Phase-Dependent Adsorption of Myelin Basic Protein to Phosphatidylcholine Lipid Bilayers
Abstract
1. Introduction
2. Experimental Section
2.1. DLS and Confocal Microscopy: Measurements and Data Analysis
2.2. Differential Scanning Calorimetry: Measurements and Data Analysis
2.3. Temperature-Dependent UV-Vis Spectroscopy: Spectral Acquisition and Multivariate Analysis
2.4. CD Spectroscopy: Measurements and Data Analysis
2.5. FTIR ATR: Spectral Acquisition and Signature Analysis
3. Molecular Dynamics Simulations
4. Results and Discussion
4.1. Thermotropic Properties of MBP, DPPC, and MBP + DPPC: DSC and UV-Vis Data
4.2. Spectroscopic and Molecular Properties of MBP, DPPC, and MBP + DPPC: CD, FTIR, and Modeling Data (MD Data)
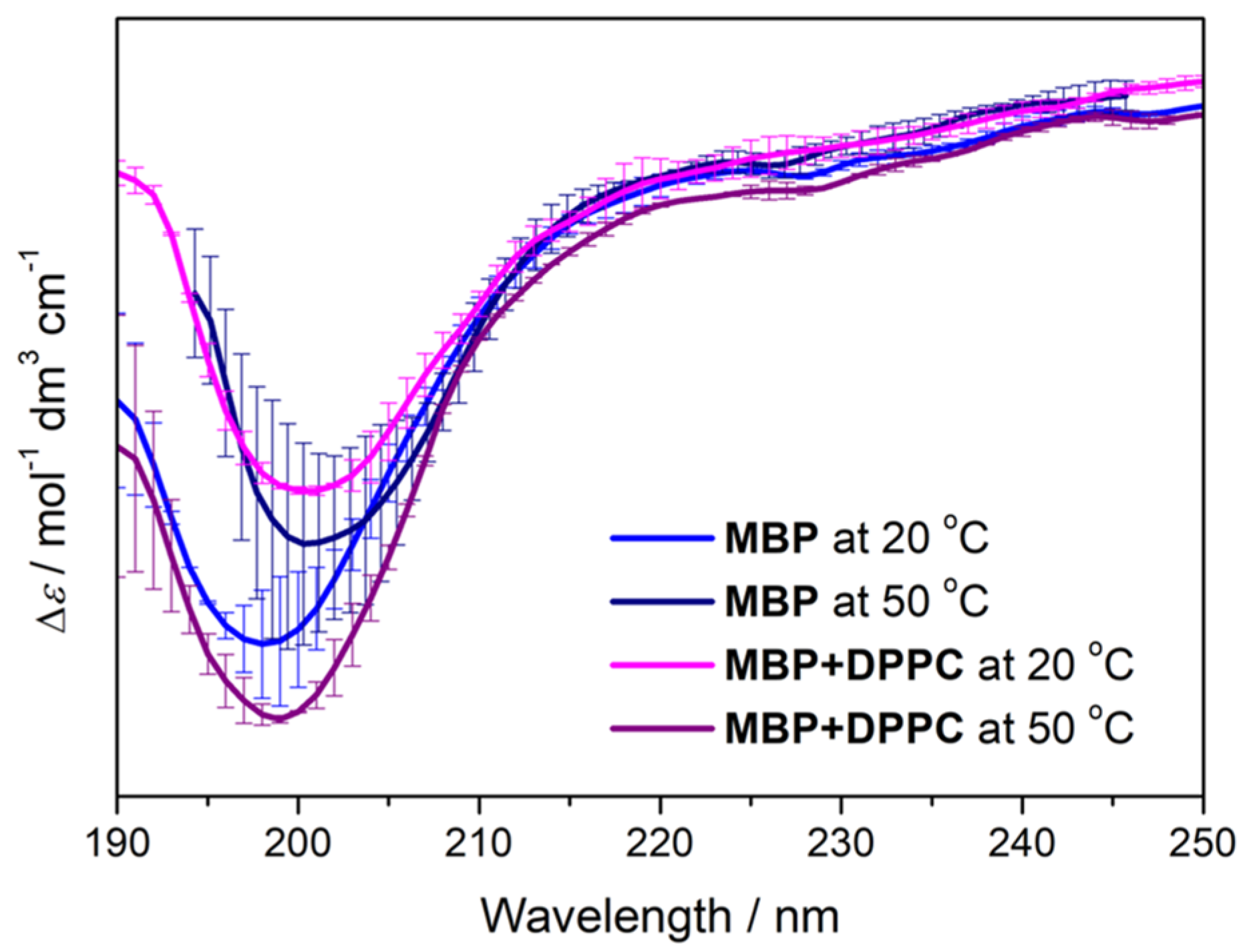
4.2.1. CD Spectra
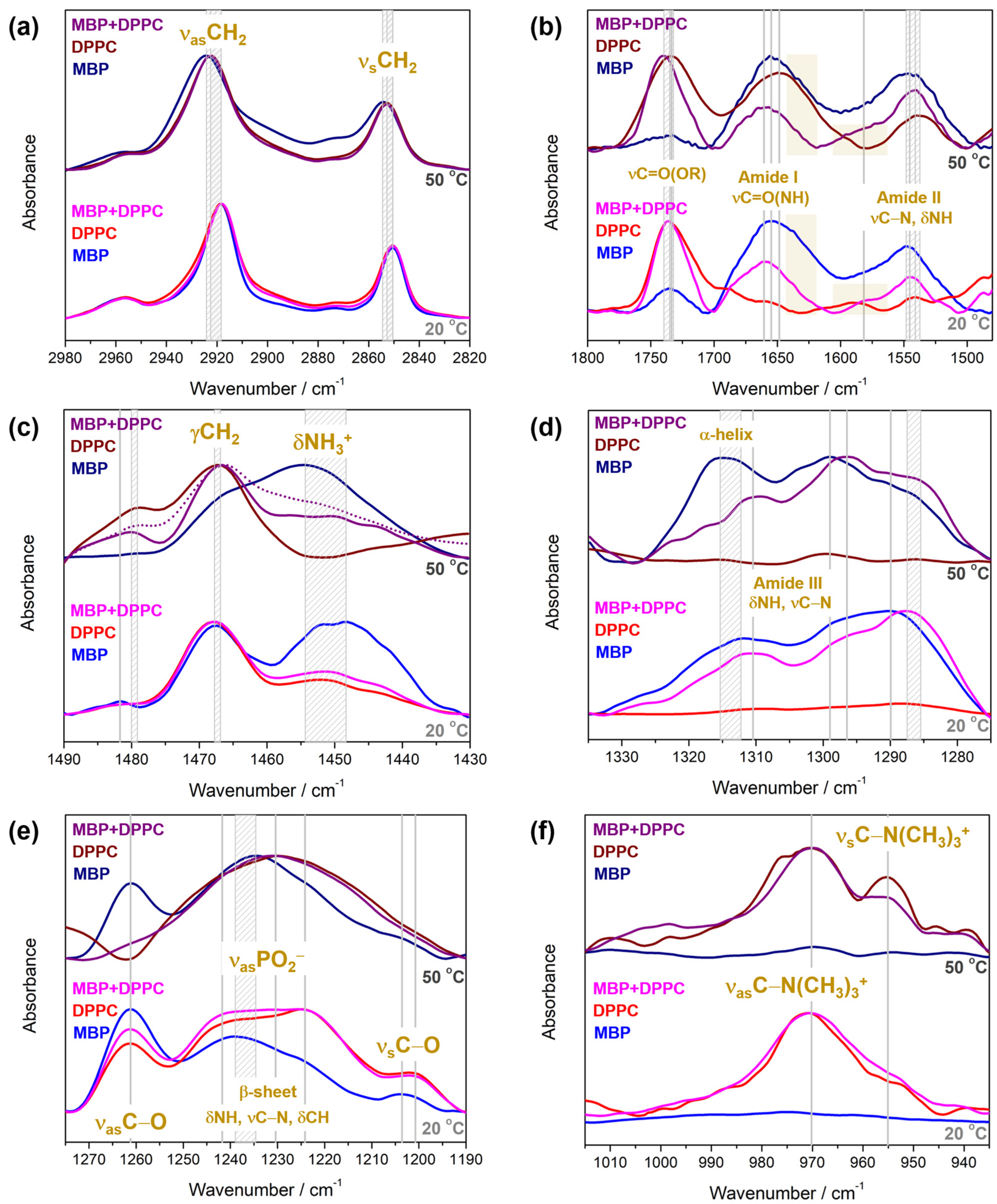
4.2.2. FTIR Spectra
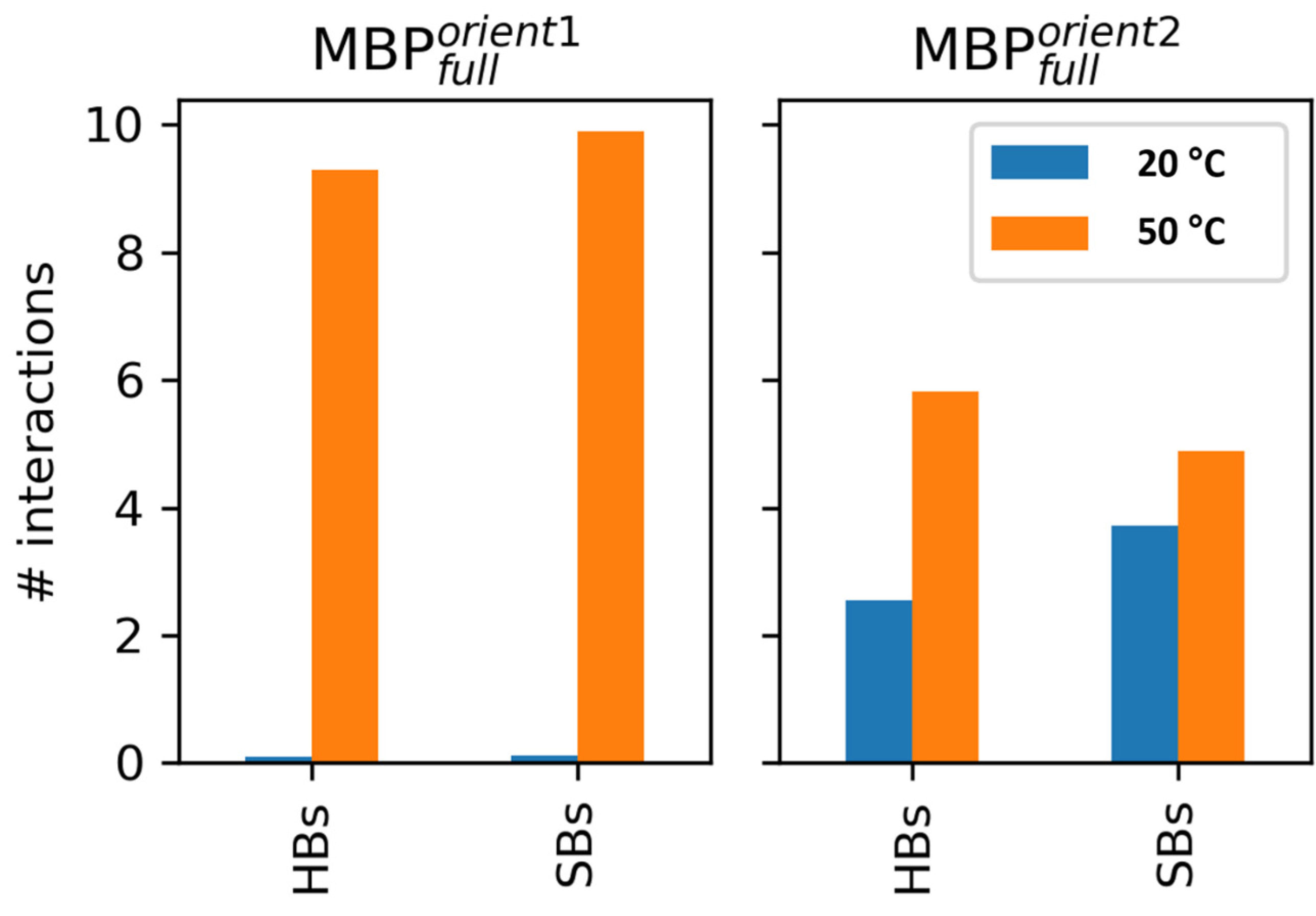
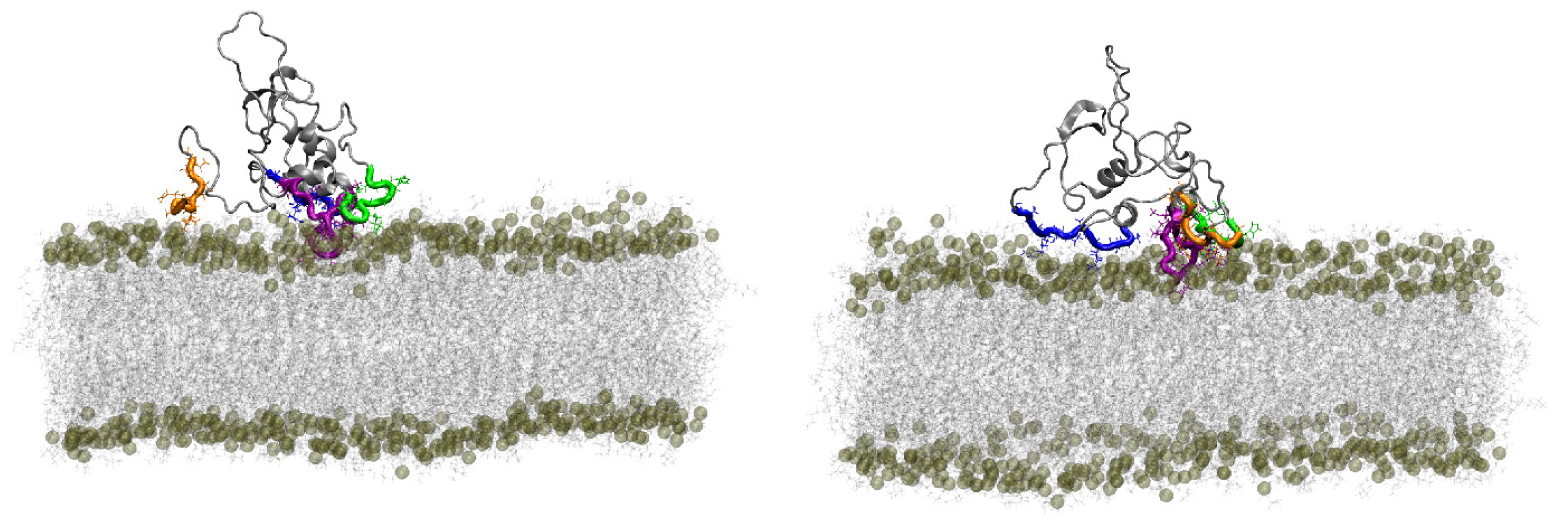
4.2.3. Modeling Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kister, A.; Kister, I. Overview of Myelin, Major Myelin Lipids, and Myelin-Associated Proteins. Front. Chem. 2023, 10, 1041961. [Google Scholar] [CrossRef] [PubMed]
- Raasakka, A.; Ruskamo, S.; Kowal, J.; Barker, R.; Baumann, A.; Martel, A.; Tuusa, J.; Myllykoski, M.; Bürck, J.; Ulrich, A.S.; et al. Membrane Association Landscape of Myelin Basic Protein Portrays Formation of the Myelin Major Dense Line. Sci. Rep. 2017, 7, 4974. [Google Scholar] [CrossRef] [PubMed]
- Jahn, O.; Tenzer, S.; Werner, H.B. Myelin Proteomics: Molecular Anatomy of an Insulating Sheath. Mol. Neurobiol. 2009, 40, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Simons, K. Cell Membranes: A Subjective Perspective. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 2569–2572. [Google Scholar] [CrossRef]
- Smith, R. The Basic Protein of CNS Myelin: Its Structure and Ligand Binding. J. Neurochem. 1992, 59, 1589–1608. [Google Scholar] [CrossRef]
- Palavicini, J.P.; Wang, C.; Chen, L.; Ahmar, S.; Higuera, J.D.; Dupree, J.L.; Han, X. Novel Molecular Insights into the Critical Role of Sulfatide in Myelin Maintenance/Function. J. Neurochem. 2016, 139, 40–54. [Google Scholar] [CrossRef]
- Martinsen, V.; Kursula, P. Multiple Sclerosis and Myelin Basic Protein: Insights into Protein Disorder and Disease. Amino Acids 2022, 54, 99–109. [Google Scholar] [CrossRef]
- Raasakka, A.; Jones, N.C.; Hoffmann, S.V.; Kursula, P. Ionic Strength and Calcium Regulate Membrane Interactions of Myelin Basic Protein and the Cytoplasmic Domain of Myelin Protein Zero. Biochem. Biophys. Res. Commun. 2019, 511, 7–12. [Google Scholar] [CrossRef]
- Boggs, J.M. Myelin Basic Protein: A Multifunctional Protein. Cell. Mol. Life Sci. 2006, 63, 1945–1961. [Google Scholar] [CrossRef]
- Harauz, G.; Ishiyama, N.; Hill, C.M.D.; Bates, I.R.; Libich, D.S.; Farès, C. Myelin Basic Protein—Diverse Conformational States of an Intrinsically Unstructured Protein and Its Roles in Myelin Assembly and Multiple Sclerosis. Micron 2004, 35, 503–542. [Google Scholar] [CrossRef]
- Di Gioacchino, M.; Bianconi, A.; Burghammer, M.; Ciasca, G.; Bruni, F.; Campi, G. Myelin Basic Protein Dynamics from Out-of-Equilibrium Functional State to Degraded State in Myelin. Biochim. Biophys. Acta-Biomembr. 2020, 1862, 183256. [Google Scholar] [CrossRef] [PubMed]
- Quarles, R.H. Myelin Lipids and Proteins: Structure, Function, and Roles in Neurological Disorders. Handb. Contemp. Neuropharmacol. 2007, 591–620. [Google Scholar] [CrossRef]
- Beniac, D.R.; Luckevich, M.D.; Czarnota, G.J.; Tompkins, T.A.; Ridsdale, R.A.; Ottensmeyer, F.P.; Moscarello, M.A.; Harauz, G. Three-Dimensional Structure of Myelin Basic Protein. I. Reconstruction via Angular Reconstitution of Randomly Oriented Single Particles. J. Biol. Chem. 1997, 272, 4261–4268. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Neugebauer, U.; Bürck, J.; Myllykoski, M.; Baumgärtel, P.; Popp, J.; Kursula, P. Charge Isomers of Myelin Basic Protein: Structure and Interactions with Membranes, Nucleotide Analogues, and Calmodulin. PLoS ONE 2011, 6, e19915. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Querejeta, I.; Alberro, A.; Muñoz-Culla, M.; Mäger, I.; Otaegui, D. Therapeutic Potential of Extracellular Vesicles for Demyelinating Diseases; Challenges and Opportunities. Front. Mol. Neurosci. 2018, 11, 434. [Google Scholar] [CrossRef] [PubMed]
- Psenicka, M.W.; Smith, B.C.; Tinkey, R.A.; Williams, J.L. Connecting Neuroinflammation and Neurodegeneration in Multiple Sclerosis: Are Oligodendrocyte Precursor Cells a Nexus of Disease? Front. Cell. Neurosci. 2021, 15, 654284. [Google Scholar] [CrossRef]
- Seth, A.; Anunciado, D.; Tian, J.; Vu, D.M.; Gnanakaran, S. Deducing Conformational Variability of Intrinsically Disordered Proteins from Infrared Spectroscopy with Bayesian Statistics. Chem. Phys. 2013, 422, 143–155. [Google Scholar] [CrossRef]
- Uversky, V.N. Paradoxes and Wonders of Intrinsic Disorder: Stability of Instability. Intrinsically Disord. Proteins 2017, 5, e1327757. [Google Scholar] [CrossRef]
- Uversky, V.N.; Finkelstein, A.V. Life in Phases: Intra-and Inter-Molecular Phase Transitions in Protein Solutions. Biomolecules 2019, 9, 842. [Google Scholar] [CrossRef]
- Bondos, S.E.; Dunker, A.K.; Uversky, V.N. Intrinsically Disordered Proteins Play Diverse Roles in Cell Signaling. Cell Commun. Signal. 2022, 20, 20. [Google Scholar] [CrossRef]
- Li, D.W.; Xie, M.; Brüschweiler, R. Quantitative Cooperative Binding Model for Intrinsically Disordered Proteins Interacting with Nanomaterials. J. Am. Chem. Soc. 2020, 142, 10730–10738. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.V.; Rakitina, T.V.; Ziganshin, R.H.; Arapidi, G.P.; Saratov, G.A.; Kudriaeva, A.A.; Belogurov, A.A. Comprehensive Atlas of the Myelin Basic Protein Interaction Landscape. Biomolecules 2021, 11, 1628. [Google Scholar] [CrossRef]
- Morris, O.M.; Torpey, J.H.; Isaacson, R.L. Intrinsically Disordered Proteins: Modes of Binding with Emphasis on Disordered Domains. Open Biol. 2021, 11, 210222. [Google Scholar] [CrossRef]
- Keniry, M.A.X.A.; Smith, R. Circular dichroic analysis of the secondary structure of myelin basic protein and derived peptides bound to detergents and to lipid vesicles. Biochim. Biophys. Acta 1979, 578, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Polverini, E.; Fasano, A.; Zito, F.; Riccio, P.; Cavatorta, P. Conformation of Bovine Myelin Basic Protein Purified with Bound Lipids. Eur. Biophys. J. 1999, 28, 351–355. [Google Scholar] [CrossRef]
- Bates, I.R.; Feix, J.B.; Boggs, J.M.; Harauz, G. An Immunodominant Epitope of Myelin Basic Protein Is an Amphipathic α-Helix. J. Biol. Chem. 2004, 279, 5757–5764. [Google Scholar] [CrossRef] [PubMed]
- Moskaitis, J.E.; Shriver, L.C.; Campagnoni, A.T. The Association of Myelin Basic Protein with Itself and Other Proteins. Neurochem. Res. 1987, 12, 409–417. [Google Scholar] [CrossRef]
- Lee, D.W.; Banquy, X.; Kristiansen, K.; Kaufman, Y.; Boggs, J.M.; Israelachvili, J.N. Lipid Domains Control Myelin Basic Protein Adsorption and Membrane Interactions between Model Myelin Lipid Bilayers. Proc. Natl. Acad. Sci. USA 2014, 111, E768–E775. [Google Scholar] [CrossRef]
- Rispoli, P.; Carzino, R.; Svaldo-Lanero, T.; Relini, A.; Cavalleri, O.; Fasano, A.; Liuzzi, G.M.; Carlone, G.; Riccio, P.; Gliozzi, A.; et al. A Thermodynamic and Structural Study of Myelin Basic Protein in Lipid Membrane Models. Biophys. J. 2007, 93, 1999–2010. [Google Scholar] [CrossRef][Green Version]
- Gorbenko, G.P.; Ioffe, V.M.; Kinnunen, P.K.J. Binding of Lysozyme to Phospholipid Bilayers: Evidence for Protein Aggregation upon Membrane Association. Biophys. J. 2007, 93, 140–153. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining Information about Protein Secondary Structures in Aqueous Solution Using Fourier Transform IR Spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Natalello, A.; Ami, D.; Doglia, S.M. Fourier Transform Infrared Spectroscopy of Intrinsically Disordered Proteins: Measurement Procedures and Data Analyses. In Intrinsically Disordered Protein Analysis: Volume 1, Methods and Experimental Tools, Methods in Molecular Biology; Uversky, V.N.U., Dunker, A.K., Eds.; Springer Science + Business Media LLC: Berlin/Heidelberg, Germany, 2012; Volume 895, pp. 229–244. ISBN 9781617799273. [Google Scholar]
- Lorenz-Fonfria, V.A. Infrared Difference Spectroscopy of Proteins: From Bands to Bonds. Chem. Rev. 2020, 120, 3466–3576. [Google Scholar] [CrossRef] [PubMed]
- Di Foggia, M.; Taddei, P.; Torreggiani, A.; Dettin, M.; Tinti, A. Self-assembling peptides for biomedical applications: Ir and raman spectroscopies for the study of secondary structure. Proteomics Res. J. 2011, 2, 231–272. [Google Scholar]
- Barth, A. Infrared Spectroscopy of Proteins. Biochim. Biophys. Acta-Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed]
- Schultz, Z.D.; Levin, I.W. Vibrational Spectroscopy of Biomembranes. Annu. Rev. Anal. Chem. 2011, 4, 343–366. [Google Scholar] [CrossRef] [PubMed]
- Tamm, L.K.; Tatulian, S.A. Infrared Spectroscopy of Proteins and Peptides in Lipid Bilayers. Q. Rev. Biophys. 1997, 30, 365–429. [Google Scholar] [CrossRef] [PubMed]
- De Meutter, J.; Goormaghtigh, E. Evaluation of Protein Secondary Structure from FTIR Spectra Improved after Partial Deuteration. Eur. Biophys. J. 2021, 50, 613–628. [Google Scholar] [CrossRef]
- Aggarwal, S.; Snaidero, N.; Pähler, G.; Frey, S.; Sánchez, P.; Zweckstetter, M.; Janshoff, A.; Schneider, A.; Weil, M.T.; Schaap, I.A.T.; et al. Myelin Membrane Assembly Is Driven by a Phase Transition of Myelin Basic Proteins into a Cohesive Protein Meshwork. PLoS Biol. 2013, 11, e1001577. [Google Scholar] [CrossRef]
- Min, Y.; Kristiansen, K.; Boggs, J.M.; Husted, C.; Zasadzinski, J.A.; Israelachvili, J. Interaction Forces and Adhesion of Supported Myelin Lipid Bilayers Modulated by Myelin Basic Protein. Proc. Natl. Acad. Sci. USA 2009, 106, 3154–3159. [Google Scholar] [CrossRef]
- Polverini, E.; Coll, E.P.; Tieleman, D.P.; Harauz, G. Conformational Choreography of a Molecular Switch Region in Myelin Basic Protein-Molecular Dynamics Shows Induced Folding and Secondary Structure Type Conversion upon Threonyl Phosphorylation in Both Aqueous and Membrane-Associated Environments. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 674–683. [Google Scholar] [CrossRef]
- Schmitt, S.; Cantuti Castelvetri, L.; Simons, M. Metabolism and Functions of Lipids in Myelin. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2015, 1851, 999–1005. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically Disordered Proteins and Their Environment: Effects of Strong Denaturants, Temperature, PH, Counter Ions, Membranes, Binding Partners, Osmolytes, and Macromolecular Crowding. Protein J. 2009, 28, 305–325. [Google Scholar] [CrossRef]
- Tantos, A.; Friedrich, P.; Tompa, P. Cold Stability of Intrinsically Disordered Proteins. FEBS Lett. 2009, 583, 465–469. [Google Scholar] [CrossRef]
- Mazurenko, S.; Kunka, A.; Beerens, K.; Johnson, C.M.; Damborsky, J.; Prokop, Z. Exploration of Protein Unfolding by Modelling Calorimetry Data from Reheating. Sci. Rep. 2017, 7, 16321. [Google Scholar] [CrossRef]
- Thole, J.F.; Waudby, C.A.; Pielak, G.J. Disordered Proteins Mitigate the Temperature Dependence of Site-Specific Binding Free Energies. J. Biol. Chem. 2023, 299, 102984. [Google Scholar] [CrossRef]
- Tompa, P. Structure and Function of Intrinscially Disordered Proteins; Chapman & Hall/CRC Press: Boca Raton, FL, USA, 2010; ISBN 9781420078923. [Google Scholar]
- Frid, K.; Einstein, O.; Friedman-Levi, Y.; Binyamin, O.; Ben-Hur, T.; Gabizon, R. Aggregation of MBP in Chronic Demyelination. Ann. Clin. Transl. Neurol. 2015, 2, 711–721. [Google Scholar] [CrossRef]
- Krugmann, B.; Radulescu, A.; Appavou, M.S.; Koutsioubas, A.; Stingaciu, L.R.; Dulle, M.; Förster, S.; Stadler, A.M. Membrane Stiffness and Myelin Basic Protein Binding Strength as Molecular Origin of Multiple Sclerosis. Sci. Rep. 2020, 10, 16691. [Google Scholar] [CrossRef]
- Maleš, P.; Brkljača, Z.; Domazet Jurašin, D.; Bakarić, D. New Spirit of an Old Technique: Characterization of Lipid Phase Transitions via UV/Vis Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 272, 121013. [Google Scholar] [CrossRef]
- Maleš, P.; Brkljača, Z.; Crnolatac, I.; Bakarić, D. Application of MCR-ALS with EFA on FT-IR Spectra of Lipid Bilayers in the Assessment of Phase Transition Temperatures: Potential for Discernment of Coupled Events. Colloids Surf. B Biointerfaces 2021, 201, 111645. [Google Scholar] [CrossRef]
- Heimburg, T. Thermal Biophysics of Membranes; Wiley-VCH Verlag GmbH, Ed.; Wiley-VCH Verlag GmbH: Weinheim, Germany, 2007; ISBN 9783527404711. [Google Scholar]
- Lewis, R.N.A.H.; Mannock, D.A.; McElhaney, R.N. Differential Scanning Calorimetry in the Study of Lipid Phase Transitions in Model and Biological Membranes. In Methods in Membrane Lipids; Dopico, A.M., Ed.; Humana Press: Totowa, NJ, USA, 2006; pp. 171–195. ISBN 9780415475976. [Google Scholar]
- Menges, F. Spectragryph—Optical Spectroscopy Software. Version 1.2.15. 2015. Available online: https://www.effemm2.de/spectragryph/ (accessed on 5 October 2023).
- Maleš, P.; Pem, B.; Petrov, D.; Domazet Jurašin, D.; Bakarić, D. Deciphering the Origin of the Melting Profile of Unilamellar Phosphatidylcholine Liposomes by Measuring the Turbidity of Its Suspensions. Soft Matter 2022, 18, 6703–6715. [Google Scholar] [CrossRef]
- Jaumot, J.; Gargallo, R.; De Juan, A.; Tauler, R. A Graphical User-Friendly Interface for MCR-ALS: A New Tool for Multivariate Curve Resolution in MATLAB. Chemom. Intell. Lab. Syst. 2005, 76, 101–110. [Google Scholar] [CrossRef]
- Fabian, H.; Mäntele, W. Infrared Spectroscopy of Proteins Biochemical Applications Infrared Spectroscopy of Proteins. In Handbook of Vibrational Spectroscopy; John Wiley & Sons: Hoboken, NJ, USA, 2006; pp. 3399–3425. ISBN 9780470027325. [Google Scholar]
- Cobb, J.S.; Zai-Rose, V.; Correia, J.J.; Janorkar, A.V. FT-IR Spectroscopic Analysis of the Secondary Structures Present during the Desiccation Induced Aggregation of Elastin-Like Polypeptide on Silica. ACS Omega 2020, 5, 8403–8413. [Google Scholar] [CrossRef]
- Pašalić, L.; Pem, B.; Domazet Jurašin, D.; Vazdar, M.; Bakarić, D. Interaction of Guanidinium and Ammonium Cations with Phosphatidylcholine and Phosphatidylserine Lipid Bilayers—Calorimetric, Spectroscopic and Molecular Dynamics Simulations Study. Biochim. Biophys. Acta-Biomembr. 2023, 1865, 184122. [Google Scholar] [CrossRef]
- Fu, F.-N.; DeOliveira, D.B.; Trumble, W.R.; Sarkar, H.K.; Singh, B.R. Secondary Structure Estimation of Proteins Using the Amide III Region of Fourier Transform Infrared Spectroscopy: Application to Analyze Calcium-Binding-Induced Structural Changes in Calsequestrin. Appl. Spectrosc. 1994, 48, 1432–1441. [Google Scholar] [CrossRef]
- Cai, S.; Singh, B.R. Identification of β-Turn and Random Coil Amide III Infrared Bands for Secondary Structure Estimation of Proteins. Biophys. Chem. 1999, 80, 7–20. [Google Scholar] [CrossRef]
- Lewis, R.N.A.H.; McElhaney, R.N. Membrane Lipid Phase Transitions and Phase Organization Studied by Fourier Transform Infrared Spectroscopy. Biochim. Biophys. Acta-Biomembr. 2013, 1828, 2347–2358. [Google Scholar] [CrossRef]
- Grdadolnik, J.; Hadži, D. FT Infrared and Raman Investigation of Saccharide-Phosphatidylcholine Interactions Using Novel Structure Probes. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 1998, 54, 1989–2000. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Ahmed, M.A.M.; De Avila, M.; Polverini, E.; Bessonov, K.; Bamm, V.V.; Harauz, G. Solution Nuclear Magnetic Resonance Structure and Molecular Dynamics Simulations of a Murine 18.5 KDa Myelin Basic Protein Segment (S72-S107) in Association with Dodecylphosphocholine Micelles. Biochemistry 2012, 51, 7475–7487. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D.J.; de la Groot, B.; Grubmueller, H.; et al. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Nosé, S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Battisti, A.; Ciasca, G.; Grottesi, A.; Tenenbaum, A. Thermal Compaction of the Intrinsically Disordered Protein Tau: Entropic, Structural, and Hydrophobic Factors. Phys. Chem. Chem. Phys. 2017, 19, 8435–8446. [Google Scholar] [CrossRef]
- Maleš, P.; Butumović, M.; Erceg, I.; Brkljača, Z.; Bakarić, D. Influence of DPPE Surface Undulations on Melting Temperature Determination: UV/Vis Spectroscopic and MD Study. Biochim. Biophys. Acta-Biomembr. 2023, 1865, 184072. [Google Scholar] [CrossRef]
- Mabrey, S.; Sturtevant, J.M. Investigation of Phase Transitions of Lipids and Lipid Mixtures by High Sensitivity Differential Scanning Calorimetry. Proc. Natl. Acad. Sci. USA 1976, 73, 3862–3866. [Google Scholar] [CrossRef]
- Koynova, R.; Caffrey, M. Phases and Phase Transitions of the Phosphatidylcholines. Biochim. Biophys. Acta 1998, 1376, 91–145. [Google Scholar] [CrossRef]
- Na, J.H.; Lee, W.K.; Yu, Y.G. How Do We Study the Dynamic Structure of Unstructured Proteins: A Case Study on Nopp140 as an Example of a Large, Intrinsically Disordered Protein. Int. J. Mol. Sci. 2018, 19, 381. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Moussong, É.; Murvai, N.; Tantos, Á.; Tőke, O.; Réfrégiers, M.; Wien, F.; Kardos, J. Disordered–Ordered Protein Binary Classification by Circular Dichroism Spectroscopy. Front. Mol. Biosci. 2022, 9, 863141. [Google Scholar] [CrossRef] [PubMed]
- Kjaergaard, M.; Nørholm, A.B.; Hendus-Altenburger, R.; Pedersen, S.F.; Poulsen, F.M.; Kragelund, B.B. Temperature-Dependent Structural Changes in Intrinsically Disordered Proteins: Formation of α-Helices or Loss of Polyproline II? Protein Sci. 2010, 19, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.F. Induced Circular Dichroism. Chem. Phys. Lett. 1975, 32, 201–203. [Google Scholar] [CrossRef]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization Spectroscopy Methods in the Determination of Interactions of Small Molecules with Nucleic Acids—Tutorial. Beilstein J. Org. Chem. 2017, 14, 84–105. [Google Scholar] [CrossRef]
- Pašalić, L.; Pem, B.; Bakarić, D. Lamellarity-Driven Differences in Surface Structural Features of DPPS Lipids: Spectroscopic, Calorimetric and Computational Study. Membranes 2023, 13, 83. [Google Scholar] [CrossRef]
- Chatterley, A.S.; Laity, P.; Holland, C.; Weidner, T.; Woutersen, S.; Giubertoni, G. Broadband Multidimensional Spectroscopy Identifies the Amide II Vibrations in Silkworm Films. Molecules 2022, 27, 6275. [Google Scholar] [CrossRef]
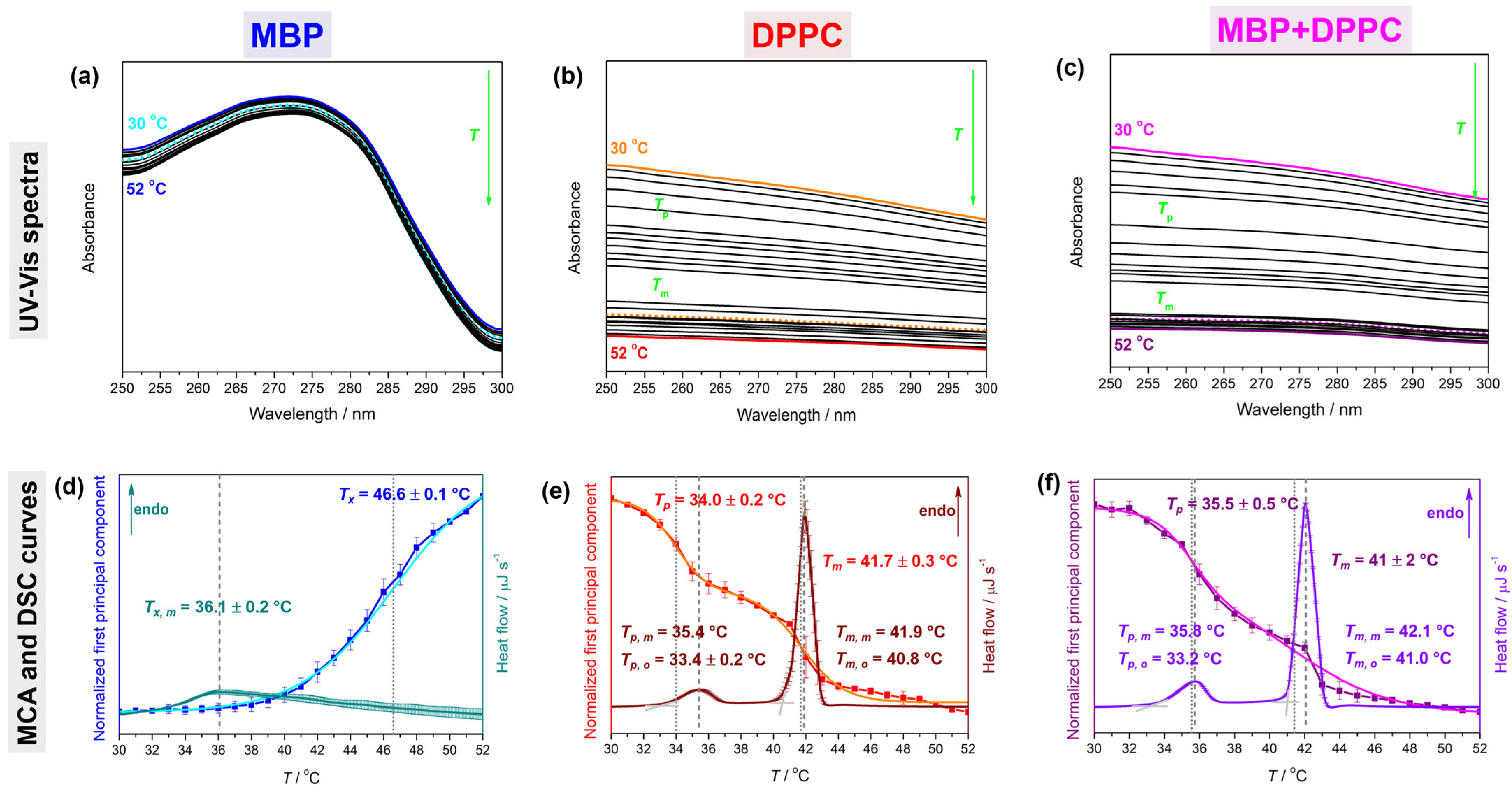

| System | Tpt a | ΔHcal c | ΔS d,e | |||
|---|---|---|---|---|---|---|
| DSC b | UV-Vis | |||||
| Tp, o/m | Tm, o/m | Tp | Tm | |||
| MBP f | 36.1 ± 0.2 | 46.6 ± 0.1 | 8.6 ± 0.1 | 27.7 ± 0.2 | ||
| DPPC | 33.4 ± 0.2/35.4 | 40.8/41.9 | 34.0 ± 0.2 | 41.7 ± 0.3 | 26.9 ± 0.2 | 85.4 ± 0.6 |
| MBP + DPPC | 33.2/35.8 | 41.0/42.1 | 35.5 ± 0.5 | 41 ± 2 | 62 ± 1 | 197 ± 4 |
| Orientation 2, T = 20 °C | Orientation 1, T = 50 °C | Orientation 2, T = 50 °C | ||||||
|---|---|---|---|---|---|---|---|---|
| Residue | Average # of HBs | Average # of Contacts | Residue | Average # of HBs | Average # of Contacts | Residue | Average # of HBs | Average # of Contacts |
| 129ARG | 1.0 | 177.1 | 158ARG | 2.7 | 247.4 | 129ARG | 1.9 | 202.8 |
| 51LYS | 0.7 | 92.1 | 161ARG | 2.2 | 329.3 | 169ARG | 1.1 | 113.4 |
| 4LYS | 0.1 | 21.2 | 162SER | 1.0 | 93.3 | 168ARG | 0.9 | 122.9 |
| 1ALA | 0.1 | 24.6 | 160SER | 0.8 | 211.1 | 112ARG | 0.7 | 62.4 |
| 128GLY | 0.1 | 45.1 | 169ARG | 0.8 | 127.9 | 104LYS | 0.4 | 50.3 |
| 134LYS | 0.1 | 14.2 | 47ARG | 0.8 | 125.8 | 134LYS | 0.4 | 53.9 |
| 2ALA | 0.1 | 10.8 | 168ARG | 0.5 | 81.7 | 161ARG | 0.3 | 27.7 |
| 169ARG | 0.1 | 15.6 | 159ASP | 0.5 | 143.8 | 109SER | 0.2 | 25.6 |
| 131SER | 0.1 | 14.9 | 5ARG | 0.4 | 30.9 | 138LYS | 0.2 | 26.8 |
| 168ARG | 0.1 | 5.8 | 141LYS | 0.3 | 59.2 | 128GLY | 0.1 | 44.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maleš, P.; Brkljača, Z.; Crnolatac, I.; Petrov, D.; Bakarić, D. Phase-Dependent Adsorption of Myelin Basic Protein to Phosphatidylcholine Lipid Bilayers. Membranes 2024, 14, 15. https://doi.org/10.3390/membranes14010015
Maleš P, Brkljača Z, Crnolatac I, Petrov D, Bakarić D. Phase-Dependent Adsorption of Myelin Basic Protein to Phosphatidylcholine Lipid Bilayers. Membranes. 2024; 14(1):15. https://doi.org/10.3390/membranes14010015
Chicago/Turabian StyleMaleš, Petra, Zlatko Brkljača, Ivo Crnolatac, Dražen Petrov, and Danijela Bakarić. 2024. "Phase-Dependent Adsorption of Myelin Basic Protein to Phosphatidylcholine Lipid Bilayers" Membranes 14, no. 1: 15. https://doi.org/10.3390/membranes14010015
APA StyleMaleš, P., Brkljača, Z., Crnolatac, I., Petrov, D., & Bakarić, D. (2024). Phase-Dependent Adsorption of Myelin Basic Protein to Phosphatidylcholine Lipid Bilayers. Membranes, 14(1), 15. https://doi.org/10.3390/membranes14010015







