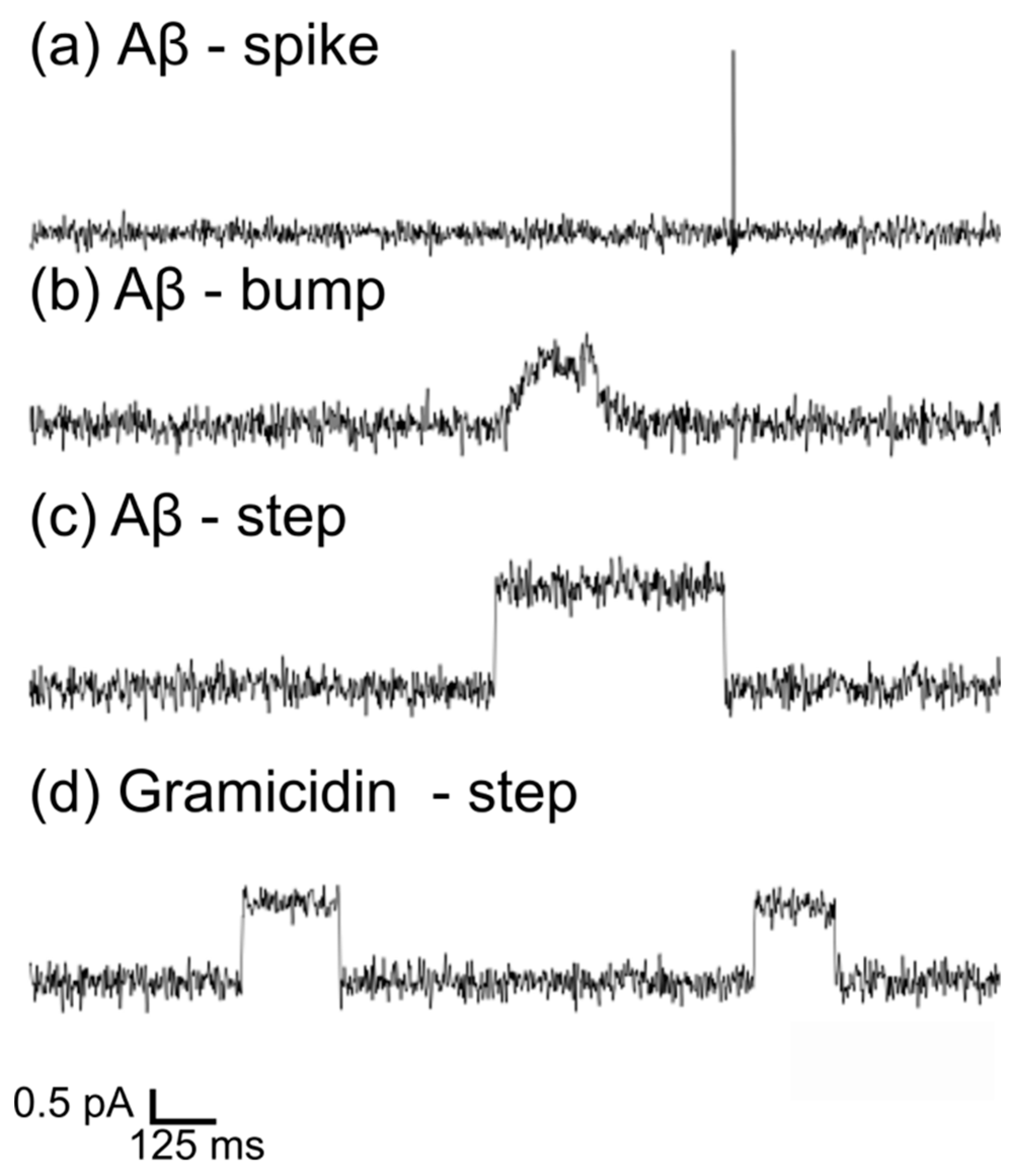
3.2. BLM Electrophysiology for Aβ1-42 and Gramicidin–Membrane Interaction
3.3. Interactions of Aβ1-42 Oligomers with Suspended Lipid Membranes
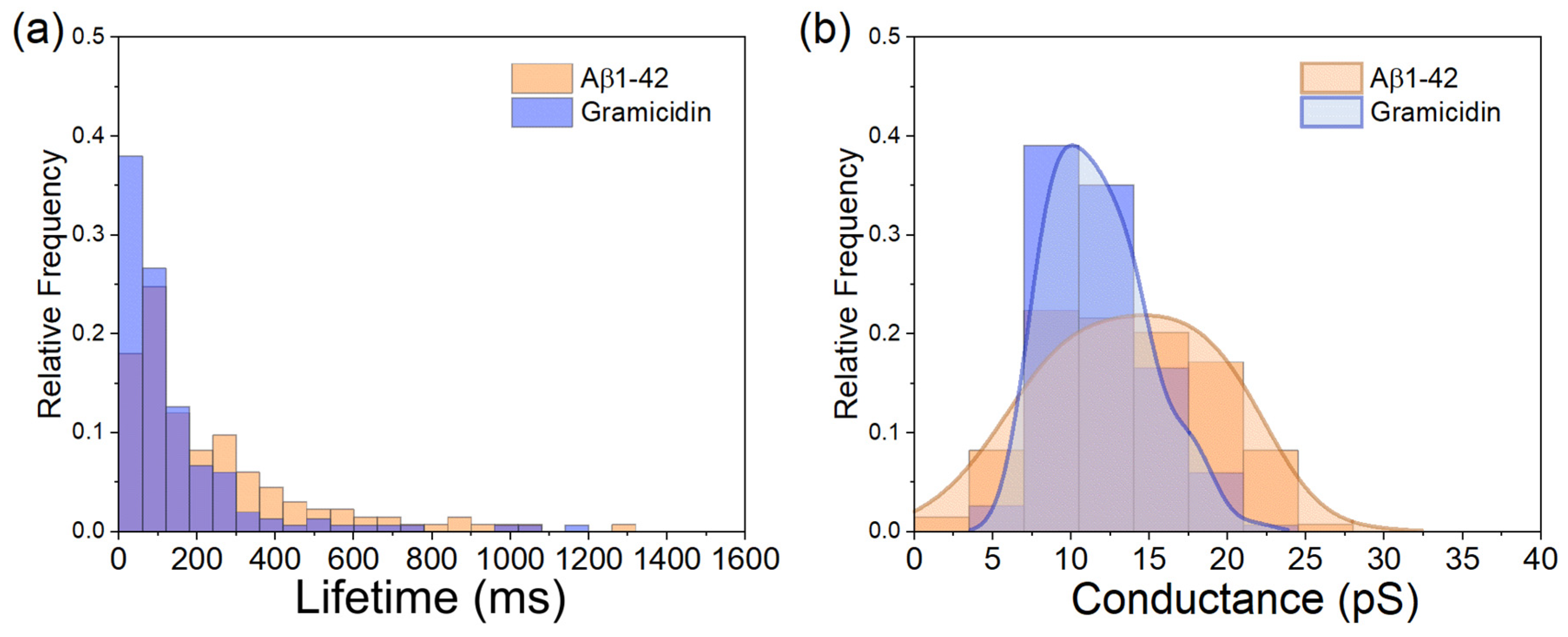
3.4. Comparison of Aβ1-42-Induced Step-like Current with Gramicidin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatia, R.; Lin, H.; Lal, R. Fresh and globular amyloid beta protein (1-42) induces rapid cellular degeneration: Evidence for AbetaP channel-mediated cellular toxicity. FASEB J. 2000, 14, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Siniscalco, D.; Francius, G.; Tarek, M.; Bali, S.K.; Laprévote, O.; Malaplate, C.; Oster, T.; Pauron, L.; Quilès, F. Molecular Insights for Alzheimer’s Disease: An Unexplored Storyline on the Nanoscale Impact of Nascent Aβ1–42 toward the Lipid Membrane. ACS Appl. Mater. Interfaces 2023, 15, 17507–17517. [Google Scholar] [CrossRef]
- Xu, Y.; Filice, C.T.; Leonenko, Z. Protective effect of trehalose sugar on amyloid-membrane interactions using BLM electrophysiology. Biophys. J. 2024, 123, 1690–1704. [Google Scholar] [CrossRef]
- Fernandez-Perez, E.J.; Peters, C.; Aguayo, L.G. Membrane Damage Induced by Amyloid Beta and a Potential Link with Neuroinflammation. Curr. Pharm. Des. 2016, 22, 1295–1304. [Google Scholar] [CrossRef]
- Drolle, E.; Hane, F.; Lee, B.; Leonenko, Z. Atomic force microscopy to study molecular mechanisms of amyloid fibril formation and toxicity in Alzheimer’s disease. Drug Metab. Rev. 2014, 46, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Viles, J.H. Imaging Amyloid-β Membrane Interactions: Ion-Channel Pores and Lipid-Bilayer Permeability in Alzheimer’s Disease. Angew. Chem. Int. Ed. 2023, 62, e202215785. [Google Scholar] [CrossRef]
- Serra-Batiste, M.; Ninot-Pedrosa, M.; Bayoumi, M.; Gairí, M.; Maglia, G.; Carulla, N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. USA 2016, 113, 10866–10871. [Google Scholar] [CrossRef]
- Bode, D.C.; Freeley, M.; Nield, J.; Palma, M.; Viles, J.H. Amyloid-β oligomers have a profound detergent-like effect on lipid membrane bilayers, imaged by atomic force and electron microscopy. J. Biol. Chem. 2019, 294, 7566–7572. [Google Scholar] [CrossRef] [PubMed]
- Arispe, N.; Pollard, H.B.; Rojas, E. Giant multilevel cation channels formed by Alzheimer disease amyloid beta-protein [A beta P-(1-40)] in bilayer membranes. Proc. Natl. Acad. Sci. USA 1993, 90, 10573–10577. [Google Scholar] [CrossRef]
- Kayed, R.; Sokolov, Y.; Edmonds, B.; McIntire, T.M.; Milton, S.C.; Hall, J.E.; Glabe, C.G. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 2004, 279, 46363–46366. [Google Scholar] [CrossRef]
- Mattson, M.P.; Cheng, B.; Davis, D.; Bryant, K.; Lieberburg, I.; Rydel, R.E. beta-Amyloid peptides destabilize calcium homeostasis and render human cortical neurons vulnerable to excitotoxicity. J. Neurosci. 1992, 12, 376–389. [Google Scholar] [CrossRef]
- Arispe, N.; Rojas, E.; Pollard, H.B. Alzheimer disease amyloid beta protein forms calcium channels in bilayer membranes: Blockade by tromethamine and aluminum. Proc. Natl. Acad. Sci. USA 1993, 90, 567–571. [Google Scholar] [CrossRef]
- Jang, H.; Connelly, L.; Arce, F.T.; Ramachandran, S.; Lal, R.; Kagan, B.L.; Nussinov, R. Alzheimer’s disease: Which type of amyloid-preventing drug agents to employ? Phys. Chem. Chem. Phys. 2013, 15, 8868–8877. [Google Scholar] [CrossRef]
- Moir, R.D.; Lathe, R.; Tanzi, R.E. The antimicrobial protection hypothesis of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 1602–1614. [Google Scholar] [CrossRef]
- Gosztyla, M.L.; Brothers, H.M.; Robinson, S.R. Alzheimer’s Amyloid-β is an Antimicrobial Peptide: A Review of the Evidence. J. Alzheimer’s Dis. 2018, 62, 1495–1506. [Google Scholar] [CrossRef]
- Shafrir, Y.; Durell, S.R.; Anishkin, A.; Guy, H.R. Beta-barrel models of soluble amyloid beta oligomers and annular protofibrils. Proteins 2010, 78, 3458–3472. [Google Scholar] [CrossRef]
- Jang, H.; Zheng, J.; Nussinov, R. Models of β-Amyloid Ion Channels in the Membrane Suggest That Channel Formation in the Bilayer Is a Dynamic Process. Biophys. J. 2007, 93, 1938–1949. [Google Scholar] [CrossRef]
- Jang, H.; Arce, F.T.; Ramachandran, S.; Capone, R.; Azimova, R.; Kagan, B.L.; Nussinov, R.; Lal, R. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer’s Disease and Down syndrome. Proc. Natl. Acad. Sci. USA 2010, 107, 6538–6543. [Google Scholar] [CrossRef]
- Liang, R.; Torres-Flores, A.P.; Qi, S.; Khursheed, A.; Tian, Y.; Szwedziak, P.; Baker, M.D.; Volkov, V.A.; Darbari, V.C.; Viles, J.H. Structural architecture of amyloid-β oligomers, curvilinear protofibrils and annular assemblies, imaged by cryo-EM and cryo-ET. bioRxiv 2024. bioRxiv:2024.03.01.582902. [Google Scholar] [CrossRef]
- Bode, D.C.; Baker, M.D.; Viles, J.H. Ion Channel Formation by Amyloid-β42 Oligomers but Not Amyloid-β40 in Cellular Membranes. J. Biol. Chem. 2017, 292, 1404–1413. [Google Scholar] [CrossRef]
- Cizas, P.; Budvytyte, R.; Morkuniene, R.; Moldovan, R.; Broccio, M.; Lösche, M.; Niaura, G.; Valincius, G.; Borutaite, V. Size-dependent neurotoxicity of beta-amyloid oligomers. Arch. Biochem. Biophys. 2010, 496, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Liang, R.; Kumar, A.; Szwedziak, P.; Viles, J.H. 3D-visualization of amyloid-β oligomer interactions with lipid membranes by cryo-electron tomography. Chem. Sci. 2021, 12, 6896–6907. [Google Scholar] [CrossRef] [PubMed]
- Lendel, C.; Bjerring, M.; Dubnovitsky, A.; Kelly, R.T.; Filippov, A.; Antzutkin, O.N.; Nielsen, N.C.; Härd, T. A Hexameric Peptide Barrel as Building Block of Amyloid-β Protofibrils. Angew. Chem. Int. Ed. 2014, 53, 12756–12760. [Google Scholar] [CrossRef]
- Ciudad, S.; Puig, E.; Botzanowski, T.; Meigooni, M.; Arango, A.S.; Do, J.; Mayzel, M.; Bayoumi, M.; Chaignepain, S.; Maglia, G.; et al. Aβ(1-42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat. Commun. 2020, 11, 3014. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.D.; Dubos, R.J. Fractionation of the bactericidal agent from cultures of a soil Bacillus. J. Biol. Chem. 1940, 132, 791–792. [Google Scholar] [CrossRef]
- Pavithrra, G.; Rajasekaran, R. Gramicidin Peptide to Combat Antibiotic Resistance: A Review. Int. J. Pept. Res. Ther. 2020, 26, 191–199. [Google Scholar] [CrossRef]
- O’Connell, A.M.; Koeppe, R.E.; Andersen, O.S. Kinetics of Gramicidin Channel Formation in Lipid Bilayers: Transmembrane Monomer Association. Science 1990, 250, 1256–1259. [Google Scholar] [CrossRef]
- Lum, K.; Ingólfsson, H.I.; Koeppe, R.E.; Andersen, O.S. Exchange of Gramicidin between Lipid Bilayers: Implications for the Mechanism of Channel Formation. Biophys. J. 2017, 113, 1757–1767. [Google Scholar] [CrossRef]
- Kelkar, D.A.; Chattopadhyay, A. The gramicidin ion channel: A model membrane protein. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1768, 2011–2025. [Google Scholar] [CrossRef]
- Stine, W.B.; Dahlgren, K.N.; Krafft, G.A.; LaDu, M.J. In Vitro Characterization of Conditions for Amyloid-β Peptide Oligomerization and Fibrillogenesis*. J. Biol. Chem. 2003, 278, 11612–11622. [Google Scholar] [CrossRef]
- Winterhalter, M. Black lipid membranes. Curr. Opin. Colloid Interface Sci. 2000, 5, 250–255. [Google Scholar] [CrossRef]
- Kageyama, H.; Ma, T.; Sato, M.; Komiya, M.; Tadaki, D.; Hirano-Iwata, A. New Aspects of Bilayer Lipid Membranes for the Analysis of Ion Channel Functions. Membranes 2022, 12, 863. [Google Scholar] [CrossRef] [PubMed]
- Pintre, I.C.; Webb, S.J. Chapter Three-Binding and Reactivity at Bilayer Membranes. In Advances in Physical Organic Chemistry; Williams, I.H., Williams, N.H., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 47, pp. 129–183. [Google Scholar]
- Sebaaly, C.; Greige-Gerges, H.; Charcosset, C. Chapter 11-Lipid Membrane Models for Biomembrane Properties’ Investigation. In Current Trends and Future Developments on (Bio-) Membranes; Basile, A., Charcosset, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 311–340. [Google Scholar]
- Saigo, N.; Izumi, K.; Kawano, R. Electrophysiological Analysis of Antimicrobial Peptides in Diverse Species. ACS Omega 2019, 4, 13124–13130. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; McRae, D.M.; Leonenko, Z. Quantitative Analysis of the Influence of Trehalose on Amyloid-β Binding to Membranes by Localized Surface Plasmon Resonance Spectroscopy. ACS Omega 2025, 10, 12872–12879. [Google Scholar] [CrossRef]
- Abramov, A.Y.; Ionov, M.; Pavlov, E.; Duchen, M.R. Membrane cholesterol content plays a key role in the neurotoxicity of β-amyloid: Implications for Alzheimer’s disease. Aging Cell 2011, 10, 595–603. [Google Scholar] [CrossRef]
- Heron, A.J.; Thompson, J.R.; Cronin, B.; Bayley, H.; Wallace, M.I. Simultaneous Measurement of Ionic Current and Fluorescence from Single Protein Pores. J. Am. Chem. Soc. 2009, 131, 1652–1653. [Google Scholar] [CrossRef]
- Baaken, G.; Sondermann, M.; Schlemmer, C.; Rühe, J.; Behrends, J.C. Planar microelectrode-cavity array for high-resolution and parallel electrical recording of membrane ionic currents. Lab Chip 2008, 8, 938–944. [Google Scholar] [CrossRef]
- Nishio, M.; Shoji, A.; Sugawara, M. Planar Lipid Bilayers Containing Gramicidin A as a Molecular Sensing System Based on an Integrated Current. Anal. Sci. 2012, 28, 661–667. [Google Scholar] [CrossRef]
- Kondrashov, O.V.; Galimzyanov, T.R.; Molotkovsky, R.J.; Batishchev, O.V.; Akimov, S.A. Membrane-Mediated Lateral Interactions Regulate the Lifetime of Gramicidin Channels. Membranes 2020, 10, 368. [Google Scholar] [CrossRef]
- Smeets, R.M.M.; Keyser, U.F.; Krapf, D.; Wu, M.-Y.; Dekker, N.H.; Dekker, C. Salt Dependence of Ion Transport and DNA Translocation through Solid-State Nanopores. Nano Lett. 2006, 6, 89–95. [Google Scholar] [CrossRef]
- Wallace, B.A.; Ravikumar, K. The gramicidin pore: Crystal structure of a cesium complex. Science 1988, 241, 182–187. [Google Scholar] [CrossRef]
- Finkelstein, A.; Andersen, O.S. The gramicidin a channel: A review of its permeability characteristics with special reference to the single-file aspect of transport. J. Membr. Biol. 1981, 59, 155–171. [Google Scholar] [CrossRef]
- Österlund, N.; Moons, R.; Ilag, L.L.; Sobott, F.; Gräslund, A. Native Ion Mobility-Mass Spectrometry Reveals the Formation of β-Barrel Shaped Amyloid-β Hexamers in a Membrane-Mimicking Environment. J. Am. Chem. Soc. 2019, 141, 10440–10450. [Google Scholar] [CrossRef]
- Sulatskaya, A.I.; Kosolapova, A.O.; Bobylev, A.G.; Belousov, M.V.; Antonets, K.S.; Sulatsky, M.I.; Kuznetsova, I.M.; Turoverov, K.K.; Stepanenko, O.V.; Nizhnikov, A.A. β-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis. Int. J. Mol. Sci. 2021, 22, 11316. [Google Scholar] [CrossRef] [PubMed]
- Capone, R.; Quiroz, F.G.; Prangkio, P.; Saluja, I.; Sauer, A.M.; Bautista, M.R.; Turner, R.S.; Yang, J.; Mayer, M. Amyloid-beta-induced ion flux in artificial lipid bilayers and neuronal cells: Resolving a controversy. Neurotox. Res. 2009, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Quist, A.; Doudevski, I.; Lin, H.; Azimova, R.; Ng, D.; Frangione, B.; Kagan, B.; Ghiso, J.; Lal, R. Amyloid ion channels: A common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA 2005, 102, 10427–10432. [Google Scholar] [CrossRef]
- Lin, H.A.I.; Bhatia, R.; Lal, R. Amyloid β protein forms ion channels: Implications for Alzheimer’s disease pathophysiology. FASEB J. 2001, 15, 2433–2444. [Google Scholar] [CrossRef] [PubMed]
- Lashuel, H.A.; Hartley, D.; Petre, B.M.; Walz, T.; Lansbury, P.T. Amyloid pores from pathogenic mutations. Nature 2002, 418, 291. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Hartley, D.M.; Petre, B.M.; Wall, J.S.; Simon, M.N.; Walz, T.; Lansbury, P.T., Jr. Mixtures of wild-type and a pathogenic (E22G) form of Abeta40 in vitro accumulate protofibrils, including amyloid pores. J. Mol. Biol. 2003, 332, 795–808. [Google Scholar] [CrossRef]
- Lee, J.; Kim, Y.H.; Arce, F.T.; Gillman, A.L.; Jang, H.; Kagan, B.L.; Nussinov, R.; Yang, J.; Lal, R. Amyloid β Ion Channels in a Membrane Comprising Brain Total Lipid Extracts. ACS Chem. Neurosci. 2017, 8, 1348–1357. [Google Scholar] [CrossRef]
- Drolle, E.; Negoda, A.; Hammond, K.; Pavlov, E.; Leonenko, Z. Changes in lipid membranes may trigger amyloid toxicity in Alzheimer’s disease. PLoS ONE 2017, 12, e0182194. [Google Scholar] [CrossRef] [PubMed]
- Kourie, J.I.; Henry, C.L.; Farrelly, P. Diversity of amyloid beta protein fragment [1-40]-formed channels. Cell. Mol. Neurobiol. 2001, 21, 255–284. [Google Scholar] [CrossRef] [PubMed]
- Capone, R.; Jang, H.; Kotler, S.A.; Kagan, B.L.; Nussinov, R.; Lal, R. Probing Structural Features of Alzheimer’s Amyloid-β Pores in Bilayers Using Site-Specific Amino Acid Substitutions. Biochemistry 2012, 51, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Soscia, S.J.; Kirby, J.E.; Washicosky, K.J.; Tucker, S.M.; Ingelsson, M.; Hyman, B.; Burton, M.A.; Goldstein, L.E.; Duong, S.; Tanzi, R.E.; et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS ONE 2010, 5, e9505. [Google Scholar] [CrossRef]
- Kagan, B.L.; Jang, H.; Capone, R.; Arce, F.T.; Ramachandran, S.; Lal, R.; Nussinov, R. Antimicrobial Properties of Amyloid Peptides. Mol. Pharm. 2012, 9, 708–717. [Google Scholar] [CrossRef]

| Signal Type | Number of Membranes (N) | Sample Size (n) | Peak Conductance (pS) | Mean Conductance (pS) | Lifetimes (ms) |
|---|---|---|---|---|---|
| Spikes from oligomeric Aβ solution | 4 | 27 | 31.50 ± 24.34 | 13.38 ± 10.28 | 9.04 ± 4.58 |
| Bumps from oligomeric Aβ solution | 6 | 31 | 15.92 ± 4.34 | 8.79 ± 3.34 | 321.30 ± 426.47 |
| Steps from oligomeric Aβ solution | 7 | 137 | 18.85 ± 7.98 | 14.40 ± 7.64 | 277.20 ± 371.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Bukhteeva, I.; Potsiluienko, Y.; Leonenko, Z. Amyloid β 1-42 Can Form Ion Channels as Small as Gramicidin in Model Lipid Membranes. Membranes 2025, 15, 204. https://doi.org/10.3390/membranes15070204
Xu Y, Bukhteeva I, Potsiluienko Y, Leonenko Z. Amyloid β 1-42 Can Form Ion Channels as Small as Gramicidin in Model Lipid Membranes. Membranes. 2025; 15(7):204. https://doi.org/10.3390/membranes15070204
Chicago/Turabian StyleXu, Yue, Irina Bukhteeva, Yurii Potsiluienko, and Zoya Leonenko. 2025. "Amyloid β 1-42 Can Form Ion Channels as Small as Gramicidin in Model Lipid Membranes" Membranes 15, no. 7: 204. https://doi.org/10.3390/membranes15070204
APA StyleXu, Y., Bukhteeva, I., Potsiluienko, Y., & Leonenko, Z. (2025). Amyloid β 1-42 Can Form Ion Channels as Small as Gramicidin in Model Lipid Membranes. Membranes, 15(7), 204. https://doi.org/10.3390/membranes15070204






