The Evaluation of DHPMs as Biotoxic Agents on Pathogen Bacterial Membranes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Media
2.2. Agarose-Native Gel Electrophoresis
2.3. Statistical Analysis
2.4. Experimental Chemistry
3. Microorganisms and Media
4. Experimental Section
4.1. General Procedure for the Synthesis of P-fluorophenyl-substituted 3,4-dihydropyrimidine-2 (1H)-one Derivatives 4–6
4.2. Product 4a: Ethyl 4-(4-fluorophenyl)-6-methyl-1,2,3,4-tetrahydro-2-oxopyrimidine-5-carboxylate
4.3. Product 4b: Methyl 4-(4-fluorophenyl)-6-methyl-1,2,3,4-tetrahydro-2-oxopyrimidine-5-carboxylate
4.4. Product 4c: iso-Butyl 4-(4-fluorophenyl)-6-methyl-1,2,3,4-tetrahydro-2-oxopyrimidine-5-carboxylate
4.5. Product 4d: Ethyl 4-(4-fluorophenyl)-6-propyl-1,2,3,4-tetrahydro-2-oxopyrimidine-5-carboxylate
4.6. Product 4e: Ethyl 4-(4-fluorophenyl)-6-phenyl-1,2,3,4-tetrahydro-2-oxopyrimidine-5-carboxylate
4.7. Product 4h: 4-(4-fluorophenyl)-1,2,3,4-tetrahydro-6-methyl-2-oxo-N-phenylpyrimi-dinecarboxamide
4.8. Product 4i: 4-(4-fluorophenyl)-1,2,3,4-tetrahydro-6-methyl-2-oxopyrimidine-5-carbo-xamide
4.9. Product 5a: Ethyl 4-(4-fluorophenyl)-6-methyl-1,2,3,4-tetrahydro-2-thioxopyrimi-dine-5-carboxylate
4.10. Product 5b: Methyl 4-(4-fluorophenyl)-6-methyl-1,2,3,4-tetrahydro-2-thioxopyrimi-dine-5-carboxylate
4.11. Product 5e: Ethyl 4-(4-fluorophenyl)-6-phenyl-1,2,3,4-tetrahydro-2-thioxopyrimi-dine-5-carboxylate
4.12. Product 5g: Ethyl 4-(4-fluorophenyl)-6-(3-methylphenyl)-1,2,3,4-tetrahydro-2-thi-oxopyrimi-dine-5-carboxylate
4.13. Product 5h: Ethyl 4-(4-fluorophenyl)-6-(4-methylphenyl)-1,2,3,4-tetrahydro-2-thioxo-pyrimidine-5-carboxylate
4.14. Product 5j: 4-(4-fluorophenyl)-1,2,3,4-tetrahydro-6-methyl-N-phenyl-2-thioxo-pyrimidine-5-carboxamide
4.15. Product 5k: 4-(4-fluorophenyl)-1,2,3,4-tetrahydro-6-methyl-2-thioxopyrimidine-5-carboxamide
4.16. Product 6a: Ethyl 1,2,3,4-tetrahydro-6-methyl-2-oxo-4-phenylpyrimidine-5-carboxy-late
4.17. Product 6b: Ethyl 1,2,3,4-tetrahydro-6-methyl-2-thioxo-4-phenylpyrimidine-5-carbo-xylate
5. Results and Discussion
5.1. Chemistry
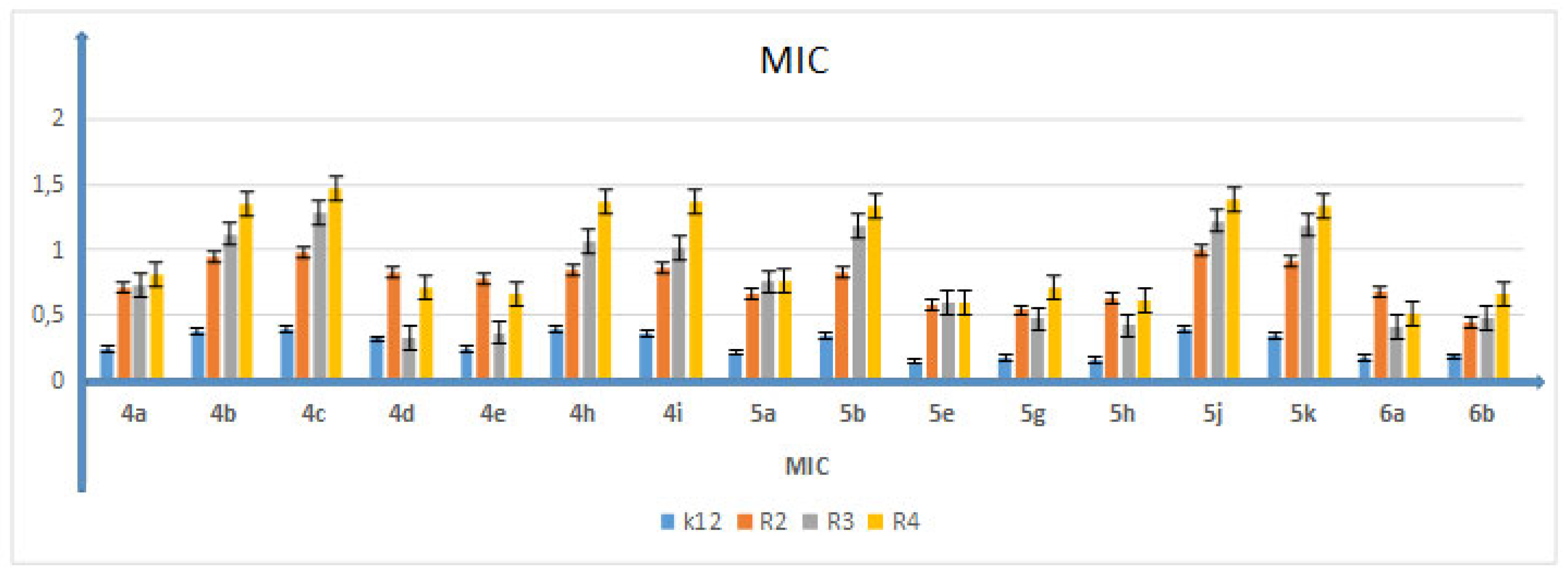
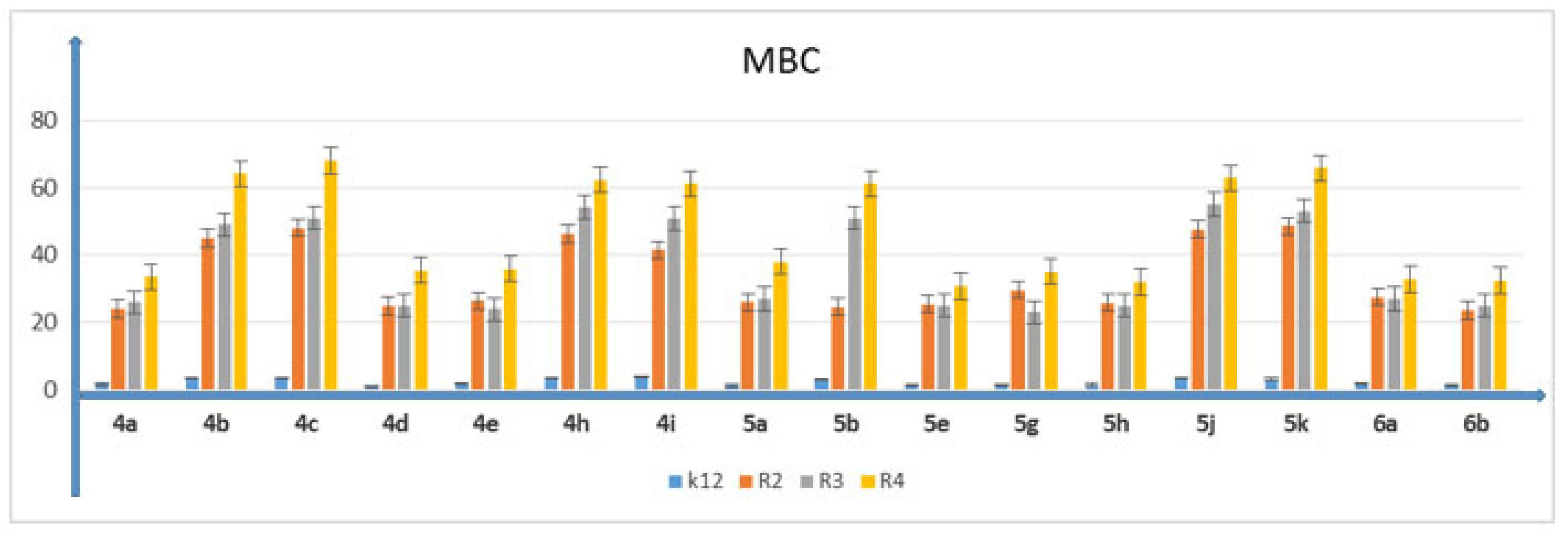
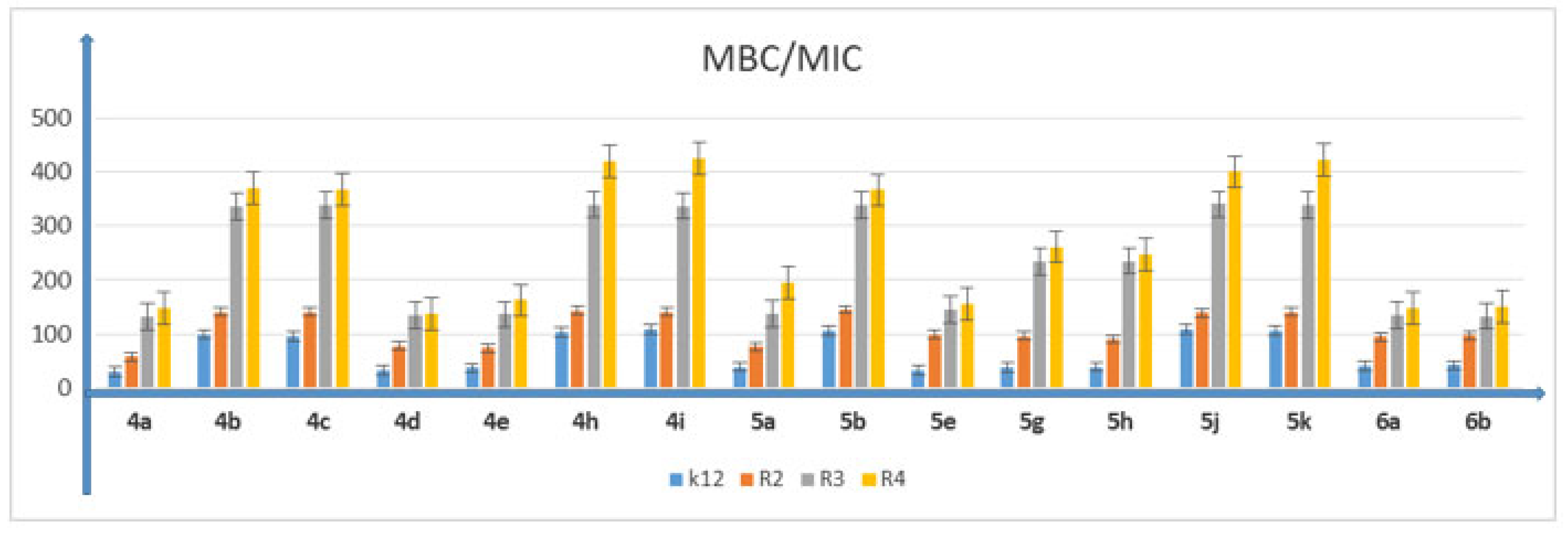
5.2. Cytotoxic Studies of the Library of Peptidomimetics
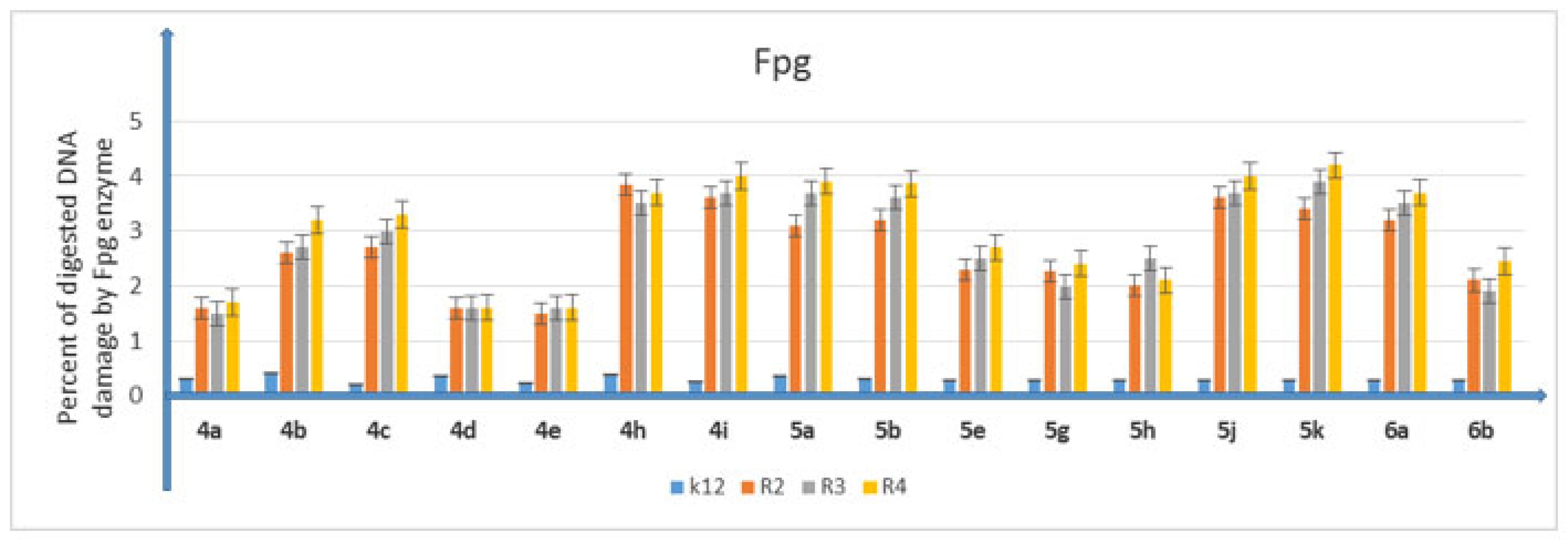
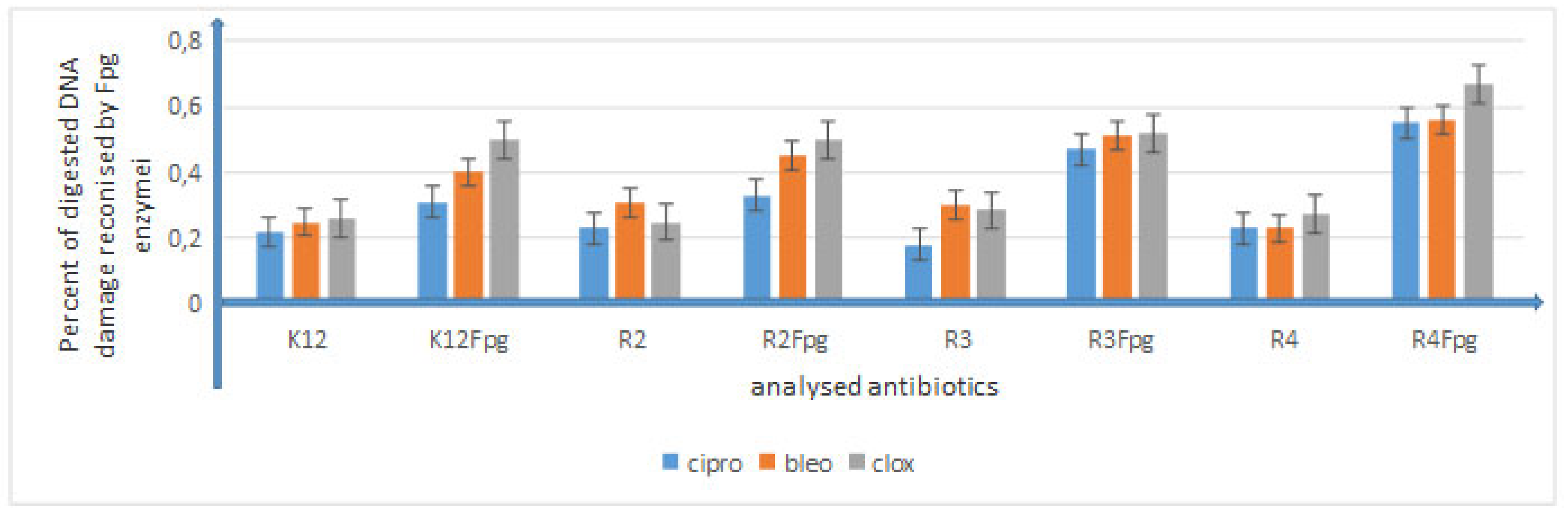
5.3. Analysis of Bacterial DNA Isolated from E. coli R2–R4 Strains Modified with 3,4-dihydropyrimidin-2(1H)-ones (DHPMs)
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MIC | minimum inhibitory concentration |
| MBC | minimum bactericidal concentration |
| Oc | open circle |
| Ccc | covalently closed circle |
| BER | base excision repair |
| Fpg DNA | formamidopyrimidine glycosylase |
References
- Deepa; Yadav, G.D.; Aalam, M.J.; Chaudhary, P.; Singh, S. Synthesis of Dihydropyrimidinones (DHPMs) and Hexahydro Xanthene Catalyzed by 1,4-Diazabicyclo [2.2.2] Octane Triflate Under Solvent-Free Condition. Curr. Org. Synth. 2019, 16, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.H.S.; Masson, F.T.; Simeoni, L.A.; Homem-De-Mello, M. Biological activity of dihydropyrimidinone (DHPM) derivatives: A systematic review. Eur. J. Med. Chem. 2018, 143, 1779–1789. [Google Scholar] [CrossRef] [PubMed]
- Biginelli, P. The urea-aldehyde derivatives of acetoacetic esters. Gazz. Chim. Ital. 1893, 23, 360–416. [Google Scholar]
- Rogerio, K.; Vitório, F.; Kümmerle, A.; Graebin, C. Reações multicomponentes: Um breve histórico e a versatilidade destas reações na síntese de moléculas bioativas. Rev. Virtual Quim. 2016, 8, 1934–1962. [Google Scholar] [CrossRef]
- Sweet, F.; Fissekis, J.D. Synthesis of 3,4-dihydro-2(1H)-pyrimidinones and the mechanism of the Biginelli reaction. J. Am. Chem. Soc. 1973, 95, 8741–8749. [Google Scholar] [CrossRef]
- Kappe, C.O. A Reexamination of the Mechanism of the Biginelli Dihydropyrimidine Synthesis. Support for an N-Acyliminium Ion Intermediate1. J. Org. Chem. 1997, 62, 7201–7204. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.M.; Guido, B.C.; Nobrega, C.C.; Corrêa, J.R.; Silva, R.G.; de Oliveira, H.C.B.; Gomes, A.F.; Gozzo, F.C.; Neto, B.A.D. The Biginelli Reaction with an Imidazolium-Tagged Recyclable Iron Catalyst: Kinetics, Mechanism, and Antitumoral Activity. Chem. Eur. J. 2013, 19, 4156–4168. [Google Scholar] [CrossRef] [PubMed]
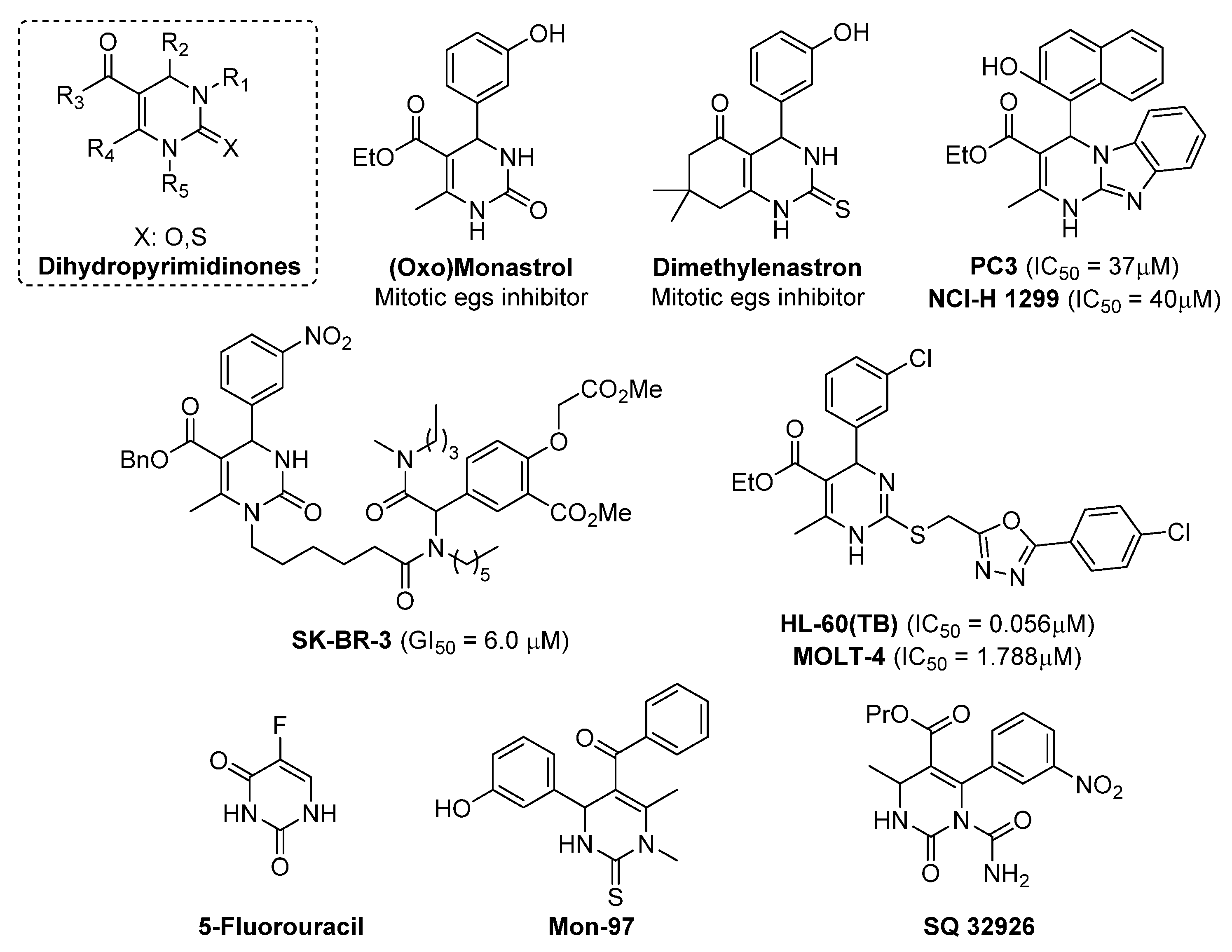
- Mayer, T.U.; Kapoor, T.M.; Haggarty, S.J.; King, R.W.; Schreiber, S.L.; Mitchison, T.J. Small Molecule Inhibitor of Mitotic Spindle Bipolarity Identified in a Phenotype-Based Screen. Science 1999, 286, 971–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kappe, C. Biologically active dihydropyrimidones of the Biginelli-type—A literature survey. Eur. J. Med. Chem. 2000, 35, 1043–1052. [Google Scholar] [CrossRef]
- Kapoor, T.M.; Mayer, T.; Coughlin, M.L.; Mitchison, T.J. Probing Spindle Assembly Mechanisms with Monastrol, a Small Molecule Inhibitor of the Mitotic Kinesin, Eg5. J. Cell Biol. 2000, 150, 975–988. [Google Scholar] [CrossRef]
- Alrazi, I.M.D.; Sadakane, K.; Maruta, S. Novel photochromic inhibitor for mitotic kinesin Eg5 which forms multiple isomerization states. J. Biochem. 2021, 170, 229–237. [Google Scholar] [CrossRef] [PubMed]
- DeBonis, S.; Simorre, J.P.; Crevel, I.; Lebeau, L.; Skoufias, D.A.; Blangy, A.; Ebel, C.; Gans, P.; Cross, R.; Hackney, D.D.; et al. Interaction of the mitotic inhibitor monastrol with human kinesin Eg5. Biochemistry 2003, 42, 338–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, T.M.; Mayer, T.U.; Desai, A.; Maddox, P.; Salmon, E.D.; Schreiber, S.L.; Mitchison, T.J. Investigating the inhibition of bipolar spindle formation by monastrol, a small molecule kinesin inhibitor. Mol. Biol. Cell 1999, 10, 128A. [Google Scholar]
- Duan, L.; Wang, T.-Q.; Bian, W.; Liu, W.; Sun, Y.; Yang, B.-S. Centrin: Another target of monastrol, an inhibitor of mitotic spindle. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 137, 1086–1091. [Google Scholar] [CrossRef] [PubMed]
- Abassi, Y.A.; Xi, B.; Zhang, W.; Ye, P.; Kirstein, S.L.; Gaylord, M.R.; Feinstein, S.C.; Wang, X.; Xu, X. Kinetic Cell-Based Morphological Screening: Prediction of Mechanism of Compound Action and Off-Target Effects. Chem. Biol. 2009, 16, 712–723. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Cheng, P.; Lei, N.; Yao, J.; Sheng, C.; Zhuang, C.; Guo, W.; Liu, W.; Zhang, Y.; Dong, G.; et al. Synthesis and Biological Evaluation of Novel Homocamptothecins Conjugating with Dihydropyrimidine Derivatives as Potent Topoisomerase I Inhibitors. Arch. Pharm. 2011, 344, 726–734. [Google Scholar] [CrossRef]
- Abdou, A.M.; Botros, S.; Hassan, R.A.; Kamel, M.M.; Taber, D.F.; Taher, A.T. Useful four-carbon synthons en route to monastrol analogs. Tetrahedron 2015, 71, 139–146. [Google Scholar] [CrossRef]
- Bahekar, S.S.; Shinde, D.B. Synthesis and anti-inflammatory activity of some [4,6-(4-substituted aryl)-2-thioxo-1,2,3,4-tetrahydro-pyrimidin-5-yl]-acetic acid derivatives. Bioorg. Med. Chem. Lett. 2004, 14, 1733–1736. [Google Scholar] [CrossRef]
- Bryzgalov, A.O.; Dolgikh, M.P.; Sorokina, I.V.; Tolstikova, T.G.; Sedova, V.F.; Shkurko, O.P. Antiarrhythmic activity of 4,6-di(het)aryl-5-nitro-3,4-dihydropyrimidin-(1H)-2-ones and its effects on arterial pressure in rats. Bioorg. Med. Chem. Lett. 2006, 16, 1418–1420. [Google Scholar] [CrossRef]
- Akhaja, T.N.; Raval, J. 1,3-dihydro-2H-indol-2-ones derivatives: Design, Synthesis, in vitro antibacterial, antifungal and antitubercular study. Eur. J. Med. Chem. 2011, 46, 5573–5579. [Google Scholar] [CrossRef]
- Chhillar, A.K.; Arya, P.; Mukherjee, C.; Kumar, P.; Yadav, Y.; Sharma, A.K.; Yadav, V.; Gupta, J.; Dabur, R.; Jha, H.N.; et al. Microwave-assisted synthesis of antimicrobial dihydropyridines and tetrahydropyrimidin-2-ones: Novel compounds against aspergillosis. Bioorg. Med. Chem. 2006, 14, 973–981. [Google Scholar] [CrossRef]
- Kim, J.; Ok, T.; Park, C.; So, W.; Jo, M.; Kim, Y.; Seo, M.; Lee, D.; Jo, S.; Ko, Y.; et al. A novel 3,4-dihydropyrimidin-2(1H)-one: HIV-1 replication inhibitors with improved metabolic stability. Bioorg. Med. Chem. Lett. 2012, 22, 2522–2526. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Kaur, J.; Kumar, P.; Gupta, S.; Singh, N.; Ghosal, A.; Dutta, A.; Kumar, A.; Tripathi, R.; Siddiqi, M.I.; et al. An orally effective dihydropyrimidone (DHPM) analogue induces apoptosis-like cell death in clinical isolates of Leishmania donovani overexpressing pteridine reductase 1. Parasitol. Res. 2009, 105, 1317–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacotte, P.; Buisson, D.-A.; Ambroise, Y. Synthesis, evaluation and absolute configuration assignment of novel dihydropyrimidin-2-ones as picomolar sodium iodide symporter inhibitors. Eur. J. Med. Chem. 2013, 62, 722–727. [Google Scholar] [CrossRef]
- Acharya, B.N.; Rao, G.B.D.; Kumar, D.; Kumar, P.; Kaushik, M.P. Design, synthesis, and evaluation of dihydropyrimidinone (DHPM) based muscarinic receptor antagonist. Med. Chem. Res. 2014, 24, 1763–1775. [Google Scholar] [CrossRef]
- Dhumaskar, K.L.; Meena, S.N.; Ghadi, S.C.; Tilve, S.G. Graphite catalyzed solvent free synthesis of dihydropyrimidin-2(1H)-ones/thiones and their antidiabetic activity. Bioorg. Med. Chem. Lett. 2014, 24, 2897–2899. [Google Scholar] [CrossRef]
- Blackburn, C.; Guan, B.; Brown, J.; Cullis, C.; Condon, S.M.; Jenkins, T.J.; Peluso, S.; Ye, Y.; Gimeno, R.E.; Punreddy, S.; et al. Identification and characterization of 4-aryl-3,4-dihydropyrimidin-2(1H)-ones as inhibitors of the fatty acid transporter FATP4. Bioorg. Med. Chem. Lett. 2006, 16, 3504–3509. [Google Scholar] [CrossRef]
- Ahmad, S.; Iftikhar, F.; Ullah, F.; Sadiq, A.; Rashid, U. Rational design and synthesis of dihydropyrimidine based dual binding site acetylcholinesterase inhibitors. Bioorg. Chem. 2016, 69, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Hashim, J.; Arshad, N.; Khan, I.; Siddiqui, N.; Wadood, A.; Ali, M.; Arshad, F.; Khan, K.M.; Choudhary, M.I. Dihydropyrimidine based hydrazine dihydrochloride derivatives as potent urease inhibitors. Bioorg. Chem. 2016, 64, 85–96. [Google Scholar] [CrossRef]
- Atwal, K.S.; Swanson, B.N.; Unger, S.E.; Floyd, D.M.; Moreland, S.; Hedberg, A.; O’Reilly, B.C. Dihydropyrimidine calcium channel blockers. 3. 3-Carbamoyl-4-aryl-1,2,3,4-tetrahydro-6-methyl-5-pyrimidinecarboxylic acid esters as orally effective antihypertensive agents. J. Med. Chem. 1991, 34, 806–811. [Google Scholar] [CrossRef]
- Chikhale, R.; Thorat, S.; Pant, A.; Jadhav, A.; Thatipamula, K.C.; Bansode, R.; Bhargavi, G.; Karodia, N.; Rajasekharan, M.; Paradkar, A.; et al. Design, synthesis and pharmacological evaluation of pyrimidobenzothiazole-3-carboxylate derivatives as selective L-type calcium channel blockers. Bioorg. Med. Chem. 2015, 23, 6689–6713. [Google Scholar] [CrossRef]
- Lewis, R.W.; Mabry, J.; Polisar, J.G.; Eagen, K.; Ganem, B.; Hess, G.P. Dihydropyrimidinone Positive Modulation of δ-Subunit-Containing γ-Aminobutyric Acid Type A Receptors, Including an Epilepsy-Linked Mutant Variant. Biochemistry 2010, 49, 4841–4851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Fátima, Â.; Braga, T.C.; Neto, L.; Terra, B.S.; Oliveira, B.; da Silva, D.L.; Modolo, L. A mini-review on Biginelli adducts with notable pharmacological properties. J. Adv. Res. 2015, 6, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Prokopcová, H.; Dallinger, D.; Uray, G.; Kaan, H.Y.K.; Ulaganathan, V.; Kozielski, F.; Laggner, C.; Kappe, C.O. Structure-Activity Relationships and Molecular Docking of Novel Dihydropyrimidine-Based Mitotic Eg5 Inhibitors. ChemMedChem 2010, 5, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Holla, B.; Rao, B.; Sarojini, B.; Akberali, P. One pot synthesis of thiazolodihydropyrimidinones and evaluation of their anticancer activity. Eur. J. Med. Chem. 2004, 39, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Treptow, T.G.; Figueiró, F.; Jandrey, E.H.F.; Battastini, A.M.; Salbego, C.G.; Hoppe, J.B.; Taborda, P.S.; Rosa, S.B.; Piovesan, L.A.; Doca, C.; et al. Novel hybrid DHPM-fatty acids: Synthesis and activity against glioma cell growth in vitro. Eur. J. Med. Chem. 2015, 95, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, P.; Trzepizur, D.; Szymczak, M.; Skiba, G.; Kramkowski, K.; Ostaszewski, R. 1,2-Diarylethanols—A New Class of Compounds That Are Toxic to E. coli K12, R2–R4 Strains. Materials 2021, 14, 1025. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, P.; Madej, A.; Szymczak, M.; Ostaszewski, R. α-Amidoamids as New Replacements of Antibiotics—Research on the Chosen K12, R2–R4 E. coli Strains. Materials 2020, 13, 5169. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, P.; Borkowski, A.; Czerwonka, G.; Cłapa, T.; Cieśla, J.; Misiewicz, A.; Borowiec, M.; Szala, M. The microbial toxicity of quaternary ammonium ionic liquids is dependent on the type of lipopolysaccharide. J. Mol. Liq. 2018, 266, 540–547. [Google Scholar] [CrossRef]
- Borkowski, A.; Kowalczyk, P.; Czerwonka, G.; Cieśla, J.; Cłapa, T.; Misiewicz, A.; Szala, M.; Drabik, M. Interaction of quaternary ammonium ionic liquids with bacterial membranes—Studies with Escherichia coli R1–R4-type lipopolysaccharides. J. Mol. Liq. 2017, 246, 282–289. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Gawdzik, B.; Trzepizur, D.; Szymczak, M.; Skiba, G.; Raj, S.; Kramkowski, K.; Lizut, R.; Ostaszewski, R. δ-Lactones—A New Class of Compounds That Are Toxic to E. coli K12 and R2–R4 Strains. Materials 2021, 14, 2956. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, P.; Wilk, M.; Parul, P.; Szymczak, M.; Kramkowski, K.; Raj, S.; Skiba, G.; Sulejczak, D.; Kleczkowska, P.; Ostaszewski, R. The Synthesis and Evaluation of Aminocoumarin Peptidomimetics as Cytotoxic Agents on Model Bacterial E. coli Strains. Materials 2021, 14, 5725. [Google Scholar] [CrossRef]
- Dissanayake, D.; Wijewardana, T.; Gunawardena, G.; Poxton, I. Distribution of lipopolysaccharide core types among avian pathogenic Escherichia coli in relation to the major phylogenetic groups. Vet. Microbiol. 2008, 132, 355–363. [Google Scholar] [CrossRef]
- Maciejewska, A.; Kaszowska, M.; Jachymek, W.; Lugowski, C.; Lukasiewicz, J. Lipopolysaccharide-linked Enterobacterial Common Antigen (ECALPS) Occurs in Rough Strains of Escherichia coli R1, R2, and R4. Int. J. Mol. Sci. 2020, 21, 6038. [Google Scholar] [CrossRef] [PubMed]
- Prost, M.E.; Prost, R. Basic parameters of evaluation of the effectiveness of antibiotic therapy. OphthaTherapy 2017, 4, 233–236. [Google Scholar] [CrossRef]
- Albadi, J.; Mansournezhad, A.; Baghernehad, M.; Frozan, N. A Green Recyclable Poly(4-vinylpyridine)-Supported Copper Iodide Nanoparticles Catalyst for the Multicomponent Synthesis of 3,4-dihydropyrimidin-2(1H)-ones/thiones. J. Korean Chem. Soc. 2013, 57, 169–171. [Google Scholar] [CrossRef]
- Li, Y.; Ma, J.; Liu, Z.; Jin, D.; Jiao, G.; Guo, Y.; Wang, Q.; Zhou, J.; Sun, R. Fabrication of porous ultrathin carbon nitride nanosheet catalysts with enhanced photocatalytic activity for N- and O-heterocyclic compound synthesis. New J. Chem. 2020, 45, 365–372. [Google Scholar] [CrossRef]
- Nagarajan, S.; Shaikh, T.; Kandasamy, E. Synthesis of 1-alkyl triazolium triflate room temperature ionic liquids and their catalytic studies in multi-component Biginelli reaction. J. Chem. Sci. 2015, 127, 1539–1545. [Google Scholar] [CrossRef] [Green Version]
- Paudyal, M.P.; Wang, M.; Siitonen, J.H.; Hu, Y.; Yousufuddin, M.; Shen, H.C.; Falck, J.R.; Kürti, L. Intramolecular N-Me and N-H aminoetherification for the synthesis of N-unprotected 3-amino-O-heterocycles. Org. Biomol. Chem. 2021, 19, 557–560. [Google Scholar] [CrossRef]
- Han, B.; Han, R.-F.; Ren, Y.-W.; Duan, X.-Y.; Xu, Y.-C.; Zhang, W. Efficient aerobic oxidative dehydrogenation of dihydropyrimidinones and dihydropyrimidines. Tetrahedron 2011, 67, 5615–5620. [Google Scholar] [CrossRef]
- Kurmach, M.N.; Ryabitskiy, A.B.; Britsun, V.N. 2-Acylthioacetamides in the Biginelli Reaction. Chem. Heterocycl. Compd. 2014, 49, 1770–1776. [Google Scholar] [CrossRef]
- Braga, T.C.; Silva, T.F.; Maciel, T.M.S.; da Silva, E.C.D.; da Silva-Júnior, E.F.; Modolo, L.V.; Figueiredo, I.M.; Santos, J.C.C.; de Aquino, T.M.; de Fátima, Â. Ionic liquid-assisted synthesis of dihydropyrimidin(thi)one Biginelli adducts and investigation of their mechanism of urease inhibition. New J. Chem. 2019, 43, 15187–15200. [Google Scholar] [CrossRef]
- Bais, J.; Benedetti, F.; Berti, F.; Cerminara, I.; Drioli, S.; Funicello, M.; Regini, G.; Vidali, M.; Felluga, F. One Pot Synthesis of Micromolar BACE-1 Inhibitors Based on the Dihydropyrimidinone Scaffold and Their Thia and Imino Analogues. Molecules 2020, 25, 4152. [Google Scholar] [CrossRef] [PubMed]
- Zolfagharinia, S.; Koukabi, N.; Kolvari, E. A unique opportunity for the utilization of glass wastes as a resource for catalytic applications: Toward a cleaner environment. RSC Adv. 2016, 6, 113844–113858. [Google Scholar] [CrossRef]
- Mohamadpour, F.; Lashkari, M.; Maghsoodlou, M.T.; Heydari, R. Phthalic acid: A green, biodegradable and environmentally bening nature di-functional Brønsted acid catalyst for the one-pot synthesis of 3, 4-dihydropyrimidin-2-(1H)-one derivatives and substituted dihydro-2-oxypyroles. J. Chil. Chem. Soc. 2018, 63, 3811–3818. [Google Scholar] [CrossRef] [Green Version]
- Sari, O.; Roy, V.; Métifiot, M.; Marchand, C.; Pommier, Y.; Bourg, S.; Bonnet, P.; Schinazi, R.F.; Agrofoglio, L.A. Synthesis of dihydropyrimidine α,γ-diketobutanoic acid derivatives targeting HIV integrase. Eur. J. Med. Chem. 2015, 104, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Li, X.; Li, Y.; Xu, Y.; Zhang, Z.; Zhou, M.; Zhang, X.; Liu, Z.; Zhou, J.; Cao, C.; et al. Discovery and Development of Thiazolo[3,2-a]pyrimidinone Derivatives as General Inhibitors of Bcl-2 Family Proteins. ChemMedChem 2011, 6, 904–921. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H.; Tang, G.-M.; Yan, S.-C.; Wang, Y.-T.; Zhan, S.-J.; Zhang, E.; Sun, Y.; Jiang, Y.; Cui, Y.-Z. Cobalt-based metal coordination polymers with 4,4′-bipyridinyl groups: Highly efficient catalysis for one-pot synthesis of 3,4-dihydropyrimidin-2(1H)-ones under solvent-free conditions. Appl. Organomet. Chem. 2016, 30, 1009–1021. [Google Scholar] [CrossRef]
- Zafar, A.M.; Qureshi, S.; Khan, M.N.; Azad, M.; Munawar, M.A.; Khan, M.A. Amino Acids Catalyzed Biginelli Protocols. Asian J. Chem. 2013, 25, 3244–3246. [Google Scholar] [CrossRef]
- Godugu, K.; Yadala, V.D.S.; Pinjari, M.K.M.; Gundala, T.R.; Sanapareddy, L.R.; Nallagondu, C.G.R. Natural dolomitic limestone-catalyzed synthesis of benzimidazoles, dihydropyrimidinones, and highly substituted pyridines under ultrasound irradiation. Beilstein J. Org. Chem. 2020, 16, 1881–1900. [Google Scholar] [CrossRef]
- Khatri, C.K.; Potadar, S.M.; Chaturbhuj, G. A reactant promoted solvent free synthesis of 3,4-dihydropyrimidin-2(1H)-thione analogues using ammonium thiocyanate. Tetrahedron Lett. 2017, 58, 1778–1780. [Google Scholar] [CrossRef]
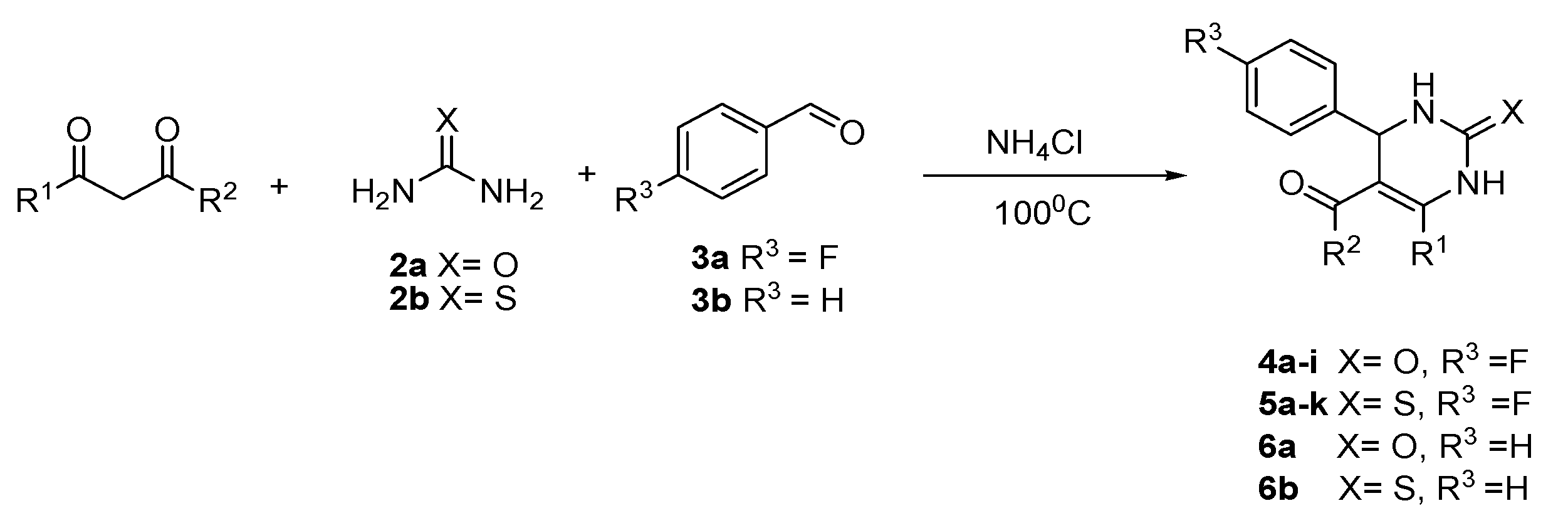

| Entry | Compound | X | R1 | R2 | R3 | Yield |
|---|---|---|---|---|---|---|
| 1 | 4a | O | Me | OEt | F | 67% |
| 2 | 4b | O | Me | OMe | F | 56% |
| 3 | 4c | O | Me | Oi-Bu | F | 63% |
| 4 | 4d | O | Pr | OEt | F | 69% |
| 5 | 4e | O | Ph | OEt | F | 82% |
| 6 | 4h | O | Me | NHPh | F | 67% |
| 7 | 4i | O | Me | NH2 | F | 57% |
| 8 | 5a | S | Me | OEt | F | 54% |
| 9 | 5b | S | Me | OMe | F | 55% |
| 10 | 5e | S | Ph | OEt | F | 54% |
| 11 | 5g | S | Ph(3-Me) | OEt | F | 58% |
| 12 | 5h | S | Ph(4-Me) | OEt | F | 63% |
| 13 | 5j | S | Me | NHPh | F | 76% |
| 14 | 5k | S | Me | NH2 | F | 61% |
| 15 | 6a | O | Me | OEt | H | 70% |
| 16 | 6b | S | Me | OEt | H | 59% |
| No. of Samples | 4b | 4c | 4h | 4i | 5b,5g | 5h | 5j,5k | Type of Test |
|---|---|---|---|---|---|---|---|---|
| K12 | * | * | * | ** | ** | * | *** | MIC |
| R2 | * | * | * | ** | ** | * | *** | MIC |
| R3 | * | * | * | ** | ** | * | *** | MIC |
| R4 | * | * | * | ** | ** | * | *** | MIC |
| K12 | * | * | ** | * | ** | ** | ** | MBC |
| R2 | ** | * | ** | * | ** | ** | ** | MBC |
| R3 | ** | * | ** | * | ** | ** | ** | MBC |
| R4 | ** | * | ** | * | ** | ** | ** | MBC |
| K12 | * | ** | * | * | * | ** | *** | MBC/MIC |
| R2 | * | ** | * | * | * | ** | *** | MBC/MIC |
| R3 | * | ** | * | * | * | ** | *** | MBC/MIC |
| R4 | * | ** | * | * | * | ** | *** | MBC/MIC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gawdzik, B.; Kowalczyk, P.; Koszelewski, D.; Brodzka, A.; Masternak, J.; Kramkowski, K.; Wypych, A.; Ostaszewski, R. The Evaluation of DHPMs as Biotoxic Agents on Pathogen Bacterial Membranes. Membranes 2022, 12, 238. https://doi.org/10.3390/membranes12020238
Gawdzik B, Kowalczyk P, Koszelewski D, Brodzka A, Masternak J, Kramkowski K, Wypych A, Ostaszewski R. The Evaluation of DHPMs as Biotoxic Agents on Pathogen Bacterial Membranes. Membranes. 2022; 12(2):238. https://doi.org/10.3390/membranes12020238
Chicago/Turabian StyleGawdzik, Barbara, Paweł Kowalczyk, Dominik Koszelewski, Anna Brodzka, Joanna Masternak, Karol Kramkowski, Aleksandra Wypych, and Ryszard Ostaszewski. 2022. "The Evaluation of DHPMs as Biotoxic Agents on Pathogen Bacterial Membranes" Membranes 12, no. 2: 238. https://doi.org/10.3390/membranes12020238
APA StyleGawdzik, B., Kowalczyk, P., Koszelewski, D., Brodzka, A., Masternak, J., Kramkowski, K., Wypych, A., & Ostaszewski, R. (2022). The Evaluation of DHPMs as Biotoxic Agents on Pathogen Bacterial Membranes. Membranes, 12(2), 238. https://doi.org/10.3390/membranes12020238