Bovine Serum Albumin Rejection by an Open Ultrafiltration Membrane: Characterization and Modeling
Abstract
1. Introduction
2. Theoretical Background
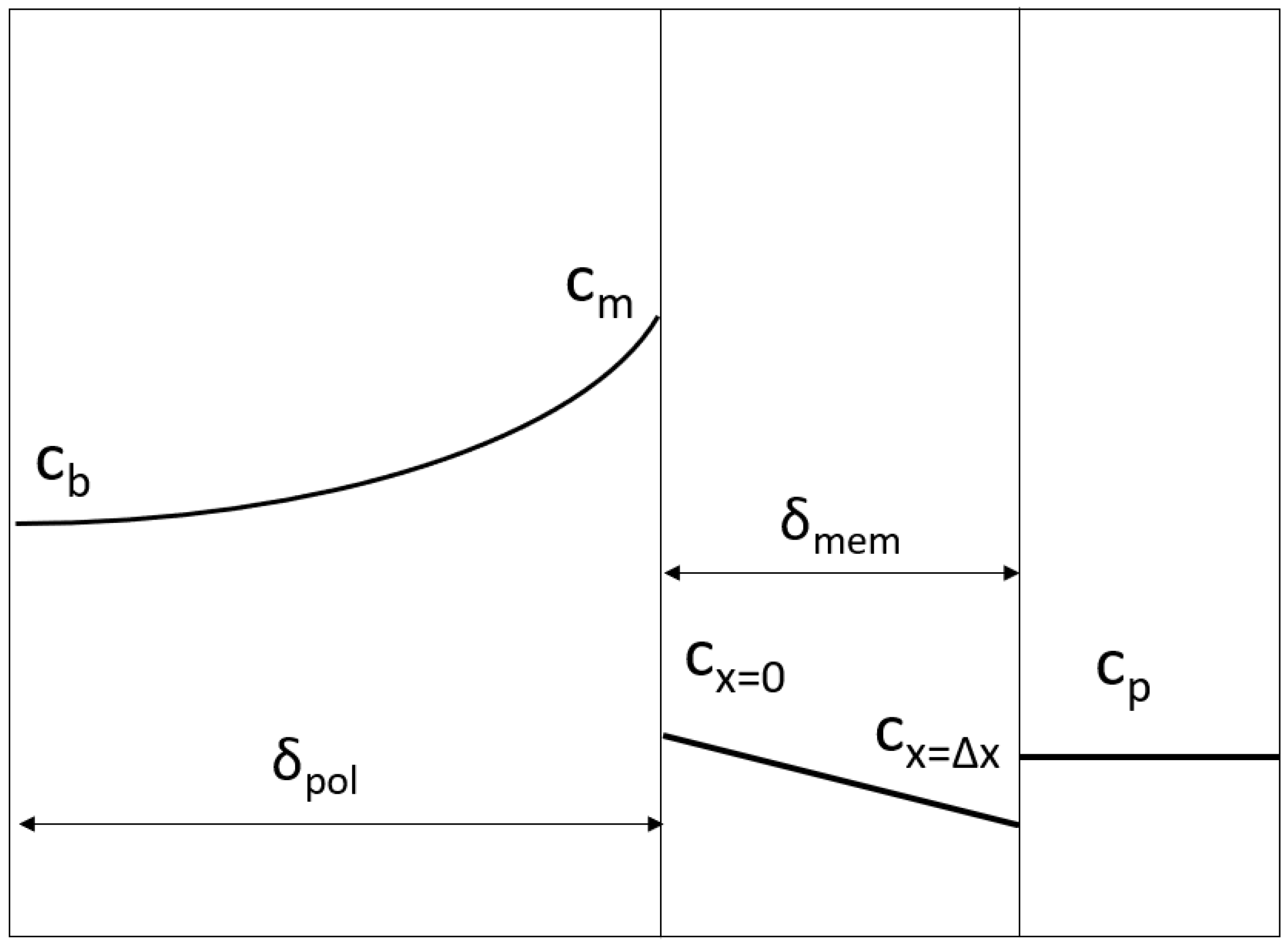
2.1. Generalized Solute Retention Equation
2.2. Mass Transfer Coefficient in the Polarization Layer
3. Materials and Methods
3.1. Materials
3.2. Membrane and Setup
3.3. Experiment
3.4. Analytical Methods
3.5. Fitting Equations
4. Results and Discussion
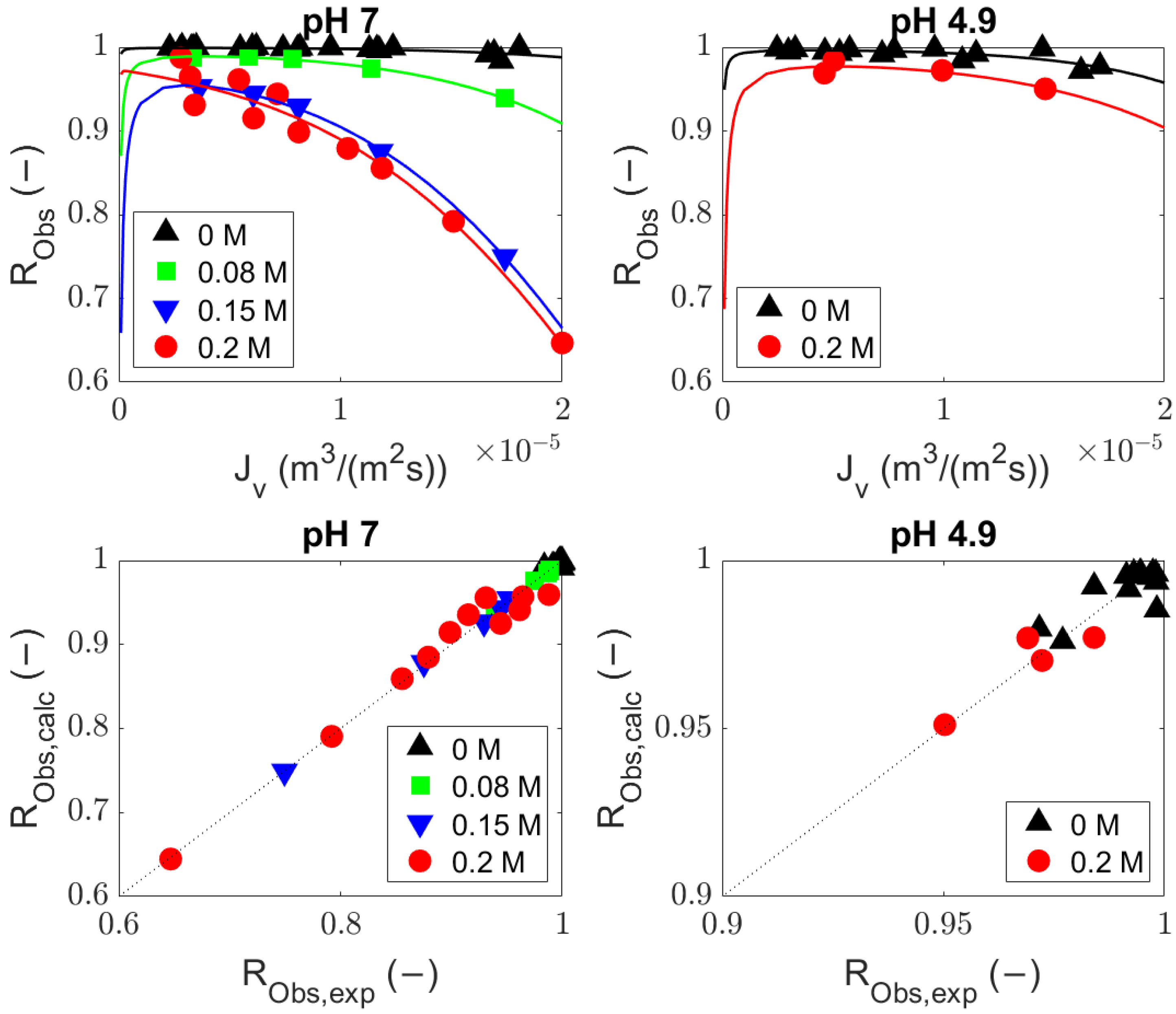
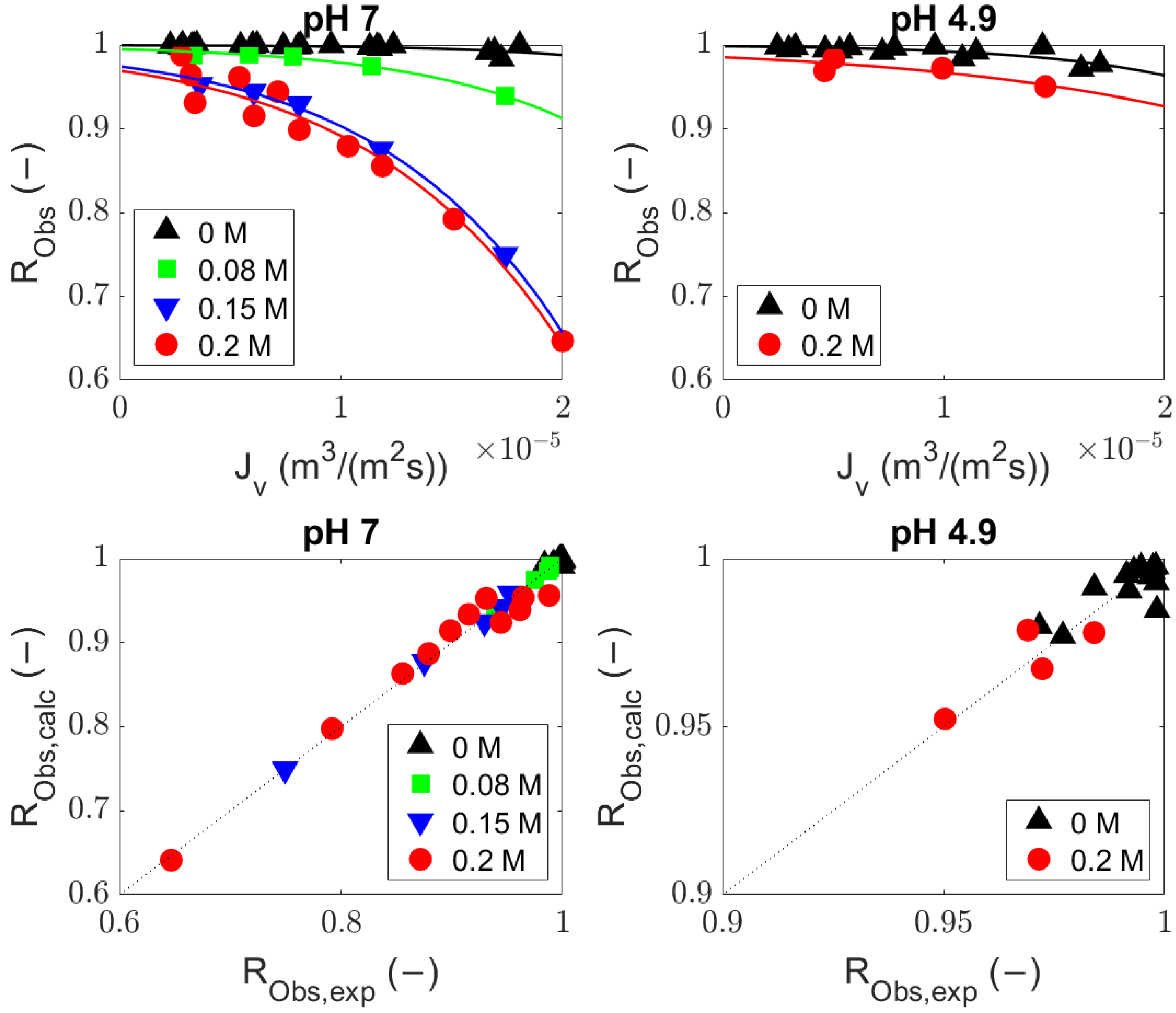
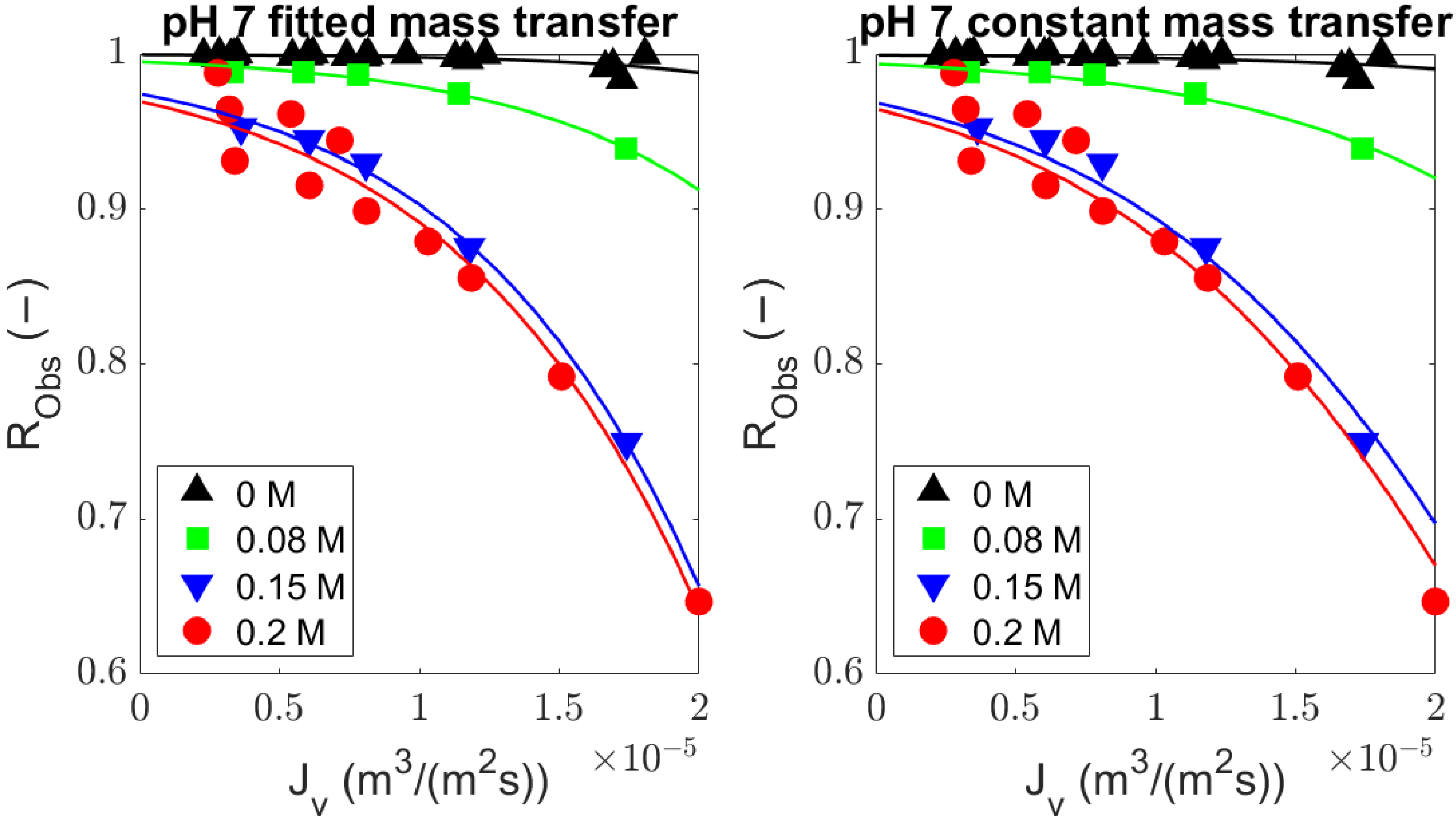
4.1. Comparison of the Generalized Solute Rejection Equation with and without Diffusion
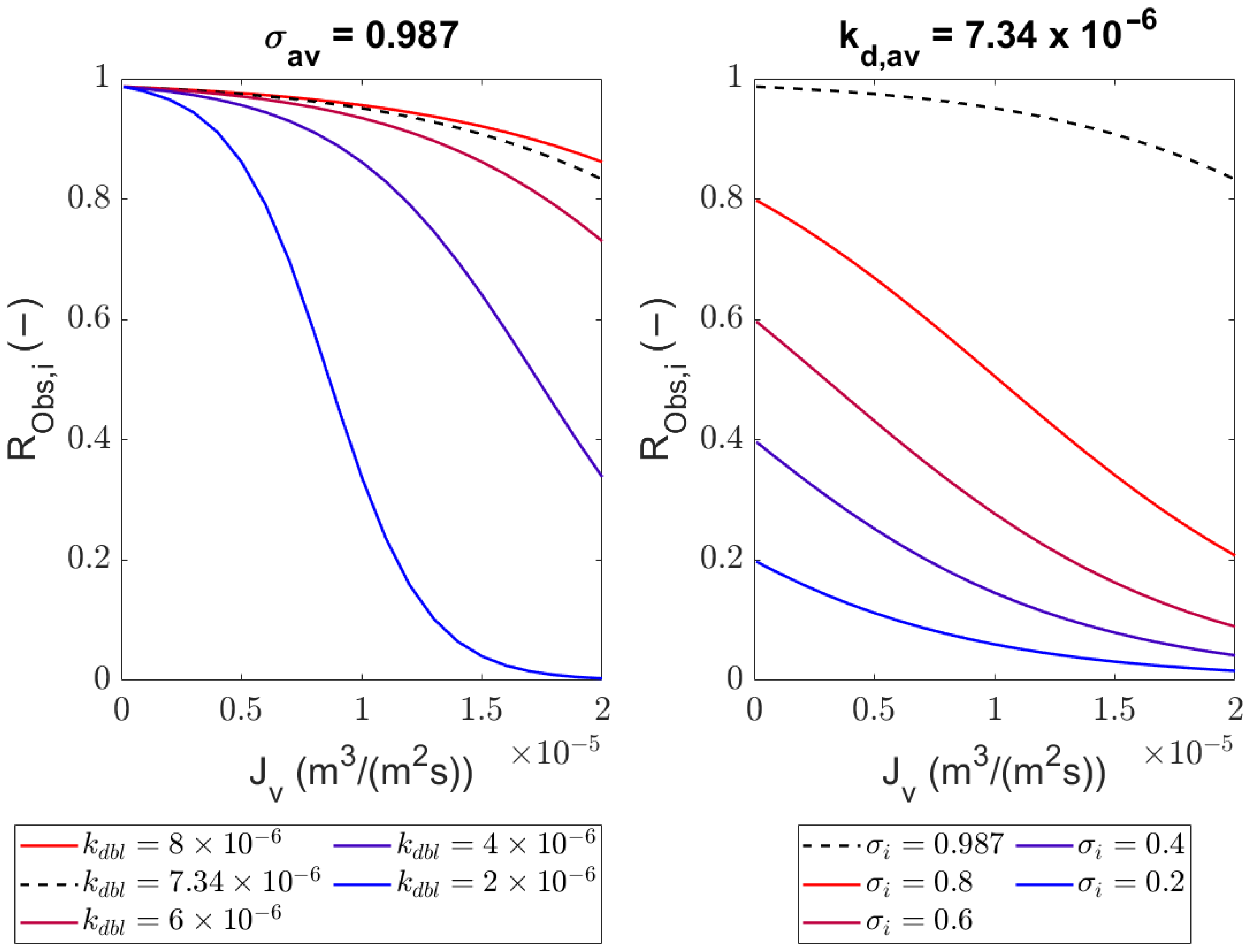
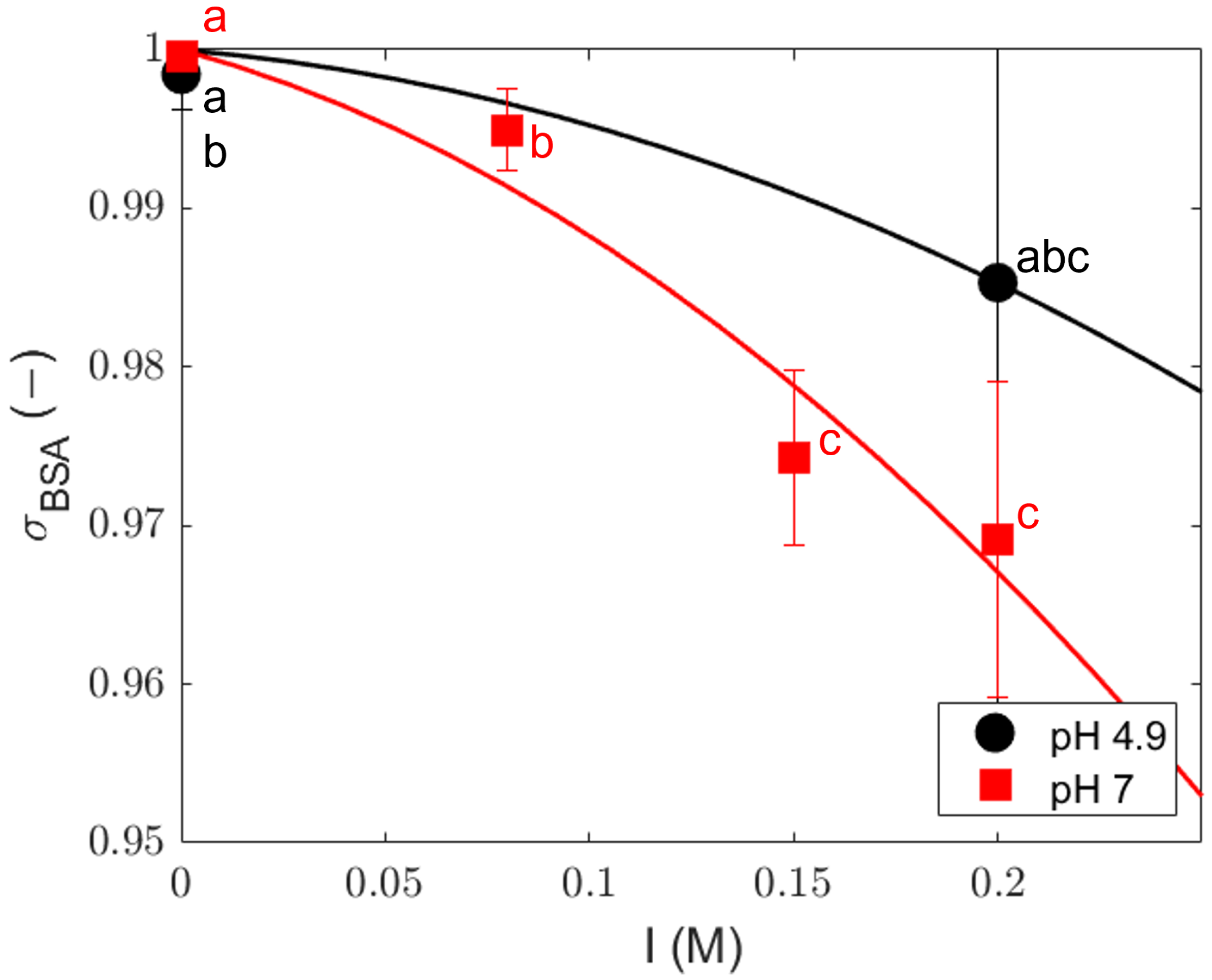
4.2. Sieving Coefficients
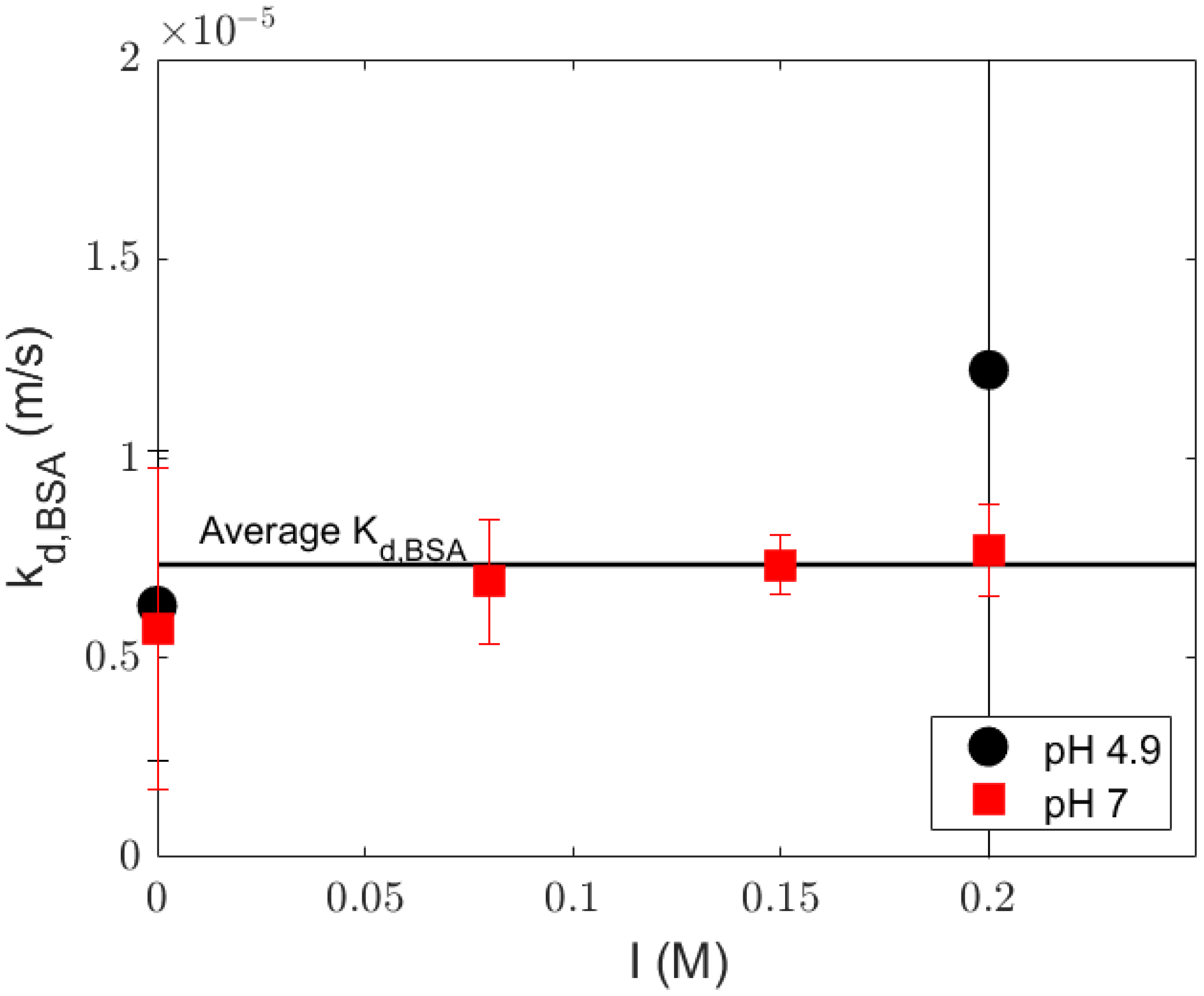
4.3. Mass Transfer Coefficient in the Polarization Layer
| Sherwood Relation | (m/s) | Adj-r2 | Source |
|---|---|---|---|
| Average fitted mass transfer | 7.34 | 0.74 | Spiral wound 1812 UF Protein |
| (this work) | |||
| Schock and Miquel [25] | 4.27 | 0.50 | Spiral wound 2540 RO and UF salt |
| Graetz–Leveque equation [16] | 2.32 | 0.02 | Heat–mass transfer analogy [30] |
| Harriot–Hamilton equation [16] | 1.97 | −2.65 | Turbulent flow in pipe [31] |
| Bandini and Morelli [26] | 3.15 | 0.03 | Spiral wound 1812 NF dextrose |
| Shi et al. [27] | 2.67 | 0.12 | Spiral wound 1812 organic solvent NF |
4.4. Utilization of the General Rejection Model for Describing Protein Transport through UF
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Nomenclatures
| Abbreviations: | |
| BSA | Bovine serum albumin |
| ED | Electrodialysis |
| HPLC | High performance liquid chromatography |
| MWCO | Molecular weight cut off |
| NF | Nanofiltration |
| RO | Reverse Osmosis |
| UF | Ultrafiltration |
| Symbols: | |
| A | Dimensionless fitted parameter for Sherwood relation [-] |
| Concentration in retentate bulk [mol m−3] | |
| Concentration at membrane interface at liquid phase [mol m−3] | |
| Concentration in the membrane [mol m−3] | |
| Concentration in permeate [mol m−3] | |
| Concentration at membrane pore entrance [mol m−3] | |
| Concentration at membrane pore exit [mol m−3] | |
| D | Diffusion coefficient [m2 s−1] |
| Diffusion coefficient in the membrane [m2 s−1] | |
| Degree of freedom [-] | |
| Hydraulic diameter [m] | |
| h | height of channel [m] |
| Solute flux [mol m−2 s−1] | |
| Convective hindrance factor [-] | |
| Diffusive hindrance factor [-] | |
| Friction factor [-] | |
| Mass transfer coefficient at concentration polarization layer [m s−1] | |
| Mass transfer coefficient in the membrane [m s−1] | |
| L | Length of membrane module [m] |
| m | Dimensionless fitted parameter for Sherwood relation [-] |
| n | Dimensionless fitted parameter for Sherwood relation [-] |
| Peclet number at concentration polarization layer [-] | |
| Peclet number in the membrane [-] | |
| Reynolds number [-] | |
| Observed rejection [-] | |
| Peclet number at concentration polarization layer [-] | |
| Schmidt number [-] | |
| Sherwood number [-] | |
| Sum of squared residuals [-] | |
| Specific surface of the spacer [m−1] | |
| Cross flow velocity [m s−1] | |
| Volumetric flux [m s−1] | |
| x | coordinate for flow direction [m] |
| Greek letters: | |
| Difference in location [m] | |
| Thickness of polarization layer [m] | |
| Thickness of membrane active layer [m] | |
| Spacer porosity [-] | |
| Solution viscosity [Pa s] | |
| Partition coefficient [-] | |
| Solution density [kg m−3] | |
| Sieving coefficient [-] |
Appendix A. Derivation of the Starov Equation
Appendix A.1. Derivation of the General Solute Rejection Equation
Appendix A.2. Derivation of the Solute Rejection Equation without Diffusion through the Membrane
Appendix B. Data Table
Appendix B.1. BSA Solution Volumetric Flux and Concentration Data at Different Condition
| pH | Ionic | TMP | Flux | SD Flux | SD | SD | ||
|---|---|---|---|---|---|---|---|---|
| Str (M) | (bar) | (m3 m−2 s−1) | (mg mL−1) | (mg mL−1) | ||||
| 4.9 | 0 | 0.2 | 3.30 × 10−6 | 8.17 × 10−8 | 3.26 | 4.20 × 10−2 | 8.55 × 10−3 | 3.54 × 10−4 |
| 4.9 | 0 | 0.2 | 2.99 × 10−6 | 3.72 × 10−8 | 2.98 | 1.10 × 10−2 | 1.60 × 10−2 | 7.07 × 10−5 |
| 4.9 | 0 | 0.2 | 2.48 × 10−6 | 3.21 × 10−8 | 3.58 | 1.97 × 10−2 | 7.05 × 10−3 | 7.07 × 10−5 |
| 4.9 | 0 | 0.5 | 5.76 × 10−6 | 6.86 × 10−8 | 3.29 | 1.04 × 10−2 | 9.10 × 10−3 | 4.24 × 10−4 |
| 4.9 | 0 | 0.5 | 5.30 × 10−6 | 5.97 × 10−8 | 3.04 | 1.13 × 10−3 | 2.12 × 10−2 | 2.12 × 10−4 |
| 4.9 | 0 | 0.5 | 4.64 × 10−6 | 1.06 × 10−7 | 3.55 | 4.17 × 10−3 | 1.95 × 10−2 | 1.41 × 10−4 |
| 4.9 | 0 | 0.8 | 7.78 × 10−6 | 4.42 × 10−8 | 3.30 | 6.36 × 10−4 | 1.09 × 10−2 | 1.41 × 10−4 |
| 4.9 | 0 | 0.8 | 7.23 × 10−6 | 6.49 × 10−8 | 3.02 | 1.20 × 10−3 | 2.60 × 10−2 | 2.12 × 10−4 |
| 4.9 | 0 | 1.5 | 1.15 × 10−5 | 4.66 × 10−8 | 3.36 | 9.40 × 10−3 | 2.70 × 10−2 | 4.24 × 10−4 |
| 4.9 | 0 | 1.5 | 1.09 × 10−5 | 3.99 × 10−8 | 2.99 | 6.93 × 10−3 | 4.75 × 10−2 | 8.49 × 10−4 |
| 4.9 | 0 | 1.5 | 9.60 × 10−6 | 6.30 × 10−8 | 3.70 | 1.07 × 10−2 | 6.95 × 10−3 | 2.12 × 10−4 |
| 4.9 | 0 | 3 | 1.71 × 10−5 | 4.31 × 10−8 | 3.41 | 1.34 × 10−3 | 7.84 × 10−2 | 5.66 × 10−4 |
| 4.9 | 0 | 3 | 1.62 × 10−5 | 3.80 × 10−8 | 3.13 | 9.90 × 10−4 | 8.86 × 10−2 | 0.00 |
| 4.9 | 0 | 3 | 1.45 × 10−5 | 3.30 × 10−8 | 3.67 | 9.83 × 10−3 | 6.35 × 10−3 | 7.07 × 10−5 |
| 4.9 | 0.2 | 0.2 | 4.62 × 10−6 | 2.72 × 10−6 | 2.92 | 1.15 × 10−2 | 9.04 × 10−2 | 4.95 × 10−4 |
| 4.9 | 0.2 | 0.5 | 5.04 × 10−6 | 4.76 × 10−8 | 3.78 | 1.83 × 10−2 | 6.02 × 10−2 | 2.83 × 10−4 |
| 4.9 | 0.2 | 1.5 | 9.95 × 10−6 | 5.82 × 10−8 | 3.77 | 1.30 × 10−2 | 1.04 × 10−1 | 7.07 × 10−4 |
| 4.9 | 0.2 | 3 | 1.46 × 10−5 | 7.83 × 10−8 | 3.84 | 2.26 × 10−2 | 1.91 × 10−1 | 1.41 × 10−4 |
| 7 | 0 | 0.2 | 3.37 × 10−6 | 2.98 × 10−8 | 2.92 | 8.20 × 10−3 | 3.55 × 10−3 | 7.07 × 10−5 |
| 7 | 0 | 0.2 | 2.77 × 10−6 | 5.35 × 10−8 | 3.57 | 3.24 × 10−2 | 9.15 × 10−3 | 7.07 × 10−5 |
| 7 | 0 | 0.2 | 2.30 × 10−6 | 4.83 × 10−8 | 3.65 | 5.49 × 10−2 | 2.85 × 10−3 | 3.54 × 10−4 |
| 7 | 0 | 0.2 | 3.27 × 10−6 | 4.34 × 10−8 | 3.43 | 5.22 × 10−2 | 5.50 × 10−3 | 0.00 |
| 7 | 0 | 0.2 | 3.50 × 10−6 | 5.27 × 10−8 | 3.53 | 5.40 × 10−2 | 0.00 | 0.00 |
| 7 | 0 | 0.2 | 2.85 × 10−6 | 3.86 × 10−8 | 3.54 | 4.98 × 10−2 | 0.00 | 0.00 |
| 7 | 0 | 0.5 | 6.00 × 10−6 | 4.47 × 10−8 | 3.00 | 1.57 × 10−2 | 3.70 × 10−3 | 1.41 × 10−4 |
| 7 | 0 | 0.5 | 5.47 × 10−6 | 7.81 × 10−8 | 3.75 | 4.27 × 10−2 | 5.40 × 10−3 | 1.41 × 10−4 |
| 7 | 0 | 0.5 | 6.02 × 10−6 | 6.15 × 10−8 | 3.44 | 4.24 × 10−4 | 9.50 × 10−4 | 7.07 × 10−5 |
| 7 | 0 | 0.5 | 6.20 × 10−6 | 6.88 × 10−8 | 3.52 | 6.07 × 10−2 | 0.00 | 0.00 |
| 7 | 0 | 0.8 | 8.07 × 10−6 | 4.47 × 10−8 | 2.99 | 1.29 × 10−2 | 5.90 × 10−3 | 1.41 × 10−4 |
| 7 | 0 | 0.8 | 7.41 × 10−6 | 5.57 × 10−8 | 3.86 | 6.91 × 10−2 | 5.95 × 10−3 | 7.07 × 10−5 |
| 7 | 0 | 0.8 | 8.30 × 10−6 | 8.09 × 10−8 | 3.50 | 8.63 × 10−3 | 0.00 | 0.00 |
| 7 | 0 | 0.8 | 8.17 × 10−6 | 7.27 × 10−8 | 3.45 | 4.74 × 10−2 | 2.00 × 10−3 | 1.41 × 10−4 |
| 7 | 0 | 1.5 | 1.17 × 10−5 | 5.70 × 10−8 | 3.15 | 3.04 × 10−3 | 1.15 × 10−2 | 7.07 × 10−5 |
| 7 | 0 | 1.5 | 1.13 × 10−5 | 8.18 × 10−8 | 3.95 | 5.60 × 10−2 | 9.75 × 10−3 | 2.12 × 10−4 |
| 7 | 0 | 1.5 | 1.24 × 10−5 | 6.78 × 10−8 | 3.54 | 8.20 × 10−3 | 1.55 × 10−3 | 7.07 × 10−5 |
| 7 | 0 | 1.5 | 1.16 × 10−5 | 7.99 × 10−8 | 3.50 | 5.42 × 10−2 | 7.20 × 10−3 | 2.83 × 10−4 |
| 7 | 0 | 3 | 1.72 × 10−5 | 6.82 × 10−8 | 3.18 | 6.22 × 10−3 | 5.16 × 10−2 | 0.00 |
| 7 | 0 | 3 | 1.69 × 10−5 | 8.17 × 10−8 | 4.08 | 4.57 × 10−2 | 2.97 × 10−2 | 2.12 × 10−4 |
| 7 | 0 | 3 | 1.81 × 10−5 | 9.90 × 10−8 | 3.59 | 3.18 × 10−3 | 1.75 × 10−3 | 7.07 × 10−5 |
| 7 | 0 | 3 | 1.66 × 10−5 | 1.28 × 10−7 | 3.44 | 4.96 × 10−2 | 3.01 × 10−2 | 7.07 × 10−5 |
| 7 | 0.08 | 0.2 | 3.32 × 10−6 | 8.93 × 10−8 | 2.99 | 2.09 × 10−2 | 3.54 × 10−2 | 2.83 × 10−4 |
| 7 | 0.08 | 0.5 | 5.85 × 10−6 | 3.95 × 10−8 | 3.11 | 2.43 × 10−2 | 3.54 × 10−2 | 1.41 × 10−4 |
| 7 | 0.08 | 0.8 | 7.84 × 10−6 | 3.05 × 10−8 | 3.17 | 2.06 × 10−2 | 4.23 × 10−2 | 2.12 × 10−4 |
| 7 | 0.08 | 1.5 | 1.14 × 10−5 | 5.22 × 10−8 | 3.25 | 5.44 × 10−3 | 8.33 × 10−2 | 2.83 × 10−4 |
| 7 | 0.08 | 3 | 1.74 × 10−5 | 8.27 × 10−8 | 3.20 | 2.70 × 10−2 | 1.95 × 10−1 | 1.06 × 10−3 |
| 7 | 0.15 | 0.2 | 3.63 × 10−6 | 5.54 × 10−8 | 3.61 | 6.08 × 10−3 | 1.71 × 10−1 | 4.95 × 10−4 |
| 7 | 0.15 | 0.5 | 6.06 × 10−6 | 5.43 × 10−8 | 3.61 | 9.12 × 10−3 | 2.00 × 10−1 | 1.27 × 10−3 |
| 7 | 0.15 | 0.8 | 8.10 × 10−6 | 3.17 × 10−8 | 3.69 | 2.55 × 10−2 | 2.60 × 10−1 | 1.06 × 10−3 |
| 7 | 0.15 | 1.5 | 1.18 × 10−5 | 6.07 × 10−8 | 3.71 | 2.57 × 10−2 | 4.63 × 10−1 | 3.75 × 10−3 |
| 7 | 0.15 | 3 | 1.74 × 10−5 | 7.23 × 10−8 | 3.67 | 1.82 × 10−2 | 9.20 × 10−1 | 2.62 × 10−3 |
| 7 | 0.2 | 0.2 | 3.41 × 10−6 | 6.15 × 10−8 | 3.59 | 4.17 × 10−3 | 2.47 × 10−1 | 1.98 × 10−3 |
| 7 | 0.2 | 0.2 | 3.22 × 10−6 | 4.76 × 10−8 | 3.43 | 1.47 × 10−2 | 1.22 × 10−1 | 2.33 × 10−3 |
| 7 | 0.2 | 0.2 | 2.80 × 10−6 | 1.87 × 10−8 | 3.56 | 3.93 × 10−2 | 4.32 × 10−2 | 8.49 × 10−4 |
| 7 | 0.2 | 0.5 | 6.08 × 10−6 | 6.35 × 10−8 | 3.59 | 2.27 × 10−2 | 3.04 × 10−1 | 7.07 × 10−5 |
| 7 | 0.2 | 0.5 | 5.41 × 10−6 | 6.39 × 10−8 | 3.61 | 4.26 × 10−2 | 1.39 × 10−1 | 1.77 × 10−3 |
| 7 | 0.2 | 0.8 | 8.12 × 10−6 | 7.46 × 10−8 | 3.71 | 2.60 × 10−2 | 3.76 × 10−1 | 5.66 × 10−4 |
| 7 | 0.2 | 0.8 | 7.15 × 10−6 | 5.31 × 10−8 | 3.61 | 3.33 × 10−2 | 2.01 × 10−1 | 1.91 × 10−3 |
| 7 | 0.2 | 1.5 | 1.19 × 10−5 | 6.28 × 10−8 | 3.75 | 1.28 × 10−2 | 5.41 × 10−1 | 1.27 × 10−3 |
| 7 | 0.2 | 1.5 | 1.03 × 10−5 | 1.07 × 10−7 | 3.61 | 3.68 × 10−2 | 4.36 × 10−1 | 2.76 × 10−3 |
| 7 | 0.2 | 3 | 2.00 × 10−5 | 1.79 × 10−7 | 3.58 | 1.19 × 10−2 | 1.26 | 2.12 × 10−4 |
| 7 | 0.2 | 3 | 1.51 × 10−5 | 1.49 × 10−7 | 3.57 | 4.16 × 10−2 | 7.42 × 10−1 | 5.44 × 10−3 |
Appendix B.2. Fitted Parameters of the General Rejection and Advection-Dominated Rejection Equations
| pH | I (M) | 95% CI | 95% CI | 95% CI | Adj-r2 | df | |||
|---|---|---|---|---|---|---|---|---|---|
| 7 | 0 | 0.999 a | 1.14 × 10−4 | 5.55 × 10−6 a | 6.21 × 10−6 | 2.32 × 10−6 a | 2.24 × 10−5 | 0.387 | 19 |
| 0.08 | 0.997 a | 7.15 × 10−3 | 5.91 × 10−6 a | 3.88 × 10−6 | 4.30 × 10−6 a | 1.41 × 10−5 | 0.994 | 2 | |
| 0.15 | 0.979 b | 9.08 × 10−3 | 6.32 × 10−6 a | 1.08 × 10−6 | 2.34 × 10−6 a | 3.07 × 10−6 | 0.998 | 2 | |
| 0.2 | 0.973 b | 9.00 × 10−3 | 6.70 × 10−6 a | 9.79 × 10−7 | 5.38 × 10−8 a | 0 | 0.966 | 9 | |
| 4.9 | 0 | 0.999 ab | 0.36 | 4.00 × 10−6 a | 5.28 × 10−4 | 6.97 × 10−5 a | 0.33 | 0.532 | 11 |
| 0.2 | 0.999 a | 6.39 × 10−5 | 5.28 × 10−6 a | 3.39 × 10−6 | 1.29 × 10−4 a | 1.70 × 10−4 | 0.704 | 2 |
| pH | I (M) | 95% CI | 95% CI | Adj-r2 | df | ||
|---|---|---|---|---|---|---|---|
| 7 | 0 | 0.999 a | 7.32 × 10−4 | 5.71 × 10−6 a | 4.03 × 10−6 | 0.415 | 20 |
| 0.08 | 0.995 b | 2.60 × 10−3 | 6.78 × 10−6 a | 1.55 × 10−6 | 0.986 | 3 | |
| 0.15 | 0.977 c | 5.90 × 10−3 | 6.62 × 10−6 a | 7.45 × 10−7 | 0.997 | 3 | |
| 0.2 | 0.973 c | 9.00 × 10−3 | 6.69 × 10−6 a | 9.78 × 10−7 | 0.962 | 9 | |
| 4.9 | 0 | 0.998 ab | 2.30 × 10−3 | 6.27 × 10−6 a | 3.85 × 10−6 | 0.549 | 12 |
| 0.2 | 0.985 abc | 2.64 × 10−2 | 1.19 × 10−5 a | 2.22 × 10−5 | 0.591 | 2 |
| pH | I (M) | 95% CI | Adj-r2 | df | |
|---|---|---|---|---|---|
| 7 | 0 | 0.999 a | 2.70 × 10−4 | 0.420 | 21 |
| 0.08 | 0.994 b | 8.43 × 10−4 | 0.978 | 4 | |
| 0.15 | 0.969 c | 4.10 × 10−3 | 0.981 | 4 | |
| 0.2 | 0.965 c | 4.40 × 10−3 | 0.954 | 10 | |
| 4.9 | 0 | 0.998 a | 7.33 × 10−4 | 0.570 | 13 |
| 0.2 | 0.992 b | 4.00 × 10−3 | 0.547 | 3 |
References
- Aguirre-Montesdeoca, V.; Janssen, A.E.; Van der Padt, A.; Boom, R. Modelling ultrafiltration performance by integrating local (critical) fluxes along the membrane length. J. Membr. Sci. 2019, 578, 111–125. [Google Scholar] [CrossRef]
- Bellara, S.R.; Cui, Z. A Maxwell-Stefan approach to modelling the cross-flow ultrafiltration of protein solutions in tubular membranes. Chem. Eng. Sci. 1998, 53, 2153–2166. [Google Scholar] [CrossRef]
- Noordman, T.; Ketelaar, T.; Donkers, F.; Wesselingh, J. Concentration and desalination of protein solutions by ultrafiltration. Chem. Eng. Sci. 2002, 57, 693–703. [Google Scholar] [CrossRef]
- Cheang, B.; Zydney, A.L. Separation of α-lactalbumin and β-lactoglobulin using membrane ultrafiltration. Biotechnol. Bioeng. 2003, 83, 201–209. [Google Scholar] [CrossRef]
- Saksena, S.; Zydney, A.L. Effect of solution pH and ionic strength on the separation of albumin from immunoglobulins (IgG) by selective filtration. Biotechnol. Bioeng. 1994, 43, 960–968. [Google Scholar] [CrossRef]
- Nyström, M.; Aimar, P.; Luque, S.; Kulovaara, M.; Metsämuuronen, S. Fractionation of model proteins using their physiochemical properties. Colloids Surfaces A Physicochem. Eng. Asp. 1998, 138, 185–205. [Google Scholar] [CrossRef]
- Ratnaningsih, E.; Reynard, R.; Khoiruddin, K.; Wenten, I.G.; Boopathy, R. Recent Advancements of UF-Based Separation for Selective Enrichment of Proteins and Bioactive Peptides—A Review. Appl. Sci. 2021, 11, 1078. [Google Scholar] [CrossRef]
- Song, Y.; Feng, L.; Basuray, S.; Sirkar, K.K.; Wickramasinghe, S. Hemoglobin-BSA separation and purification by internally staged ultrafiltration. Sep. Purif. Technol. 2023, 312, 123363. [Google Scholar] [CrossRef]
- Cui, Z. Protein separation using ultrafiltration—An example of multi-scale complex systems. China Particuol. 2005, 3, 343–348. [Google Scholar] [CrossRef]
- Pohl, C.; Polimeni, M.; Indrakumar, S.; Streicher, W.; Peters, G.H.; Nørgaard, A.; Lund, M.; Harris, P. Electrostatics drive oligomerization and aggregation of human interferon alpha-2a. J. Phys. Chem. B 2021, 125, 13657–13669. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Prajapati, R.; Chatterjee, S.; Mukherjee, T.K. Concentration-Dependent Reversible Self-Oligomerization of Serum Albumins through Intermolecular β-Sheet Formation. Langmuir 2014, 30, 14894–14904. [Google Scholar] [CrossRef] [PubMed]
- Biesheuvel, P.; Porada, S.; Elimelech, M.; Dykstra, J. Tutorial review of reverse osmosis and electrodialysis. J. Membr. Sci. 2022, 647, 120221. [Google Scholar] [CrossRef]
- Rohani, M.M.; Zydney, A.L. Role of electrostatic interactions during protein ultra fi ltration. Adv. Colloid Interface Sci. 2010, 160, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.; Stewart, W.; Lightfoot, E. Transport Phenomena; J. Wiley: New York, NY, USA, 2002. [Google Scholar]
- Starov, V.; Churaev, N. Separation of electrolyte solutions by reverse osmosis. Adv. Colloid Interface Sci. 1993, 43, 145–167. [Google Scholar] [CrossRef]
- van den Berg, G.B.; Rácz, I.G.; Smolders, C.A. Mass transfer coefficients in cross-flow ultrafiltration. J. Membr. Sci. 1989, 47, 25–51. [Google Scholar] [CrossRef]
- Aguirre Montesdeoca, V.; Van der Padt, A.; Boom, R.; Janssen, A.E. Modelling of membrane cascades for the purification of oligosaccharides. J. Membr. Sci. 2016, 520, 712–722. [Google Scholar] [CrossRef]
- Dechadilok, P.; Deen, W.M. Electrostatic and electrokinetic effects on hindered convection in pores. J. Colloid Interface Sci. 2009, 338, 135–144. [Google Scholar] [CrossRef]
- Ferry, J.D. Statistical Evaluation of Sieve constants in Ultrafiltration. J. Gen. Physiol. 1936, 20, 95–104. [Google Scholar] [CrossRef]
- Koen, T.J.M. Properties of Nanofiltration Membranes; Model Development and Industrial Application. Ph.D. Thesis, Technische Universiteit Eindhoven, Eindhoven, The Netherlands, 2001. [Google Scholar]
- Analystics, F. Hydrodynamic Radius Converter. 2023. Available online: https://www.fluidic.com/toolkit/hydrodynamic-radius-converter/ (accessed on 27 July 2023).
- Bowen, W.; Welfoot, J.S. Modelling the performance of membrane nanofiltration—critical assessment and model development. Chem. Eng. Sci. 2002, 57, 1121–1137. [Google Scholar] [CrossRef]
- Smith, F.G.; Deen, W.M. Electrostatic effects on the partitioning of spherical colloids between dilute bulk solution and cylindrical pores. J. Colloid Interface Sci. 1983, 91, 571–590. [Google Scholar] [CrossRef]
- Rabe, M.; Verdes, D.; Seeger, S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011, 162, 87–106. [Google Scholar] [CrossRef]
- Schock, G.; Miquel, A. Mass transfer and pressure loss in spiral wound modules. Desalination 1987, 64, 339–352. [Google Scholar] [CrossRef]
- Bandini, S.; Morelli, V. Mass transfer in 1812 spiral wound modules: Experimental study in dextrose-water nanofiltration. Sep. Purif. Technol. 2018, 199, 84–96. [Google Scholar] [CrossRef]
- Shi, B.; Marchetti, P.; Peshev, D.; Zhang, S.; Livingston, A.G. Performance of spiral-wound membrane modules in organic solvent nanofiltration – Fluid dynamics and mass transfer characteristics. J. Membr. Sci. 2015, 494, 8–24. [Google Scholar] [CrossRef]
- Gekas, V.; Hallström, B. Mass transfer in the membrane concentration polarization layer under turbulent cross flow: I. Critical literature review and adaptation of existing sherwood correlations to membrane operations. J. Membr. Sci. 1987, 30, 153–170. [Google Scholar] [CrossRef]
- Belfort, G.; Nagata, N. Fluid mechanics and cross-flow filtration: Some thoughts. Desalination 1985, 53, 57–79. [Google Scholar] [CrossRef]
- Martin, H. The generalized Lévêque equation and its practical use for the prediction of heat and mass transfer rates from pressure drop. Chem. Eng. Sci. 2002, 57, 3217–3223. [Google Scholar] [CrossRef]
- Harriott, P.; Hamilton, R. Solid-liquid mass transfer in turbulent pipe flow. Chem. Eng. Sci. 1965, 20, 1073–1078. [Google Scholar] [CrossRef]
- Yune, P.S.; Kilduff, J.E.; Belfort, G. Using co-solvents and high throughput to maximize protein resistance for poly(ethylene glycol)-grafted poly(ether sulfone) UF membranes. J. Membr. Sci. 2011, 370, 166–174. [Google Scholar] [CrossRef]
- Ulbricht, M.; Riedel, M. Ultrafiltration membrane surfaces with grafted polymer ‘tentacles’: Preparation, characterization and application for covalent protein binding. Biomaterials 1998, 19, 1229–1237. [Google Scholar] [CrossRef]
- Qamar, A.; Kerdi, S.; Ali, S.M.; Shon, H.K.; Vrouwenvelder, J.S.; Ghaffour, N. Novel hole-pillar spacer design for improved hydrodynamics and biofouling mitigation in membrane filtration. Sci. Rep. 2021, 11, 6979. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.; Francis, L.; Hashaikeh, R.; Hilal, N. 3D printed membrane-integrated spacers for enhanced antifouling in ultrafiltration. J. Appl. Polym. Sci. 2022, 139, e53019. [Google Scholar] [CrossRef]
- Li, X.; Yu, J.; Nnanna, A.A. Fouling mitigation for hollow-fiber UF membrane by sonication. Desalination 2011, 281, 23–29. [Google Scholar] [CrossRef]
- Wan, Y.; Cui, Z.; Ghosh, R. Fractionation of Proteins Using Ultrafiltration: Developments and Challenges. Dev. Chem. Eng. Miner. Process. 2005, 13, 121–136. [Google Scholar] [CrossRef]


| Membrane Specification | |
|---|---|
| Membrane name | LX |
| Model | 1812 F |
| MWCO (declared by manufacturer) | 300 kDa |
| Membrane area | 0.334 m2 |
| Spacer height | 7.8 × 10−4 m (31 mil) |
| System | Protein |
|---|---|
| Column type | TSKGel Extended G2000-G3000SWXL |
| Column size | 300 × 7.8 mm |
| Temperature | 30 °C |
| Eluent | 30% acetonitrile in Milli-Q with 0.1% trifluoroacetic acid |
| Eluent flow rate | 1.5 mL/min |
| Detector | UV (214 nm) |
| pH | Ionic Strength (M) | Adj-r2 Equation (8) | Adj-r2 Equation (9) | F Test | p Value |
|---|---|---|---|---|---|
| 7 | 0 | 0.38 | 0.42 | 0.067 | 0.80 |
| 0.08 | 0.99 | 0.98 | 5.200 | 0.15 | |
| 0.15 | 0.99 | 0.99 | 5.512 | 0.14 | |
| 0.2 | 0.95 | 0.96 | −0.000 | 1.00 | |
| 4.9 | 0 | 0.53 | 0.55 | 0.575 | 0.46 |
| 0.2 | 0.70 | 0.58 | 0.388 | 0.64 |
| pH | Ionic Strength (M) | Mean Fitted | Schock and Miquel [25] | Shi [27] | GL [16] | HH [16] | Bandini and Morelli [26] | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F Test | p Val | F Test | p Val | F Test | p Val | F Test | p Val | F Test | p Val | F Test | p Val | ||
| 7 | 0 | 0.60 | 0.45 | 0.81 | 0.38 | 4.86 | 0.04 * | 6.45 | 0.02 * | 8.50 | 0.01 * | 15.46 | 0.00 * |
| 0.08 | 1.17 | 0.35 | 36.96 | 0.01 * | 108.00 | 0.00 * | 125.01 | 0.00 * | 140.52 | 0.00 * | 263.66 | 0.00 * | |
| 0.15 | 8.06 | 0.06 | 158.55 | 0.00 * | 549.50 | 0.00 * | 654.58 | 0.00 * | 47803 | 0.00 * | 995.34 | 0.00 * | |
| 0.2 | 1.98 | 0.19 | 53.41 | 0.00 * | 182.75 | 0.00 * | 217.32 | 0.00 * | 250.31 | 0.00 * | 272.30 | 0.00 * | |
| 4.9 | 0 | 0.25 | 0.62 | 1.38 | 0.26 | 5.21 | 0.04 * | 6.37 | 0.03 * | 7.67 | 0.02 * | 16.20 | 0.00 * |
| 0.2 | 1.66 | 0.32 | 8.40 | 0.10 | 15.64 | 0.06 | 17.37 | 0.05 | 19.01 | 0.04 * | 4.93 | 0.16 | |
| Mean Fitted | Schock and Miquel [25] | Shi [27] | GL [16] | HH [16] | Bandini and Morelli [26] | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | I (M) | Adj r2 | Adj r2 | Adj r2 | Adj r2 | Adj r2 | Adj r2 | ||||||
| 7 | 0 | 0.9993 | 0.42 | 0.9999 | 0.42 | 1.0000 | 0.31 | 1.0000 | 0.26 | 1.0000 | 0.21 | 0.9975 | 0.01 |
| 0.08 | 0.9935 | 0.98 | 0.9988 | 0.86 | 0.9999 | 0.60 | 1.0000 | 0.54 | 1.0000 | 0.48 | 0.9758 | 0.04 | |
| 0.15 | 0.9686 | 0.98 | 0.9936 | 0.84 | 0.9994 | 0.47 | 0.9998 | 0.37 | 1.0000 | 0.27 | 0.8923 | 0.04 | |
| 0.2 | 0.9646 | 0.95 | 0.9933 | 0.79 | 0.9996 | 0.34 | 0.9999 | 0.22 | 1.0000 | −15.30 | 0.8903 | 0.03 | |
| 4.9 | 0 | 0.9977 | 0.57 | 0.9995 | 0.54 | 1.0000 | 0.42 | 1.0000 | 0.36 | 0.9999 | 0.32 | 0.9919 | 0.02 |
| 0.2 | 0.9916 | 0.55 | 0.9980 | −0.42 | 0.9998 | −1.41 | 0.9999 | −1.69 | 1.0002 | −1.87 | 0.9696 | 0.05 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suryawirawan, E.; Janssen, A.E.M.; Boom, R.M.; van der Padt, A. Bovine Serum Albumin Rejection by an Open Ultrafiltration Membrane: Characterization and Modeling. Membranes 2024, 14, 26. https://doi.org/10.3390/membranes14010026
Suryawirawan E, Janssen AEM, Boom RM, van der Padt A. Bovine Serum Albumin Rejection by an Open Ultrafiltration Membrane: Characterization and Modeling. Membranes. 2024; 14(1):26. https://doi.org/10.3390/membranes14010026
Chicago/Turabian StyleSuryawirawan, Eric, Anja E. M. Janssen, Remko M. Boom, and Albert van der Padt. 2024. "Bovine Serum Albumin Rejection by an Open Ultrafiltration Membrane: Characterization and Modeling" Membranes 14, no. 1: 26. https://doi.org/10.3390/membranes14010026
APA StyleSuryawirawan, E., Janssen, A. E. M., Boom, R. M., & van der Padt, A. (2024). Bovine Serum Albumin Rejection by an Open Ultrafiltration Membrane: Characterization and Modeling. Membranes, 14(1), 26. https://doi.org/10.3390/membranes14010026







