Simultaneous Recording of Remote Domain Dynamics in Membrane Proteins Using the Double-Labeled DXB/DXT Technique
Abstract
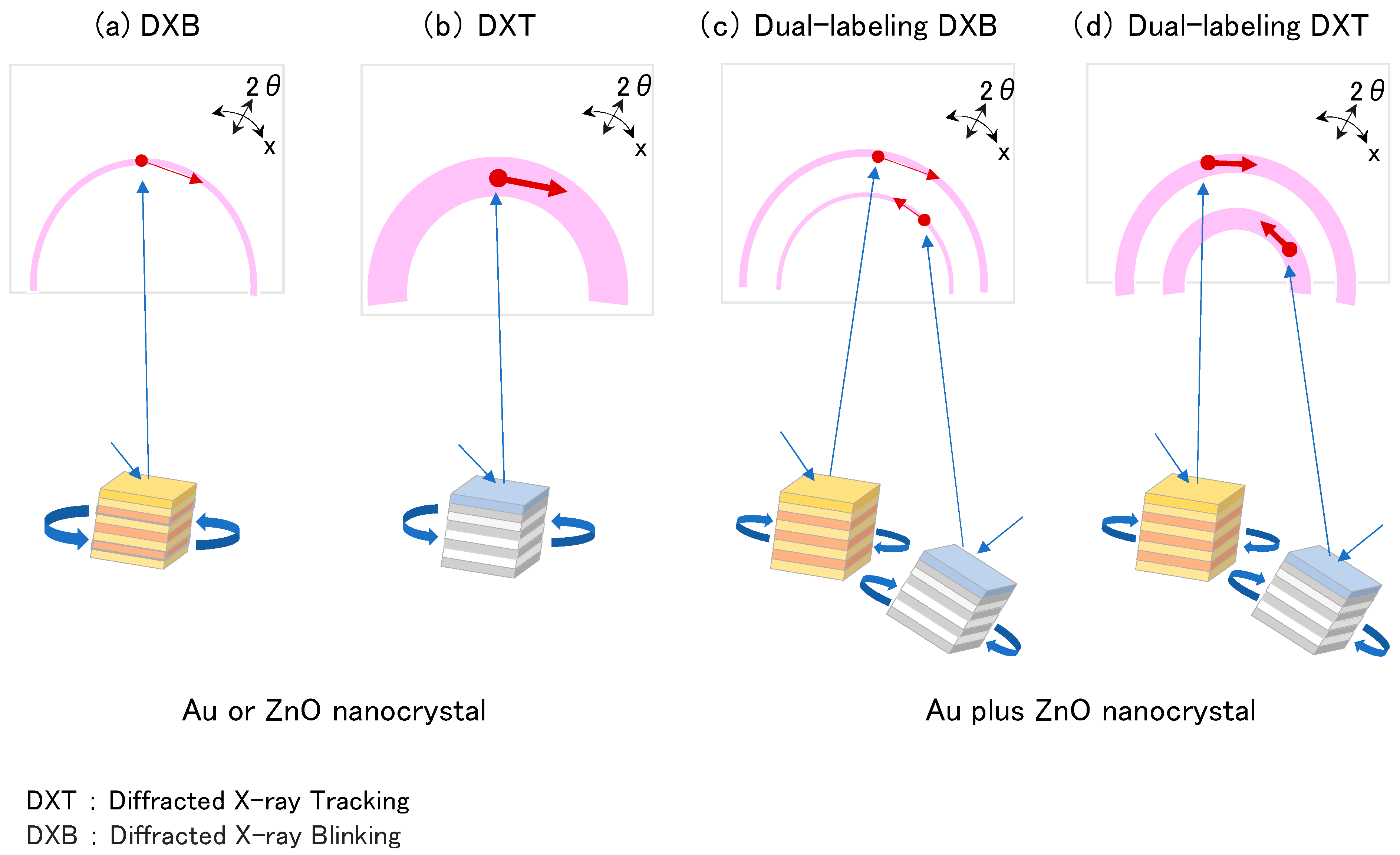
1. Introduction
2. Methods
2.1. Sample Preparation for Dual-Labeling Living-Cell DXB
2.2. Sample Preparation for Dual-Labeling DXT
2.3. DXB and DXT Recording
2.4. Data Analysis for DXB
2.5. Image Analysis for DXT
3. Results
3.1. Motion Analysis of 5-HT2A Serotonin Receptor on the Living Cells Using Dual-Labeling DXB Technique
3.2. Intracellular Domain Dynamics of TRPV1 Channel Visualized by the Dual-Labeling DXT Technique
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Adrian, M.; Dubochet, J.; Lepault, J.; McDowall, A.W. Cryo-electron microscopy of viruses. Nature 1984, 308, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Brilot, A.F.; Chen, J.Z.; Cheng, A.; Pan, J.; Harrison, S.C.; Potter, C.S.; Carragher, B.; Henderson, R.; Grigorieff, N. Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol. 2012, 177, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012, 415, 406–418. [Google Scholar] [CrossRef]
- Nannenga, B.L.; Shi, D.; Leslie, A.G.W.; Gonen, T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods 2014, 11, 927–930. [Google Scholar] [CrossRef]
- Gavira, J.A. Current trends in protein crystallization. Arch. Biochem. Biophys. 2016, 602, 3–11. [Google Scholar] [CrossRef]
- Senda, M.; Senda, T. Anaerobic crystallization of proteins. Biophys. Rev. 2018, 10, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.C.; Suzuki, Y.; Yagi, N.; Adachi, S.; Ishibashi, M.; Suda, H.; Toyota, K.; Yanagihara, M. Tracking of individual nanocrystals using diffracted X rays. Phys. Rev. E. 2000, 62, 3843. [Google Scholar] [CrossRef]
- Ohkubo, T.; Shiina, T.; Kawaguchi, K.; Sasaki, D.; Inamasu, R.; Yang, Y.; Li, Z.; Taninaka, K.; Sakaguchi, M.; Fujimura, S.; et al. Visualizing Intramolecular Dynamics of Membrane Proteins. Int. J. Mol. Sci. 2022, 23, 14539. [Google Scholar] [CrossRef]
- Sekiguchi, H.; Kuramochi, M.; Ikezaki, K.; Okamura, Y.; Yoshimura, K.; Matsubara, K.; Chang, J.W.; Ohta, N.; Kubo, T.; Mio, K.; et al. Diffracted X-ray Blinking Tracks Single Protein Motions. Sci. Rep. 2018, 8, 17090. [Google Scholar] [CrossRef]
- Mio, K.; Ishihara, M.; Fujimura, S.; Sasaki, D.; Nozawa, S.; Ichiyanagi, K.; Fukaya, R.; Adachi, S.I.; Kuramochi, M.; Sekiguchi, H.; et al. X-ray-based living-cell motion analysis of individual serotonin receptors. Biochem. Biophys. Res. Commun. 2020, 529, 306–313. [Google Scholar] [CrossRef]
- Mio, K.; Fujimura, S.; Ishihara, M.; Kuramochi, M.; Sekiguchi, H.; Kubo, T.; Sasaki, Y.C. Living-Cell Diffracted X-ray Tracking Analysis Confirmed Internal Salt Bridge Is Critical for Ligand-Induced Twisting Motion of Serotonin Receptors. Int. J. Mol. Sci. 2021, 22, 5285. [Google Scholar] [CrossRef]
- Barnes, N.M.; Ahern, G.P.; Becamel, C.; Bockaert, J.; Camilleri, M.; Chaumont-Dubel, S.; Claeysen, S.; Cunningham, K.A.; Fone, K.C.; Gershon, M.; et al. International Union of Basic and Clinical Pharmacology. CX. Classification of Receptors for 5-hydroxytryptamine; Pharmacology and Function. Pharmacol. Rev. 2021, 73, 310–520. [Google Scholar]
- Nichols, D.E.; Nichols, C.D. Serotonin receptors. Chem. Rev. 2008, 108, 1614–1641. [Google Scholar] [CrossRef] [PubMed]
- McCorvy, J.D.; Roth, B.L. Structure and function of serotonin G protein-coupled receptors. Pharmacol. Ther. 2015, 150, 129–142. [Google Scholar] [CrossRef]
- Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D.; Julius, D. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef]
- Tominaga, M.; Caterina, M.J.; Malmberg, A.B.; Rosen, T.A.; Gilbert, H.; Skinner, K.; Raumann, B.E.; Basbaum, A.I.; Julius, D. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 1998, 21, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, M.; Julius, D. Capsaicin receptor in the pain pathway. Jpn. J. Pharmacol. 2000, 83, 20–24. [Google Scholar] [CrossRef]
- Fujimura, S.; Mio, K.; Kuramochi, M.; Sekiguchi, H.; Ikezaki, K.; Mio, M.; Hengphasatporn, K.; Shigeta, Y.; Kubo, T.; Sasaki, Y.C. Agonist and Antagonist-Diverted Twisting Motions of a Single TRPV1 Channel. J. Phys. Chem. B 2020, 124, 11617–11624. [Google Scholar] [CrossRef] [PubMed]
- Mio, K.; Ohkubo, T.; Sasaki, D.; Arai, T.; Sugiura, M.; Fujimura, S.; Nozawa, S.; Sekiguchi, H.; Kuramochi, M.; Sasaki, Y.C. Real-Time Observation of Capsaicin-Induced Intracellular Domain Dynamics of TRPV1 Using the Diffracted X-ray Tracking Method. Membranes 2023, 13, 708. [Google Scholar] [CrossRef] [PubMed]
- Terpe, K. Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef]
- Fujii, Y.; Kaneko, M.; Neyazaki, M.; Nogi, T.; Kato, Y.; Takagi, J. PA tag: A versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Protein Expr. Purif. 2014, 95, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, S.; Adachi, S.I.; Takahashi, J.I.; Tazaki, R.; Guérin, L.; Daimon, M.; Tomita, A.; Sato, T.; Chollet, M.; Collet, E. Developing 100 ps-resolved X-ray structural analysis capabilities on beamline NW14A at the Photon Factory Advanced Ring. J. Synchrotron. Rad. 2007, 14, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://doi.org/10.5281/zenodo.60550 (accessed on 21 March 2024).
- Okumura, Y.; Oka, T.; Kataoka, M.; Taniguchi, Y.; Sasaki, Y.C. Picometer-scale dynamical observations of individual membrane proteins: The case of bacteriorhodopsin. Phys. Rev. E 2004, 70 Pt 1, 021917. [Google Scholar] [CrossRef] [PubMed]
- Gether, U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 2000, 21, 90–113. [Google Scholar] [CrossRef] [PubMed]
- Makita, N.; Iiri, T. Biased agonism: A novel paradigm in G protein-coupled receptor signaling observed in acquired hypocalciuric hypercalcemia. Endocr. J. 2014, 61, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Hilger, D.; Masureel, M.; Kobilka, B.K. Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 2018, 25, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Cao, E.; Liao, M.; Cheng, Y.; Julius, D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 2013, 504, 113–118. [Google Scholar] [CrossRef]
- Kwon, D.H.; Zhang, F.; Suo, Y.; Bouvette, J.; Borgnia, M.J.; Lee, S.Y. Heat-dependent opening of TRPV1 in the presence of capsaicin. Nat. Struct. Mol. Biol. 2021, 28, 554–563. [Google Scholar] [CrossRef]
- Elia, G. Biotinylation reagents for the study of cell surface proteins. Proteomics 2008, 8, 4012–4024. [Google Scholar] [CrossRef]
- RRUFF Project Database. Available online: https://rruff.info/ (accessed on 21 March 2024).
- Padlan, E.A. Anatomy of the antibody molecule. Mol. Immunol. 1994, 31, 169–217. [Google Scholar] [CrossRef]
- Maynard, J.; Georgiou, G. Antibody engineering. Annu. Rev. Biomed. Eng. 2000, 2, 339–376. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, H.; Suzuki, Y.; Nishino, Y.; Kobayashi, S.; Shimoyama, Y.; Cai, W.; Nagata, K.; Okada, M.; Ichiyanagi, K.; Ohta, N.; et al. Real time ligand-induced motion mappings of AChBP and nAChR using X-ray single molecule tracking. Sci. Rep. 2014, 4, 6384. [Google Scholar] [CrossRef] [PubMed]
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef]
- Araki, K.; Watanabe-Nakayama, T.; Sasaki, D.; Sasaki, Y.C.; Mio, K. Molecular Dynamics Mappings of the CCT/TRiC Complex-Mediated Protein Folding Cycle Using Diffracted X-ray Tracking. Int. J. Mol. Sci. 2023, 24, 14850. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mio, K.; Ohkubo, T.; Sasaki, D.; Sugiura, M.; Kawaguchi, K.; Araki, K.; Taninaka, K.; Sakaguchi, M.; Nozawa, S.; Arai, T.; et al. Simultaneous Recording of Remote Domain Dynamics in Membrane Proteins Using the Double-Labeled DXB/DXT Technique. Membranes 2024, 14, 75. https://doi.org/10.3390/membranes14040075
Mio K, Ohkubo T, Sasaki D, Sugiura M, Kawaguchi K, Araki K, Taninaka K, Sakaguchi M, Nozawa S, Arai T, et al. Simultaneous Recording of Remote Domain Dynamics in Membrane Proteins Using the Double-Labeled DXB/DXT Technique. Membranes. 2024; 14(4):75. https://doi.org/10.3390/membranes14040075
Chicago/Turabian StyleMio, Kazuhiro, Tatsunari Ohkubo, Daisuke Sasaki, Mayui Sugiura, Kayoko Kawaguchi, Kazutaka Araki, Keizaburo Taninaka, Masaki Sakaguchi, Shunsuke Nozawa, Tatsuya Arai, and et al. 2024. "Simultaneous Recording of Remote Domain Dynamics in Membrane Proteins Using the Double-Labeled DXB/DXT Technique" Membranes 14, no. 4: 75. https://doi.org/10.3390/membranes14040075
APA StyleMio, K., Ohkubo, T., Sasaki, D., Sugiura, M., Kawaguchi, K., Araki, K., Taninaka, K., Sakaguchi, M., Nozawa, S., Arai, T., & Sasaki, Y. C. (2024). Simultaneous Recording of Remote Domain Dynamics in Membrane Proteins Using the Double-Labeled DXB/DXT Technique. Membranes, 14(4), 75. https://doi.org/10.3390/membranes14040075







