Langmuir Monolayer Techniques for the Investigation of Model Bacterial Membranes and Antibiotic Biodegradation Mechanisms
Abstract
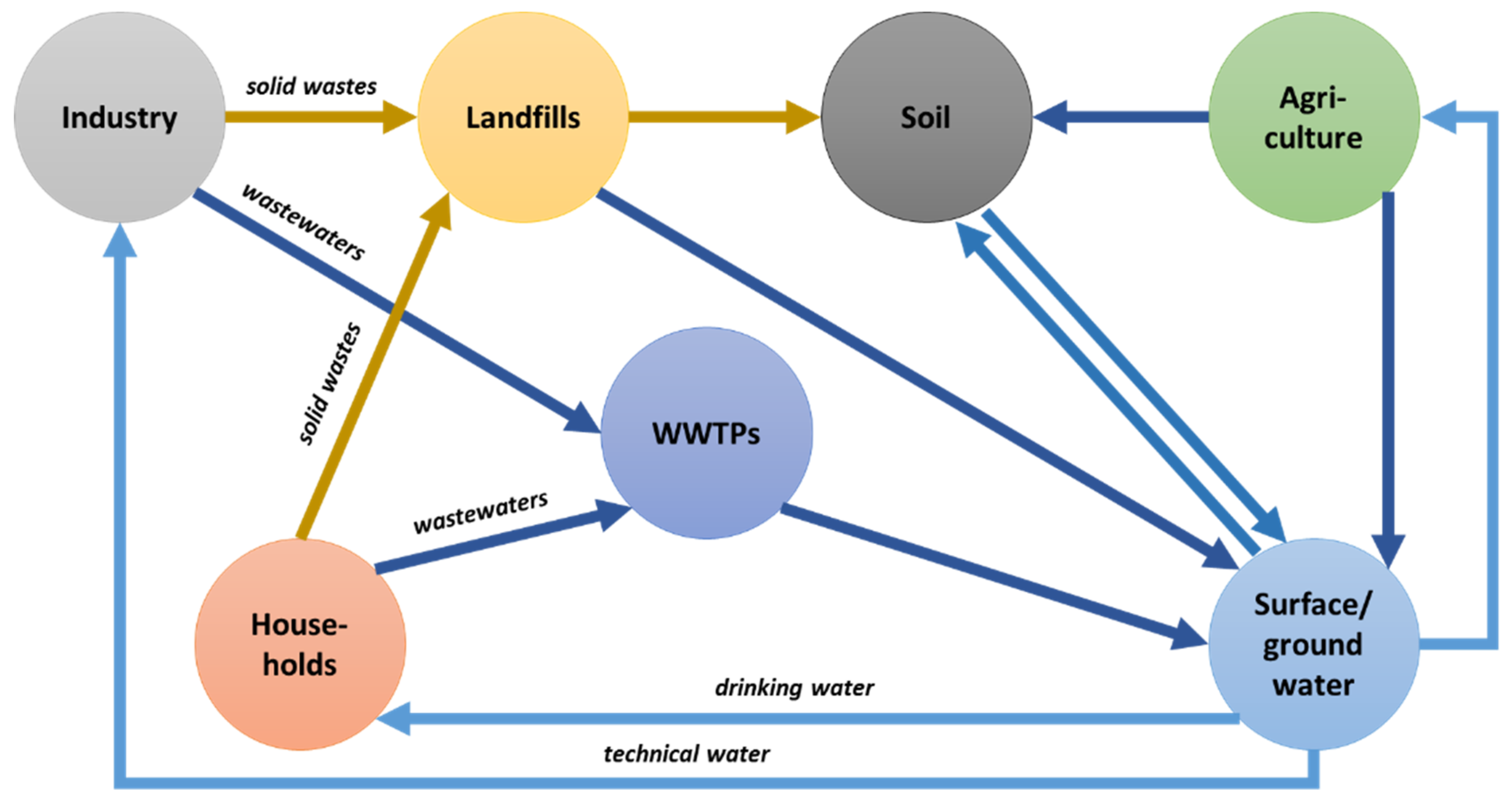
1. Antibiotics in the Natural Environments
2. Methods to Remove Antibiotics from the Environment
3. Modelling of Cellular Membrane
4. Application of the Langmuir Monolayer Technique to Form Biomimetic System
5. Langmuir Monolayer as a Model Bacterial Membrane
6. Modelling of Cellular Membrane for Antibiotic Biodegradation Studies
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davies, J. Where have all the antibiotics gone? Can. J. Infect. Dis. Med. Microbiol. 2006, 17, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Dutta, J. Ecotoxicological studies of cephalosporin antibiotics on Daphnia magna. Toxicol. Int. 2018, 25, 21–30. [Google Scholar] [CrossRef]
- Wang, N.; Xiao, W.; Niu, B.; Duan, W.; Zhou, L.; Zheng, Y. Highly efficient adsorption of fluoroquinolone antibiotics using chitosan derived granular hydrogel with 3D structure. J. Mol. Liq. 2019, 281, 307–314. [Google Scholar] [CrossRef]
- Pacholak, A.; Smułek, W.; Zgoła-Grześkowiak, A.; Kaczorek, E. Nitrofurantoin—Microbial degradation and interactions with environmental bacterial strains. Int. J. Environ. Res. Public Health 2019, 16, 1526. [Google Scholar] [CrossRef]
- Dutta, J.; Mala, A.A. Removal of antibiotic from the water environment by the adsorption technologies: A review. Water Sci. Technol. 2020, 82, 401–426. [Google Scholar] [CrossRef]
- Mohan, H.; Rajput, S.S.; Jadhav, E.B.; Sankhla, M.S.; Sonone, S.S.; Jadhav, S.; Kumar, R. Ecotoxicity, occurrence, and removal of pharmaceuticals and illicit drugs from aquatic systems. Biointerface Res. Appl. Chem. 2021, 11, 12530–12546. [Google Scholar] [CrossRef]
- Bilal, M.; Mehmood, S.; Rasheed, T.; Iqbal, H.M. Antibiotics traces in the aquatic environment: Persistence and adverse environmental impact. Curr. Opin. Environ. Sci. Health 2020, 13, 68–74. [Google Scholar] [CrossRef]
- Jjemba, P.K. Excretion and ecotoxicity of pharmaceutical and personal care products in the environment. Ecotoxicol. Environ. Saf. 2006, 63, 113–130. [Google Scholar] [CrossRef]
- Bayer, A.; Asner, R.; Schüssler, W.; Kopf, W.; Weiß, K.; Sengl, M.; Letzel, M. Behavior of sartans (antihypertensive drugs) in wastewater treatment plants. their occurrence and risk for the aquatic environment. Environ. Sci. Pollut. Res. 2014, 21, 10830–10839. [Google Scholar] [CrossRef] [PubMed]
- Galecio, J.S.; Escudero, E.; Cerón, J.J.; Crescenzo, G.; Marín, P. Pharmacokinetics of tildipirosin in ewes after intravenous. intramuscular and subcutaneous administration. Animals 2020, 10, 1332. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A. A global perspective on the use. sales, exposure pathways. occurrence, fate and effects of veterinary antibiotics (VAS) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Kołodziejska, M.; Maszkowska, J.; Białk-Bielińska, A.; Steudte, S.; Kumirska, J.; Stepnowski, P.; Stolte, S. Aquatic toxicity of four veterinary drugs commonly applied in fish farming and animal husbandry. Chemosphere 2013, 92, 1253–1259. [Google Scholar] [CrossRef]
- Henriksson, P.J.G.; Rico, A.; Troell, M.; Klinger, D.H.; Buschmann, A.H.; Saksida, S.; Chadag, M.V.; Zhang, W. Unpacking factors influencing antimicrobial use in global aquaculture and their implication for management: A review from a systems perspective. Sustain. Sci. 2018, 13, 1105–1120. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Burridge, L.; Weis, J.S.; Cabello, F.; Pizarro, J.; Bostick, K. Chemical use in salmon aquaculture: A review of current practices and possible environmental effects. Aquaculture 2010, 306, 7–23. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.-F.; Yannarell, A.C.; Maxwell, S.; Aminov, R.I. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Q. 2009, 38, 1086–1108. [Google Scholar] [CrossRef]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B Boil. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. J. 2015, 40, 277–283. [Google Scholar]
- Gozdzielewska, L.; King, C.; Flowers, P.; Mellor, D.; Dunlop, P.; Price, L. Scoping review of approaches for improving antimicrobial stewardship in livestock farmers and veterinarians. Prev. Vet. Med. 2020, 180, 105025. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Liao, Q.; Rong, H.; Zhao, M.; Luo, H.; Chu, Z.; Wang, R. Interaction between tetracycline and microorganisms during wastewater treatment: A review. Sci. Total Environ. 2021, 757, 143981. [Google Scholar] [CrossRef]
- Ozumchelouei, E.J.; Hamidian, A.H.; Zhang, Y.; Yang, M. Physicochemical properties of antibiotics: A review with an emphasis on detection in the aquatic environment. Water Environ. Res. 2020, 92, 177–188. [Google Scholar] [CrossRef]
- Boy-Roura, M.; Mas-Pla, J.; Petrovic, M.; Gros, M.; Soler, D.; Brusi, D.; Menció, A. Towards the understanding of antibiotic occurrence and transport in groundwater: Findings from the Baix Fluvià alluvial aquifer (NE Catalonia. Spain). Sci. Total Environ. 2018, 612, 1387–1406. [Google Scholar] [CrossRef]
- Nnadozie, C.F.; Odume, O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019, 254, 113067. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef]
- Cerqueira, F.; Christou, A.; Fatta-Kassinos, D.; Vila-Costa, M.; Bayona, J.M.; Piña, B. Effects of prescription antibiotics on soil- and root-associated microbiomes and resistomes in an agricultural context. J. Hazard. Mater. 2020, 400, 123208. [Google Scholar] [CrossRef] [PubMed]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the soil environment—Degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Roose-Amsaleg, C.; Laverman, A.M. Do antibiotics have environmental side-effects? Impact of synthetic antibiotics on biogeochemical processes. Environ. Sci. Pollut. Res. 2016, 23, 4000–4012. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, H.; Lehmler, H.J.; Cai, X.; Chen, J. Antibiotic pollution in marine food webs in Laizhou Bay, North China: Trophodynamics and human exposure implication. Environ. Sci. Technol. 2017, 51, 2392–2400. [Google Scholar] [CrossRef]
- Wang, S.; Ma, X.; Liu, Y.; Yi, X.; Du, G.; Li, J. Fate of antibiotics. antibiotic-resistant bacteria, and cell-free antibiotic-resistant genes in full-scale membrane bioreactor wastewater treatment plants. Bioresour. Technol. 2020, 302, 122825. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A.; Kudva, I.T.; Zhang, Q. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 2. [Google Scholar] [CrossRef]
- Reis, A.C.; Kolvenbach, B.A.; Nunes, O.C.; Corvini, P.F.X. Biodegradation of antibiotics: The new resistance determinants—Part I. New Biotechnol. 2020, 54, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Zheng, S.; Yin, D.; Wang, L.; Chen, L. Aqueous photolysis of tetracycline and toxicity of photolytic products to luminescent bacteria. Chemosphere 2008, 73, 377–382. [Google Scholar] [CrossRef]
- Langbehn, R.K.; Michels, C. Moreira Soares, antibiotics in wastewater: From its occurrence to the biological removal by environmentally conscious technologies. Environ. Pollut. 2021, 275, 116603. [Google Scholar] [CrossRef]
- García-Galán, M.J.; Blanco, S.G.; Roldán, R.L.; Díaz-Cruz, S.; Barceló, D. Ecotoxicity evaluation and removal of sulfonamides and their acetylated metabolites during conventional wastewater treatment. Sci. Total Environ. 2012, 437, 403–412. [Google Scholar] [CrossRef]
- Lyu, J.; Yang, L.; Zhang, L.; Ye, B.; Wang, L. Antibiotics in soil and water in China—A systematic review and source analysis. Environ. Pollut. 2020, 266, 115147. [Google Scholar] [CrossRef] [PubMed]
- Sodhi, K.K.; Kumar, M.; Balan, B.; Dhaulaniya, A.S.; Shree, P.; Sharma, N.; Singh, D.K. Perspectives on the antibiotic contamination, resistance, metabolomics, and systemic remediation. SN Appl. Sci. 2021, 3, 269. [Google Scholar] [CrossRef]
- Fatta-Kassinos, D.; Meric, S.; Nikolaou, A. Pharmaceutical residues in environmental waters and wastewater: Current state of knowledge and future research. Anal. Bioanal. Chem. 2011, 399, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Escher, B.I.; Baumgartner, R.; Koller, M.; Treyer, K.; Lienert, J.; McArdell, C.S. Environmental toxicology and risk assessment of pharmaceuticals from hospital wastewater. Water Res. 2011, 45, 75–92. [Google Scholar] [CrossRef]
- Al Aukidy, M.; Verlicchi, P.; Voulvoulis, N. A framework for the assessment of the environmental risk posed by pharmaceuticals originating from hospital effluents. Sci. Total Environ. 2014, 493, 54–64. [Google Scholar] [CrossRef]
- Sri, V.S.; Nagaraju, G.; Prasad, M.K. Biological treatment technologies of pharmaceuticals from hospital wastewater. Int. J. Eng. Sci. Math. 2018, 7, 1–8. [Google Scholar] [CrossRef]
- Wu, S.; Lin, Y.; Hu, Y.H. Strategies of tuning catalysts for efficient photodegradation of antibiotics in water environments: A review. J. Mater. Chem. A 2021, 9, 2592–2611. [Google Scholar] [CrossRef]
- Rathod, M.; Haldar, S.; Basha, S. Nanocrystalline cellulose for removal of tetracycline hydrochloride from water via biosorption: Equilibrium, kinetic and thermodynamic studies. Ecol. Eng. 2015, 84, 240–249. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, X.; Cao, Z.; Zhan, Y.; Shi, X.; Yang, Y.; Zhou, J.; Xu, J. Adsorption behavior and mechanism of chloramphenicols, sulfonamides, and non-antibiotic pharmaceuticals on multi-walled carbon nanotubes. J. Hazard. Mater. 2016, 310, 235–245. [Google Scholar] [CrossRef]
- Exall, K.; Balakrishnan, V.K.; Toito, J.; McFadyen, R. Impact of selected wastewater constituents on the removal of sulfonamide antibiotics via ultrafiltration and micellar enhanced ultrafiltration. Sci. Total Environ. 2013, 461, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, W.; Shi, J.; Zhu, S.; Yan, Y. Enhanced levofloxacin removal from water using zirconium (IV) loaded corn bracts. Environ. Sci. Pollut. Res. 2017, 24, 10685–10694. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.J.; Theydan, S.K. Microporous activated carbon from Siris seed pods by microwave-induced KOH activation for metronidazole adsorption. JAAP 2013, 99, 101–109. [Google Scholar] [CrossRef]
- Liu, S.; Pan, M.; Feng, Z.; Qin, Y.; Wang, Y.; Tan, L.; Sun, T. Ultra-high adsorption of tetracycline antibiotics on garlic skin-derived porous biomass carbon with high surface area. New J. Chem. 2020, 44, 1097–1106. [Google Scholar] [CrossRef]
- Lu, Z.Y.; Ma, Y.L.; Zhang, J.T.; Fan, N.S.; Huang, B.C.; Jin, R.C. A critical review of antibiotic removal strategies: Performance and mechanisms. J. Water Process Eng. 2020, 38, 101681. [Google Scholar] [CrossRef]
- Müller, E.; Schüssler, W.; Horn, H.; Lemmer, H. Aerobic biodegradation of the sulfonamide antibiotic sulfamethoxazole by activated sludge applied as co-substrate and sole carbon and nitrogen source. Chemosphere 2013, 92, 969–978. [Google Scholar] [CrossRef]
- Grandclément, C.; Seyssiecq, I.; Piram, A.; Wong-Wah-Chung, P.; Vanot, G.; Tiliacos, N.; Roche, N.; Doumenq, P. From the conventional biological wastewater treatment to hybrid processes, the evaluation of organic micropollutant removal: A review. Water Res. 2017, 111, 297–317. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides 2003, 24, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.H.; Boxer, S.G. Model membrane systems and their applications. Curr. Opin. Chem. Biol. 2007, 11, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Gaines, L.G. Insoluble Monolayers at Liquid–Gas Interfaces Interscience; Wiley-Interscience: New York, NY, USA, 1966. [Google Scholar]
- Agrawal, A.; Steigmann, D.J. Mechanics of cellular membranes. In Computational Modeling in Biomechanics; De, S., Guilak, F., Mofrad, R.K.M., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 549–566. [Google Scholar] [CrossRef]
- Pezeshkian, W.; Marrink, S.J. Simulating realistic membrane shapes. Curr. Opin. Cell Biol. 2021, 71, 103–111. [Google Scholar] [CrossRef]
- Hsu, P.-C.; Samsudin, F.; Shearer, J.; Khalid, S. It is complicated: Curvature, diffusion, and lipid sorting within the two membranes of Escherichia coli. J. Phys. Chem. Lett. 2017, 8, 5513–5518. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Neu, J.; Oster, G. Effect of protein shape on multibody interactions between membrane inclusions. Phys. Rev. E 2000, 61, 4281–4285. [Google Scholar] [CrossRef]
- Palsdottir, H.; Hunte, C. Lipids in membrane protein structures. Biochim. Biophys. Acta Biomembr. 2004, 1666, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Niemela, P.S.; Miettinen, M.S.; Monticelli, L.; Hammaren, H.; Bjelkmar, P.; Murtola, T.; Lindahl, E.; Vattulainen, I. Membrane proteins diffuse as dynamic complexes with lipids. J. Am. Chem. Soc. 2010, 132, 7574–7575. [Google Scholar] [CrossRef]
- Wydro, P. The influence of cardiolipin on phosphatidylglycerol/phosphatidylethanolamine monolayers—Studies on ternary films imitating bacterial membrane. Colloids Surf. B 2013, 106, 217–223. [Google Scholar] [CrossRef]
- Sandrino, B.; de Oliveira, J.F.A.; Nobre, T.M.; Appelt, P.; Gupta, A.; de Araujo, M.P.; Rotello, V.M.; Oliveira, O.N. Challenges in application of langmuir monolayer studies to determine the mechanisms of bactericidal activity of ruthenium complexes. Langmuir 2017, 33, 14167–14174. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef]
- Eeman, M.; Deleu, M. From biological membranes to biomimetic model membranes. Biotechnol. Agron. Soc. Environ. 2010, 14, 691–708. [Google Scholar]
- Keller, H.; Worch, R.; Schwille, P. Model Membrane Systems. In Protein-Ligand Interactions. Methods in Molecular Biology (Methods and Protocols); Williams, M., Daviter, T., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 1008. [Google Scholar] [CrossRef]
- Knobloch, J.; Suhendro, D.K.; Zieleniecki, J.L.; Shapter, J.G.; Köper, I. Membrane-drug interactions studied using model membrane systems. Saudi J. Biol. Sci. 2015, 22, 714–718. [Google Scholar] [CrossRef]
- Daraee, H.; Etemadi, A.; Kouhi, M.; Alimirzalu, S.; Akbarzadeh, A. Application of liposomes in medicine and drug delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 381–391. [Google Scholar] [CrossRef]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, É.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta (BBA)—Biomembr. 2011, 1808, 1957–1974. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, S.M. Chapter 3: Liposome techniques for synthesis of biomimetic lipid membranes. In Nanobiotechnology of Biomimetic Membranes; Martin, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Dufourc, E.J. Bicelles and nanodiscs for biophysical chemistry. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183478. [Google Scholar] [CrossRef]
- Beaugrand, M.; Arnold, A.A.; Henin, J.; Williamson, P.T.; Marcotte, I. Lipid concentration and molar ratio boundaries for the use of isotropic bicelles. Langmuir 2014, 30, 6162–6170. [Google Scholar] [CrossRef] [PubMed]
- Vácha, R.; Frenkel, D. Stability of bicelles: A simulation study. Langmuir 2014, 30, 4229–4235. [Google Scholar] [CrossRef]
- Catoire, L.J.; Warnet, X.L.; Warschawski, D.E. Micelles, bicelles, amphipols, nanodiscs, liposomes, or intact cells: The hitchhiker’s guide to the study of membrane proteins by NMR. In Membrane Proteins Production for Structural Analysis; Mus-Veteau, I., Ed.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- London, E. Membrane structure–function insights from asymmetric lipid vesicles. Acc. Chem. Res. 2019, 52, 2382–2391. [Google Scholar] [CrossRef]
- Andersson, J.; Bilotto, P.; Mears, L.L.E.; Fossatia, S.; Ramach, U.; Köper, I.; Valtiner, M.; Knoll, W. Solid-supported lipid bilayers—A versatile tool for the structural and functional characterization of membrane proteins. Methods 2020, 180, 56–68. [Google Scholar] [CrossRef]
- Girard-Egrot, A.P.; Blum, L.J. Chapter 2: Langmuir-blodgett technique for synthesis of biomimetic lipid membranes. In Nanobiotechnology of Biomimetic Membranes; Martin, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Böcking, T.; Gooding, J.J. Chapter 5: Biomimetic membranes in biosensor applications. In Nanobiotechnology of Biomimetic Membranes; Martin, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Phan, M.D.; Shin, K.; Langmuir, A. Monolayer: Ideal model membrane to study cell. J. Chem. Biol. Interfaces 2014, 2, 1–5. [Google Scholar] [CrossRef]
- Dana, R.M. The monolayer technique: A potent tool for studying the interfacial properties of antimicrobial and membrane-lytic peptides and their interactions with lipid membranes. Biochim. Biophys. Acta 1999, 1462, 109–140. [Google Scholar] [CrossRef]
- Brown, R.E.; Brockman, H.L. Using monomolecular films to characterize lipid lateral interactions. Lipid Rafts 2007, 398, 41–58. [Google Scholar] [CrossRef]
- Brezinski, G.; Moehwald, H. Langmuir monolayers to study interactions at model membrane surfaces. Adv. Colloid Interface Sci. 2003, 100–102, 563–584. [Google Scholar] [CrossRef]
- Elderdfi, M.; Sikorski, A.F. Langmuir-monolayer methodologies for characterizing protein-lipid interactions. Chem. Phys. Lipids 2018, 212, 61–72. [Google Scholar] [CrossRef]
- Kaganer, V.M.; Möhwald, H.; Dutta, P. Structura and phase transitions in Langmuir monolayers. Rev. Mod. Phys. 1999, 71, 779–819. [Google Scholar] [CrossRef]
- Davies, J.; Rideal, E. Interfacial Phenomena; Academic Press: New York, NY, USA; London, UK, 1963. [Google Scholar]
- Gagoś, M.; Arczewska, M. FTIR spectroscopic study of molecular organization of the antibiotic amphotericin B in aqueous solution and in DPPC lipid monolayers containing the sterols cholesterol and ergosterol. Eur. Biophys. J. 2012, 41, 663–673. [Google Scholar] [CrossRef]
- Rojewska, M.; Skrzypiec, M.; Prochaska, K. Surface properties and morphology of mixed POSS-DPPC monolayers at the air/water interface. Colloids Surf. B. 2017, 150, 334–343. [Google Scholar] [CrossRef]
- Hąc-Wydro, K.; Dynarowicz-Łątka, P. Biomedical applications of the Langmuir monolayer technique. In Annales Universitatis Mariae Curie-Skłodowska; Maria Curie-Skłodowska University: Lublin, Poland, 2008; Volume 63, pp. 47–60. [Google Scholar] [CrossRef]
- Rojewska, M.; Smułek, W.; Prochaska, K.; Kaczorek, E. Combined effect of nitrofurantoin and plant surfactant on bacteria phospholipid membrane. Molecules 2020, 25, 2527. [Google Scholar] [CrossRef]
- Machado, A.C.; Caseli, L. Interaction of nitrofurantoin with lipid langmuir monolayers as cellular membrane models distinguished with tensiometry and infrared spectroscopy. Colloid Surf. B 2020, 188, 110794. [Google Scholar] [CrossRef]
- Krajewska, M.; Dopierała, K.; Prochaska, K. Lipid-protein interactions in langmuir monolayers under dynamically varied conditions. J. Phys. Chem. B 2020, 124, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Dennison, S.; Harris, F.; Phoenix, D.A. Chapter three, Langmuir–Blodgett approach to investigate antimicrobial peptide–membrane interactions. APLBL 2014, 20, 83–110. [Google Scholar] [CrossRef]
- Zhao, L.; Feng, S.-S. Effects of lipid chain length on molecular interactions between paclitaxel and phospholipid within model biomembranes. J. Coll. Interf. Sci. 2004, 274, 55. [Google Scholar] [CrossRef]
- Perczyk, P.; Wójcik, A.; Hachlica, N.; Wydro, P.; Broniatowski, M. The composition of phospholipid model bacterial membranes determines their endurance to secretory phospholipase A2 attack—The role of cardiolipin. Biochim. Biophys. Acta—Biomembr. 2020, 1862, 183239. [Google Scholar] [CrossRef]
- Perczyk, P.; Broniatowski, M. Simultaneous action of microbial phospholipase C and lipase on model bacterial membranes – Modeling the processes crucial for bioaugmentation. Biochim. Biophys. Acta 2021, 1863, 183620. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.R.; Clifton, L.A.; Frazier, R.A.; Green, R.J. The role of lipid composition on the interaction between a tryptophan-rich protein and model bacterial membranes. Langmuir 2016, 32, 2050–2057. [Google Scholar] [CrossRef] [PubMed]
- Ciumac, D.; Gong, H.; Campbell, R.A.; Campana, M.; Xu, H.; Lu, J.R. Structural elucidation upon binding of antimicrobial peptides into binary mixed lipid monolayers mimicking bacterial membranes. J. Colloid Interface Sci. 2021, 598, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.G.; Almeida, A.M.; Nield, T.J.; Camacho, S.A.; Aoki, P.H.B. Modulating photochemical reactions in Langmuir monolayers of Escherichia coli lipid extract with the binding mechanisms of eosin decyl ester and toluidine blue-O photosensitizers. J. Photochem. Photobiol B Biol. 2021, 218, 112173. [Google Scholar] [CrossRef] [PubMed]
- Vandera, K.-A.; Picconi, P.; Valero, M.; González-Gaitano, G.; Woods, A.; Zain, N.M.M.; Bruce, K.D.; Clifton, L.A.; Skoda, M.W.A.; Rahman, K.M.; et al. Antibiotic-in-Cyclodextrin-in-Liposomes: Formulation development and interactions with model bacterial membranes. Mol. Pharm. 2020, 17, 2354–2369. [Google Scholar] [CrossRef]
- Tashiro, Y.; Inagaki, A.; Shimizu, M.; Ichikawa, S.; Takaya, N.; Nakajima-Kambe, T.; Uchiyama, H.; Nomura, N. Characterization of phospholipids in membrane vesicles derived from pseudomonas aeruginos. Biosci. Biotechnol. Biochem. 2011, 75, 605–607. [Google Scholar] [CrossRef]
- Mansilla, M.C.; Cybulski, L.E.; Albanesi, D.; de Mendoza, D. Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 2004, 186, 6681–6688. [Google Scholar] [CrossRef]
- De Chavigny, A.; Heacock, P.N.; Dowhan, W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J. Biol. Chem. 1991, 266, 5323–5332. [Google Scholar] [CrossRef]
- Rowlett, V.W.; Mallampalli, V.K.P.S.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. Impact of membrane phospholipid alterations in escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 2017, 199, e00849-16. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Li, J. Dynamic and morphological investigation of phospholipid monolayer hydrolysis by phospholipase C. Biochim. Biophys. Res. Commun. 2003, 300, 541–545. [Google Scholar] [CrossRef]
- Jewell, S.A.; Titball, R.W.; Huyet, J.; Naylor, C.E.; Basak, A.K.; Gologan, P.; Winlove, C.P.; Petrov, P.G. Clostridium perfringens α-toxin interaction with red cells and model membranes. Soft Matter 2015, 11, 7748–7761. [Google Scholar] [CrossRef] [PubMed]
- Urbina, P.; Collado, M.I.; Alonso, A.; Goñi, F.M.; Flores-Díazd, M.; Alape-Girónde, A.; Ruysschaerta, J.-M.; Lensink, M.F. Unexpected wide substrate specificity of C. perfringens α-toxin phospholipase C. Biochim. Biophys. Acta 2011, 1808, 2618–2627. [Google Scholar] [CrossRef]
- Hancock, R.E. The bacterial outer membrane as a drug barrier. Trends Microbial. 1997, 5, 37–42. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Raffle, V.J.; Nicas, T.I. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1981, 19, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Vaara, M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992, 56, 395–411. [Google Scholar] [CrossRef]
- Vaara, M. Outer membrane permeability barrier to azithromycin. clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1993, 37, 354. [Google Scholar] [CrossRef]
- Pages, J.-M.; James, C.E.; Winterhalter, M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [Google Scholar] [CrossRef]
- Dupuy, F.G.; Pagano, I.; Andenoro, K.; Peralta, M.F.; Elhady, Y.; Heinrich, F.; Tristram-Nagle, S. Selective interaction of colistin with lipid model membranes. Biophys. J. 2018, 114, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Clausell, A.; Garcia-Subirats, M.; Pujol, M.; Busquets, M.A.; Rabanal, F.; Cajal, Y. Gram-negative outer and inner membrane models: Insertion of cyclic cationic lipopeptides. J. Phys. Chem. B 2007, 111, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Cetuk, H.; Anishkin, A.; Scott, A.J.; Rempe, S.B.; Ernst, R.K.; Sukharev, S. Partitioning of seven different classes of antibiotics into LPS monolayers supports three different permeation mechanisms through the outer bacterial membrane. Langmuir 2021, 37, 1372–1385. [Google Scholar] [CrossRef]
- Abraham, T.; Schooling, S.R.; Beveridge, T.J.; Katsaras, J. Monolayer film behavior of lipopolysaccharide from Pseudomonas aeruginosa at the air-water interface. Biomacromolecules 2008, 9, 2799–2804. [Google Scholar] [CrossRef] [PubMed]
- Broniatowski, M.; Flasiński, M.; Zięba, K.; Miśkowiec, P. Langmuir monolayer studies of the interaction of monoamphiphilic pentacyclic triterpenes with anionic mitochondrial and bacterial membrane phospholipids—Searching for the most active terpene. Biochim. Biophys. Acta 2014, 1838, 2460–2472. [Google Scholar] [CrossRef]
- Fernandes, M.M.; Ivanova, K.; Hoyo, J.; Pérez-Rafael, S.; Francesko, A.; Tzanov, T. Nanotransformation of vancomycin overcomes the intrinsic resistance of gram-negative bacteria. ACS Appl. Mater. Interfaces 2017, 9, 15022–15030. [Google Scholar] [CrossRef]
- Knyght, I.; Clifton, L.; Saaka, Y.; Lawrence, M.J.; Barlow, D.J. Interaction of the antimicrobial peptides rhesus θ-defensin and porcine protegrin-1 with anionic phospholipid monolayers. Langmuir 2016, 32, 7403–7410. [Google Scholar] [CrossRef][Green Version]
- Rintoul, M.R.; Morero, R.D.; Dupuy, F.G. The antimicrobial peptide microcin J25 stabilizes the gel phase of bacterial model membranes. Colloids Surf. B 2015, 129, 183–190. [Google Scholar] [CrossRef]
- Mayfield, T.; le Brun, A.; Holt, S. Cell Membrane Studies Helping to Tackle Antibiotic Resistance. 16 November 2012. Available online: https://phys.org/news/2012-11-cell-membrane-tackle-antibioticresistance.html (accessed on 25 July 2021).
- Kumar, M.; Jaiswal, S.; Sodhia, K.K.; Shree, P.; Singh, D.K.; Agrawal, P.K.; Shukla, P. Antibiotics bioremediation: Perspectives on its ecotoxicity and resistance. Environ. Int. 2019, 124, 448–461. [Google Scholar] [CrossRef]



| Alpha-Proteobacteria | Beta-Proteobacteria | Gamma-Proteobacteria | Delta-Proteobacteria | Epsilon-Proteobacteria | Cyanobacteria | Actinobacteria | Spirochetes | Planctomyces | Firmicutes | |
|---|---|---|---|---|---|---|---|---|---|---|
| PG-phosphatidylglycerol | + | + | + | + | + | + | + | + | + | + |
| CL-cardiolipin | + | + | + | + | + | + | + | + | + | + |
| PS-phosphatidylserine | + | + | ||||||||
| PE-phosphatidylethanolamine | + | + | + | + | + | + | + | |||
| MMPE-monomethyl PE | + | + | ||||||||
| DMPE-dimethyl PE | + | + | ||||||||
| PT-phosphatidylthreonine | + | |||||||||
| PC-phosphatidylcholine | + | + | + | |||||||
| PA-phosphatidic acid | + | |||||||||
| GPL-glycophospholipid | + | |||||||||
| LPG-lysyl-phosphatidylglycerol | + | + | + | + | + | |||||
| APG-alanyl-phosphatidylglycerol | + | |||||||||
| LCL-lysyl-cardiolipin | + |
| Model of Mimic Biological Membrane | Characteristic of Model |
|---|---|
| Langmuir monolayer (Monomolecular insoluble lipid films)  |
|
| SLB (A flat lipid bilayer supported onto a solid surface)  |
|
| Liposomes (Lipid vesicles)  |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojewska, M.; Smułek, W.; Kaczorek, E.; Prochaska, K. Langmuir Monolayer Techniques for the Investigation of Model Bacterial Membranes and Antibiotic Biodegradation Mechanisms. Membranes 2021, 11, 707. https://doi.org/10.3390/membranes11090707
Rojewska M, Smułek W, Kaczorek E, Prochaska K. Langmuir Monolayer Techniques for the Investigation of Model Bacterial Membranes and Antibiotic Biodegradation Mechanisms. Membranes. 2021; 11(9):707. https://doi.org/10.3390/membranes11090707
Chicago/Turabian StyleRojewska, Monika, Wojciech Smułek, Ewa Kaczorek, and Krystyna Prochaska. 2021. "Langmuir Monolayer Techniques for the Investigation of Model Bacterial Membranes and Antibiotic Biodegradation Mechanisms" Membranes 11, no. 9: 707. https://doi.org/10.3390/membranes11090707
APA StyleRojewska, M., Smułek, W., Kaczorek, E., & Prochaska, K. (2021). Langmuir Monolayer Techniques for the Investigation of Model Bacterial Membranes and Antibiotic Biodegradation Mechanisms. Membranes, 11(9), 707. https://doi.org/10.3390/membranes11090707








