Effect of CM15 on Supported Lipid Bilayer Probed by Atomic Force Microscopy
Abstract
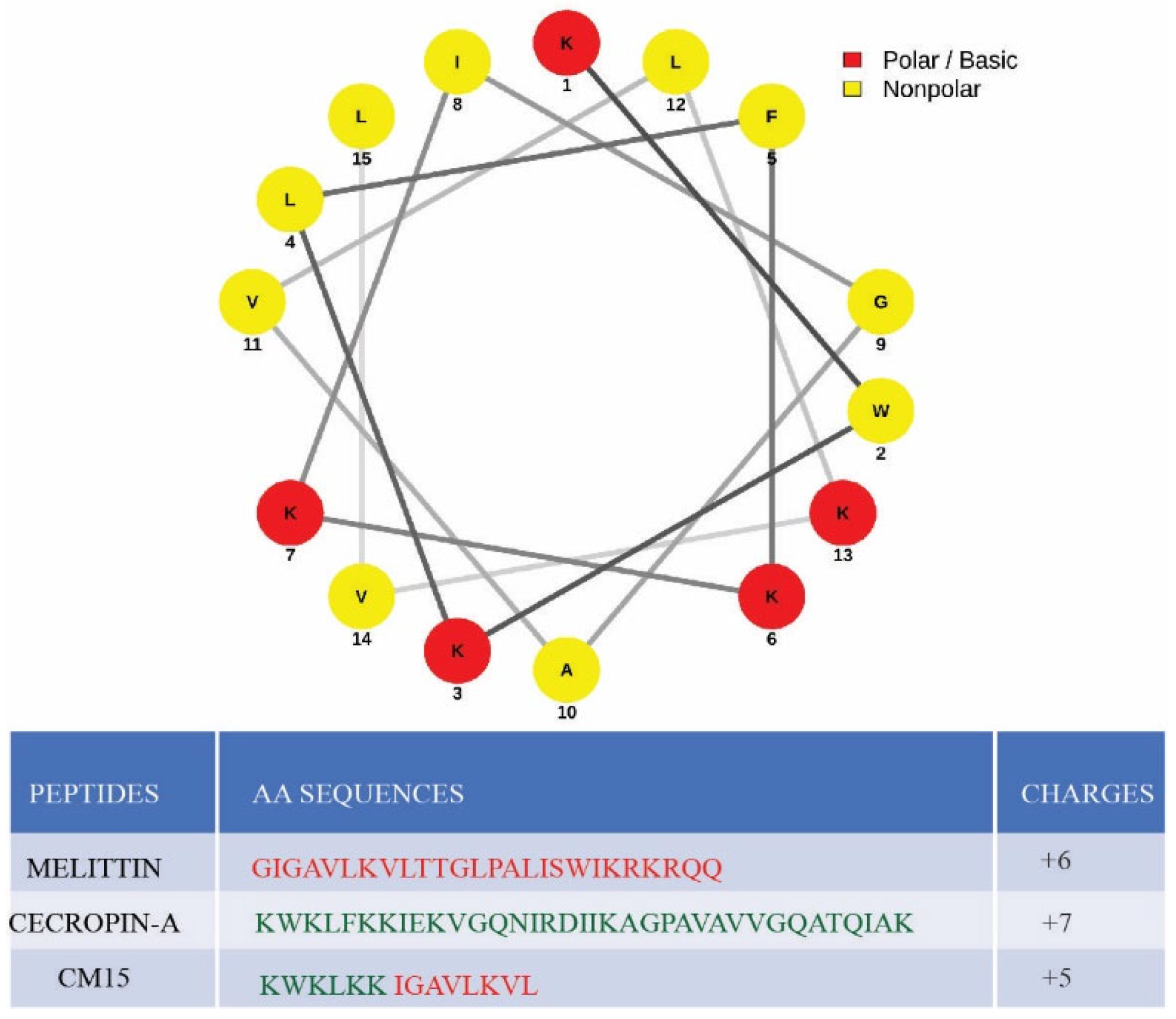
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Peptide Solution Preparation
2.3. Lipid Bilayer Preparation
2.4. AFM Imaging and Analysis
3. Results and Discussion
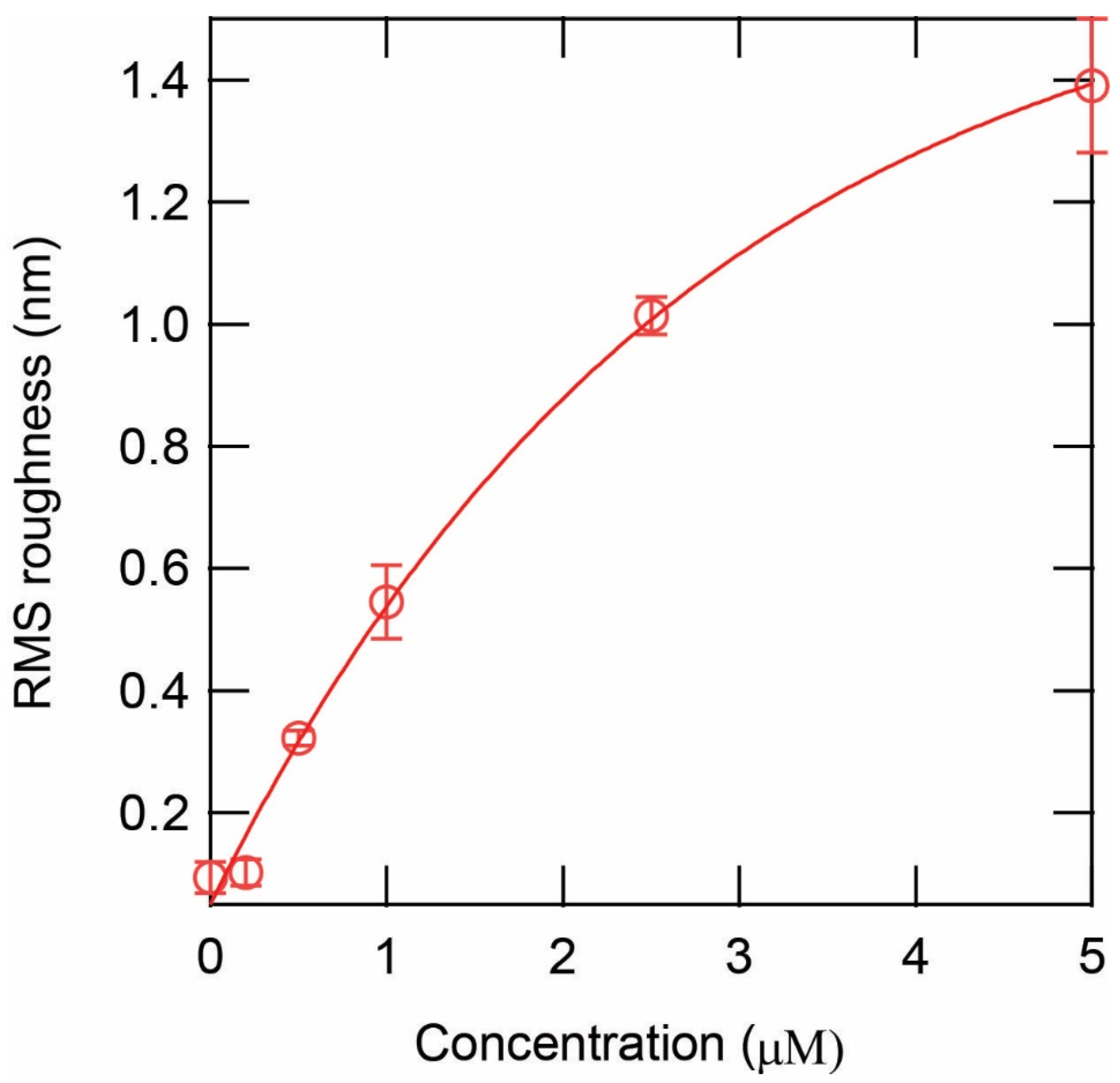
3.1. Overview
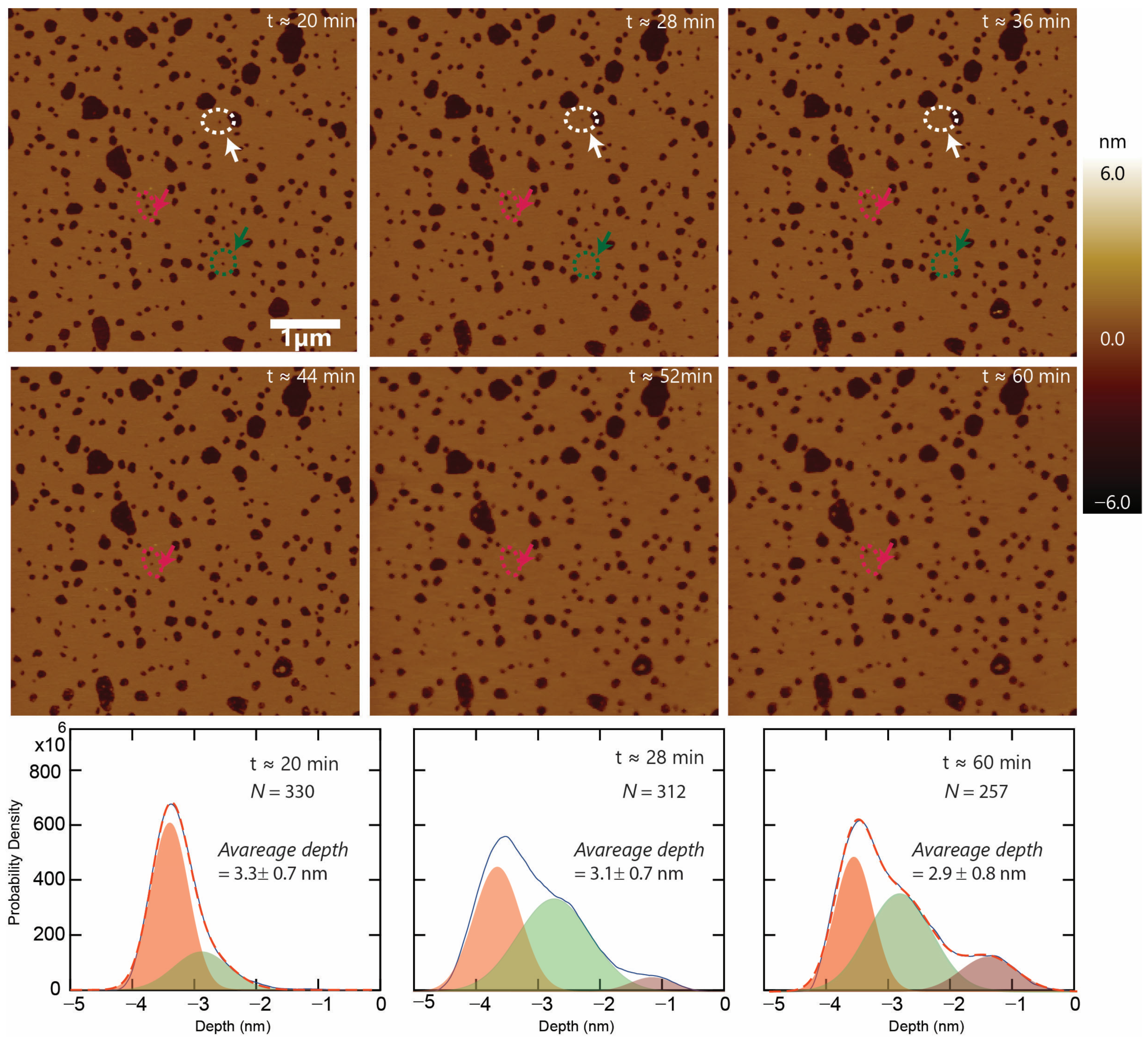
3.2. Characterizing the Defects
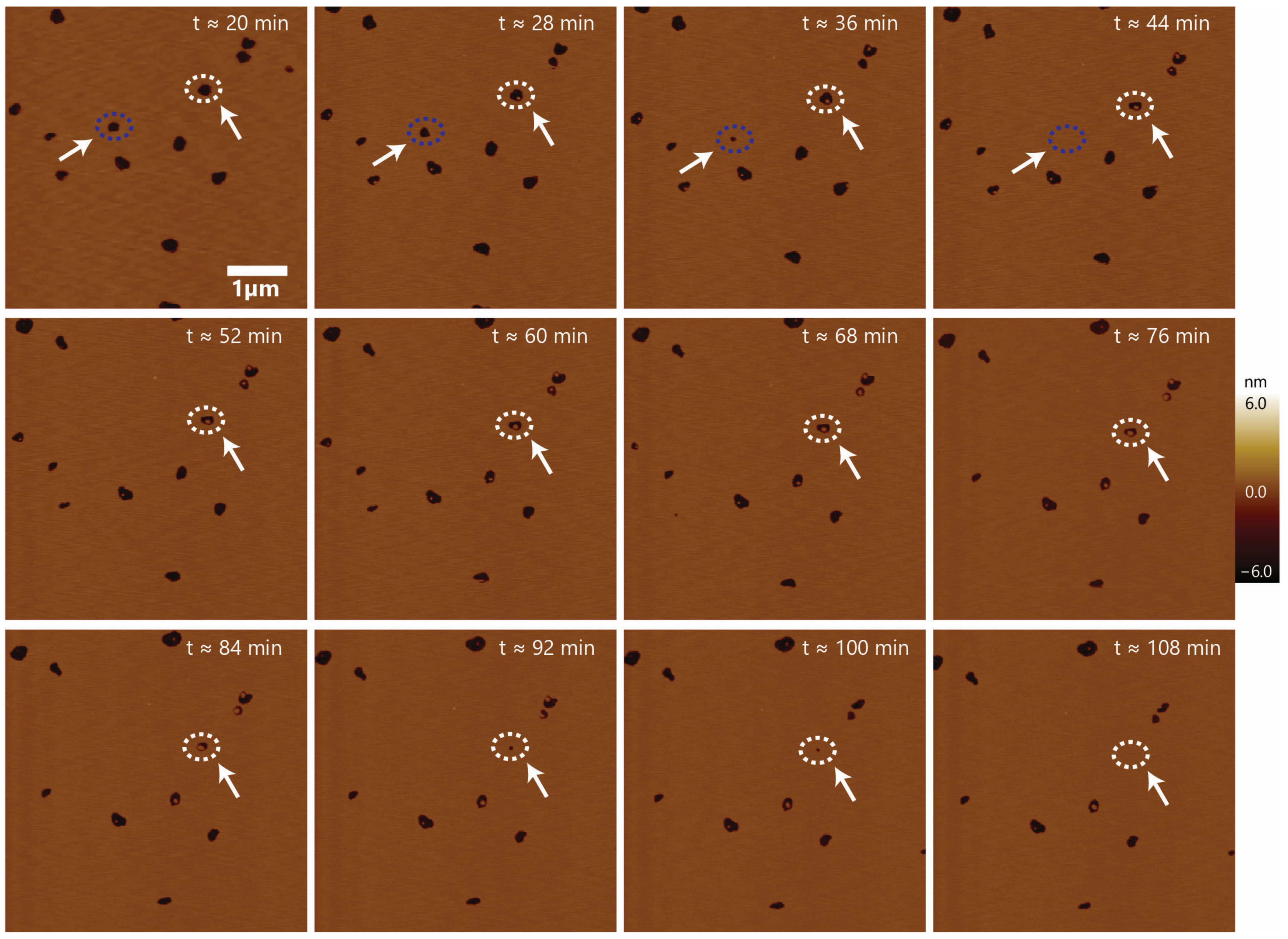
3.3. Formation of Stable Membrane-Spanning Defects and the Resealing of Some Defects
3.4. Membrane Disruption
3.5. Defect Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simon, S.A.; McIntosh, T.J. Peptide-Lipid Interactions; Academic Press: San Diego, CA, USA, 2002; Volume 52. [Google Scholar]
- Galdiero, S.; Falanga, A.; Cantisani, M.; Vitiello, M.; Morelli, G.; Galdiero, M. Peptide-Lipid Interactions: Experiments and Applications. Int. J. Mol. Sci. 2013, 14, 18758–18789. [Google Scholar] [CrossRef]
- Schaefer, K.G.; Pittman, A.E.; Barrera, F.N.; King, G.M. Atomic force microscopy for quantitative understanding of peptide-induced lipid bilayer remodeling. Methods 2022, 197, 20–29. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Faust, J.E.; Yang, P.-Y.; Huang, H.W. Action of Antimicrobial Peptides on Bacterial and Lipid Membranes: A Direct Comparison. Biophys. J. 2017, 112, 1663–1672. [Google Scholar] [CrossRef]
- Zhang, L.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Liang, Y.; Huang, Z.; Shen, X.; Zhang, Y.; Chai, Y.; Jiang, K.; Chen, Q.; Zhao, F. Global Trends in Research of Antimicrobial Peptides for the Treatment of Drug-Resistant Bacteria from 1995 to 2021: A Bibliometric Analysis. Infect. Drug Resist. 2023, 16, 4789–4806. [Google Scholar] [CrossRef]
- White, S.H.; Wimley, W.C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta 1998, 1376, 339–352. [Google Scholar] [CrossRef]
- Sabapathy, T.; Deplazes, E.; Mancera, R.L. Revisiting the Interaction of Melittin with Phospholipid Bilayers: The Effects of Concentration and Ionic Strength. Int. J. Mol. Sci. 2020, 21, 746. [Google Scholar] [CrossRef]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef]
- Wimley, W.C. How Does Melittin Permeabilize Membranes? Biophys. J. 2018, 114, 251–253. [Google Scholar] [CrossRef]
- Chen, F.-Y.; Lee, M.-T.; Huang, H.W. Evidence for Membrane Thinning Effect as the Mechanism for Peptide-Induced Pore Formation. Biophys. J. 2003, 84, 3751–3758. [Google Scholar] [CrossRef]
- Dempsey, C.E.; Sternberg, B. Reversible disc-micellization of dimyristoylphosphatidylcholine bilayers induced by melittin and [Ala-14]melittin. Biochim. Biophys. Acta Biomembr. 1991, 1061, 175–184. [Google Scholar] [CrossRef]
- Mól, A.R.; Castro, M.S.; Fontes, W. NetWheels: A web application to create high quality peptide helical wheel and net projections. BioRxiv 2018, 416347. [Google Scholar]
- Andreu, D.; Ubach, J.; Boman, A.; Wåhlin, B.; Wade, D.; Merrifield, R.B.; Boman, H.G. Shortened cecropin A-melittin hybrids. Significant size reduction retains potent antibiotic activity. FEBS Lett. 1992, 296, 190–194. [Google Scholar] [CrossRef]
- Juvvadi, P.; Vunnam, S.; Merrifield, E.L.; Boman, H.G.; Merrifield, R.B. Hydrophobic Effects on Antibacterial and Channel-forming Properties of Cecropin A–Melittin Hybrids. J. Pept. Sci. 1996, 2, 223–232. [Google Scholar] [CrossRef]
- Silva, T.; Claro, B.; Silva, B.F.B.; Vale, N.; Gomes, P.; Gomes, M.S.; Funari, S.S.; Teixeira, J.; Uhríková, D.; Bastos, M. Unravelling a Mechanism of Action for a Cecropin A-Melittin Hybrid Antimicrobial Peptide: The Induced Formation of Multilamellar Lipid Stacks. Langmuir 2018, 34, 2158–2170. [Google Scholar] [CrossRef]
- Efimova, S.S.; Medvedev, R.Y.; Chulkov, E.G.; Schagina, L.V.; Ostroumova, O.S. Regulation of the Pore-Forming Activity of Cecropin A by Local Anesthetics. Cell Tiss. Biol. 2018, 12, 331–341. [Google Scholar] [CrossRef]
- Lee, E.; Jeong, K.-W.; Lee, J.; Shin, A.; Kim, J.-K.; Lee, J.; Lee, D.G.; Kim, Y. Structure-activity relationships of cecropin-like peptides and their interactions with phospholipid membrane. BMB Rep. 2013, 46, 282–287. [Google Scholar] [CrossRef]
- Lyu, C.; Fang, F.; Li, B. Anti-Tumor Effects of Melittin and Its Potential Applications in Clinic. Curr. Protein Pept. Sci. 2019, 20, 240–250. [Google Scholar] [CrossRef]
- Zhai, Z.; Zhang, F.; Cao, R.; Ni, X.; Xin, Z.; Deng, J.; Wu, G.; Ren, W.; Yin, Y.; Deng, B. Cecropin A Alleviates Inflammation Through Modulating the Gut Microbiota of C57BL/6 Mice With DSS-Induced IBD. Front. Microbiol. 2019, 10, 1595. [Google Scholar] [CrossRef] [PubMed]
- Pistolesi, S.; Pogni, R.; Feix, J.B. Membrane Insertion and Bilayer Perturbation by Antimicrobial Peptide CM15. Biophys. J. 2007, 93, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Luo, Y.; Ma, Y.-H.; Lu, X. Interaction between Antimicrobial Peptide CM15 and a Model Cell Membrane Affected by CM15 Terminal Amidation and the Membrane Phase State. Langmuir 2021, 37, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Schlamadinger, D.E.; Wang, Y.; McCammon, J.A.; Kim, J.E. Spectroscopic and Computational Study of Melittin, Cecropin A, and the Hybrid Peptide CM15. J. Phys. Chem. B 2012, 116, 10600–10608. [Google Scholar] [CrossRef]
- Wang, Y.; Schlamadinger, D.E.; Kim, J.E.; McCammon, J.A. Comparative molecular dynamics simulations of the antimicrobial peptide CM15 in model lipid bilayers. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1402–1409. [Google Scholar] [CrossRef]
- Milani, A.; Benedusi, M.; Aquila, M.; Rispoli, G. Pore forming properties of cecropin-melittin hybrid peptide in a natural membrane. Molecules 2009, 14, 5179–5188. [Google Scholar] [CrossRef]
- Abrunhosa, F.; Faria, S.; Gomes, P.; Tomaz, I.; Pessoa, J.C.; Andreu, D.; Bastos, M. Interaction and Lipid-Induced Conformation of Two Cecropin-Melittin Hybrid Peptides Depend on Peptide and Membrane Composition. J. Phys. Chem. B 2005, 109, 17311–17319. [Google Scholar] [CrossRef]
- Sato, H.; Feix, J.B. Osmoprotection of bacterial cells from toxicity caused by antimicrobial hybrid peptide CM15. Biochemistry 2006, 45, 9997–10007. [Google Scholar] [CrossRef]
- Bhargava, K.; Feix, J.B. Membrane Binding, Structure, and Localization of Cecropin-Mellitin Hybrid Peptides: A Site-Directed Spin-Labeling Study. Biophys. J. 2004, 86, 329–336. [Google Scholar] [CrossRef]
- Chada, N.; Sigdel, K.P.; Gari, R.R.S.; Matin, T.R.; Randall, L.L.; King, G.M. Glass is a Viable Substrate for Precision Force Microscopy of Membrane Proteins. Sci. Rep. 2015, 5, 12550. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Sigdel, K.P.; Schaefer, K.G.; Mensah, G.A.K.; King, G.M.; Roberts, A.G. The effects of anthracycline drugs on the conformational distribution of mouse P-glycoprotein explains their transport rate differences. Biochem. Pharmacol. 2020, 174, 113813. [Google Scholar] [CrossRef] [PubMed]
- Pittman, A.E.; Marsh, B.P.; King, G.M. Conformations and Dynamic Transitions of a Melittin Derivative That Forms Macromolecule-Sized Pores in Lipid Bilayers. Langmuir 2018, 34, 8393–8399. [Google Scholar] [CrossRef] [PubMed]
- Hammond, K.; Ryadnov, M.G.; Hoogenboom, B.W. Atomic force microscopy to elucidate how peptides disrupt membranes. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183447. [Google Scholar] [CrossRef] [PubMed]
- Roussel, G.; Lindner, E.; White, S.H. Topology of the SecA ATPase Bound to Large Unilamellar Vesicles. J. Mol. Biol. 2022, 434, 167607. [Google Scholar] [CrossRef]
- Sigdel, K.P.; Wilt, L.A.; Marsh, B.P.; Roberts, A.G.; King, G.M. The conformation and dynamics of P-glycoprotein in a lipid bilayer investigated by atomic force microscopy. Biochem. Pharmacol. 2018, 156, 302–311. [Google Scholar] [CrossRef]
- Alessandrini, A.; Seeger, H.M.; Caramaschi, T.; Facci, P. Dynamic Force Spectroscopy on Supported Lipid Bilayers: Effect of Temperature and Sample Preparation. Biophys. J. 2012, 103, 38–47. [Google Scholar] [CrossRef]
- Soblosky, L.; Ramamoorthy, A.; Chen, Z. Membrane Interaction of Antimicrobial Peptides Using E. coli Lipid Extract as Model Bacterial Cell Membranes and SFG Spectroscopy. Chem. Phys. Lipids 2015, 187, 20–33. [Google Scholar] [CrossRef]
- Swana, K.W.; Nagarajan, R.; Camesano, T.A. Atomic Force Microscopy to Characterize Antimicrobial Peptide-Induced Defects in Model Supported Lipid Bilayers. Microorganisms 2021, 9, 1975. [Google Scholar] [CrossRef]
- Lind, T.K.; Wacklin, H.; Schiller, J.; Moulin, M.; Haertlein, M.; Pomorski, T.G.; Cárdenas, M. Formation and Characterization of Supported Lipid Bilayers Composed of Hydrogenated and Deuterated Escherichia coli Lipids. PLoS ONE 2015, 10, e0144671. [Google Scholar] [CrossRef]
- Pan, J.; Khadka, N.K. Kinetic Defects Induced by Melittin in Model Lipid Membranes: A Solution Atomic Force Microscopy Study. J. Phys. Chem. B 2016, 120, 4625–4634. [Google Scholar] [CrossRef]
- Sanganna Gari, R.R.; Frey, N.C.; Mao, C.; Randall, L.L.; King, G.M. Dynamic Structure of the Translocon SecYEG in Membrane: Direct Single Molecule Observations. J. Biol. Chem. 2013, 288, 16848–16854. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.L.; Heath, G.R.; Johnson, B.R.G.; Abdel-Rahman, M.A.; Strong, P.N.; Evans, S.D.; Miller, K. Phospholipid dependent mechanism of smp24, an α-helical antimicrobial peptide from scorpion venom. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2737–2744. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Choi, H.; Weisshaar, J.C. Melittin-Induced Permeabilization, Re-sealing, and Re-permeabilization of E. coli Membranes. Biophys. J. 2018, 114, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Guha, S.; Ghimire, J.; Wu, E.; Wimley, W.C. Mechanistic Landscape of Membrane-Permeabilizing Peptides. Chem. Rev. 2019, 119, 6040–6085. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walsh, O.D.; Choi, L.; Sigdel, K.P. Effect of CM15 on Supported Lipid Bilayer Probed by Atomic Force Microscopy. Membranes 2023, 13, 864. https://doi.org/10.3390/membranes13110864
Walsh OD, Choi L, Sigdel KP. Effect of CM15 on Supported Lipid Bilayer Probed by Atomic Force Microscopy. Membranes. 2023; 13(11):864. https://doi.org/10.3390/membranes13110864
Chicago/Turabian StyleWalsh, Olivia D., Leona Choi, and Krishna P. Sigdel. 2023. "Effect of CM15 on Supported Lipid Bilayer Probed by Atomic Force Microscopy" Membranes 13, no. 11: 864. https://doi.org/10.3390/membranes13110864
APA StyleWalsh, O. D., Choi, L., & Sigdel, K. P. (2023). Effect of CM15 on Supported Lipid Bilayer Probed by Atomic Force Microscopy. Membranes, 13(11), 864. https://doi.org/10.3390/membranes13110864





