Hydroxylated Fatty Acids: The Role of the Sphingomyelin Synthase and the Origin of Selectivity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Structure Prediction and Validation
2.2. Binding Site Definition
2.3. Molecular Docking
2.4. Molecular Dynamics Simulations
2.5. Metadynamics Simulations
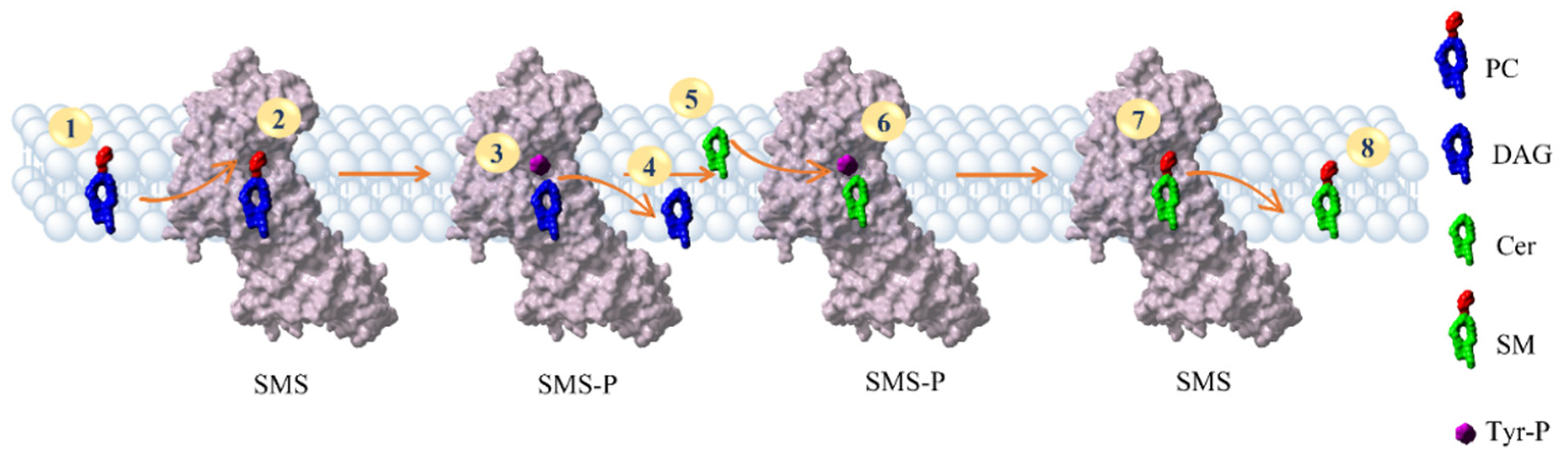
2.6. Reconstruction of the SMS Pathway
3. Results and Discussion
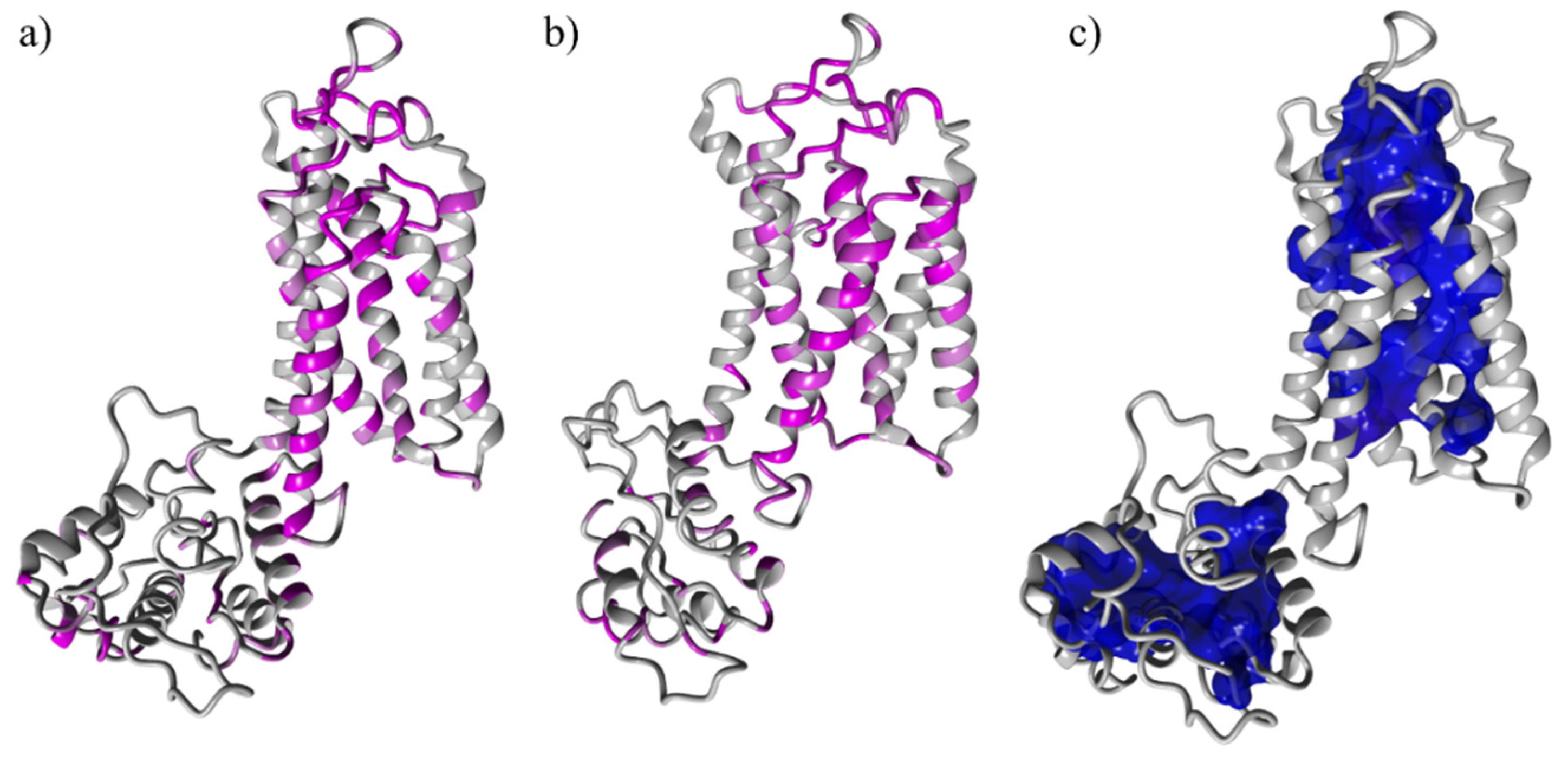
3.1. Structure Prediction and Validation: The Two Isoforms Show a High Homology Sequence
3.2. Binding Site Identification: The Transmembrane Portion Contains the Binding Site
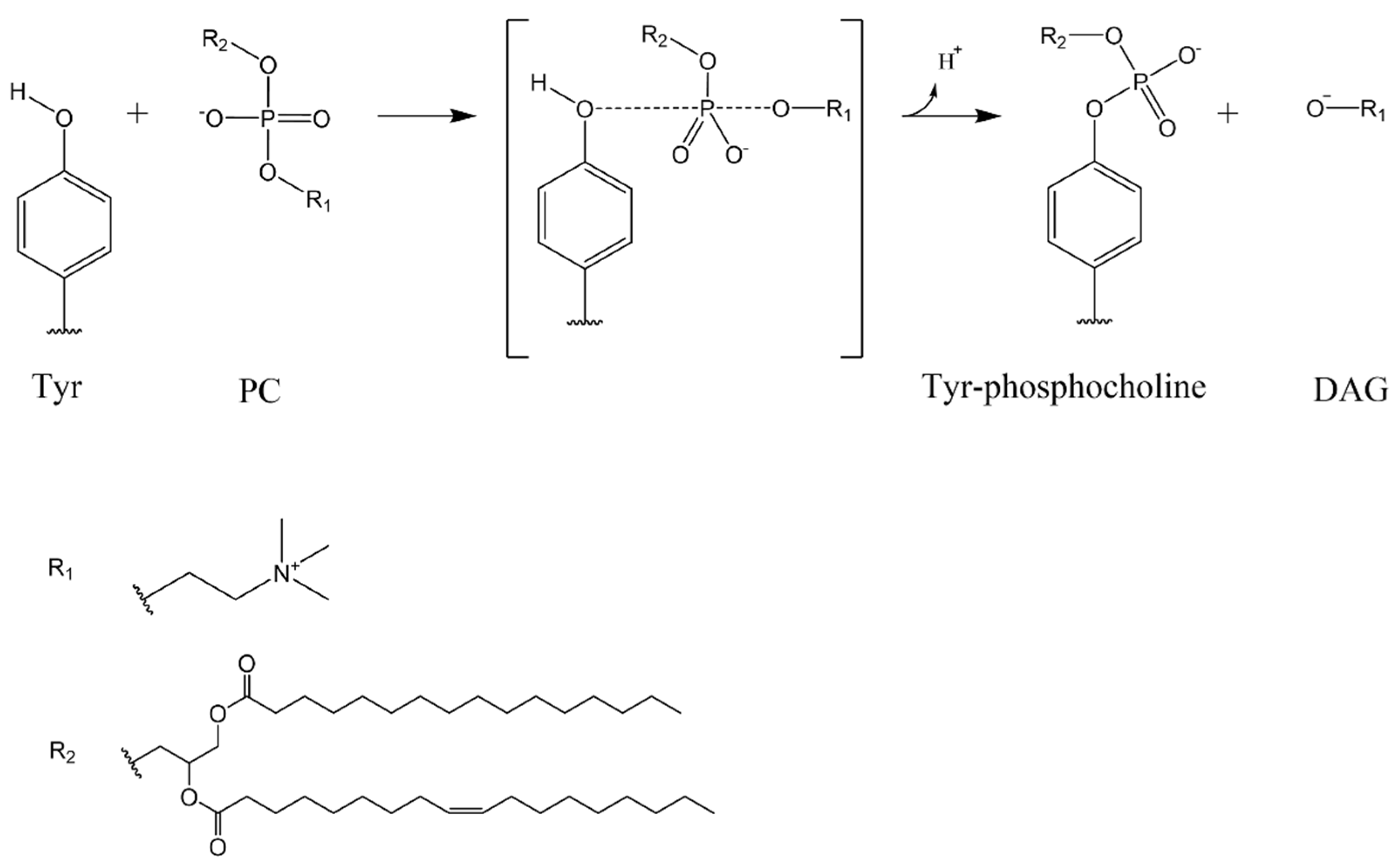
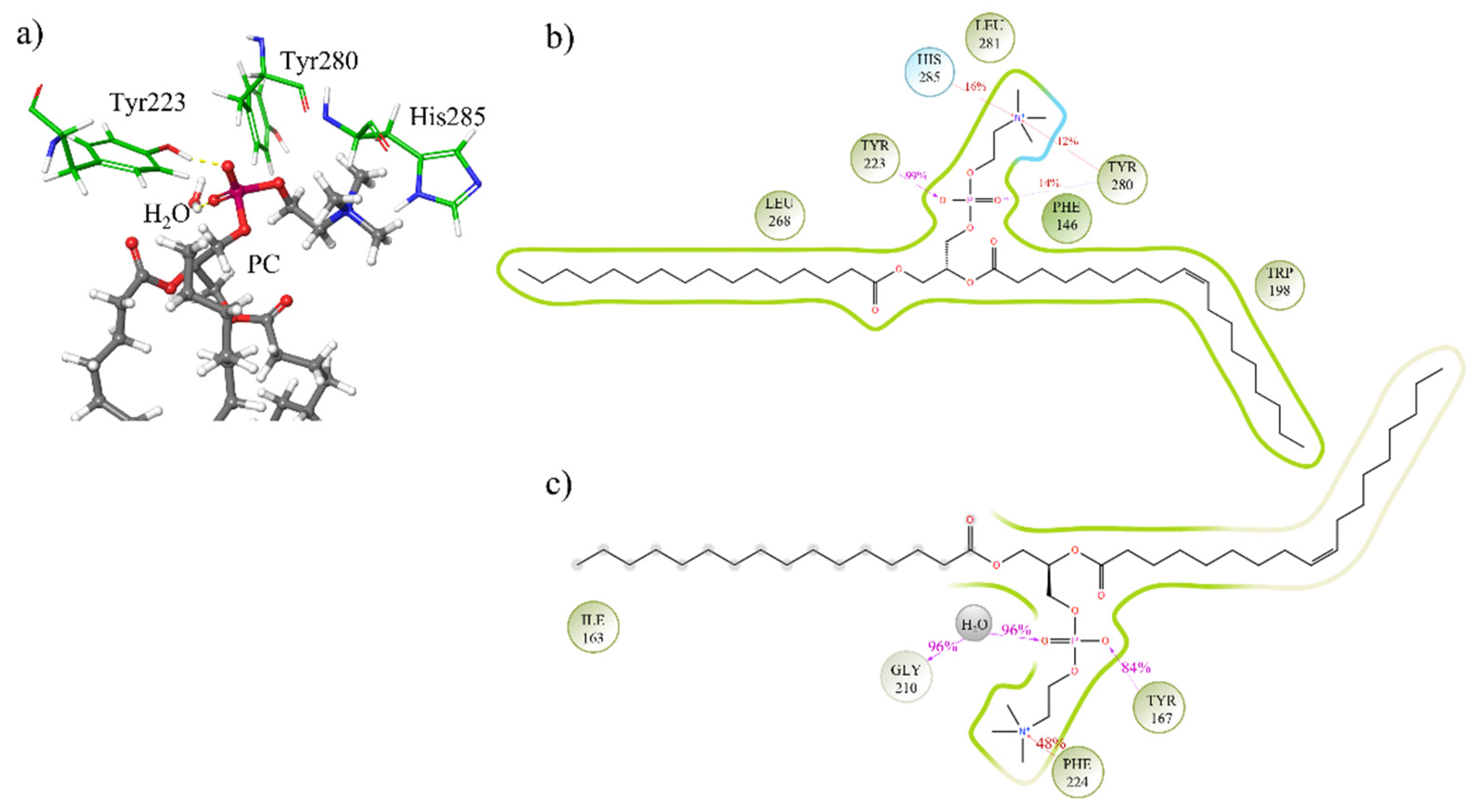
3.3. Study of the Mechanism of the Reaction: Tyrosine Is a Key Residue
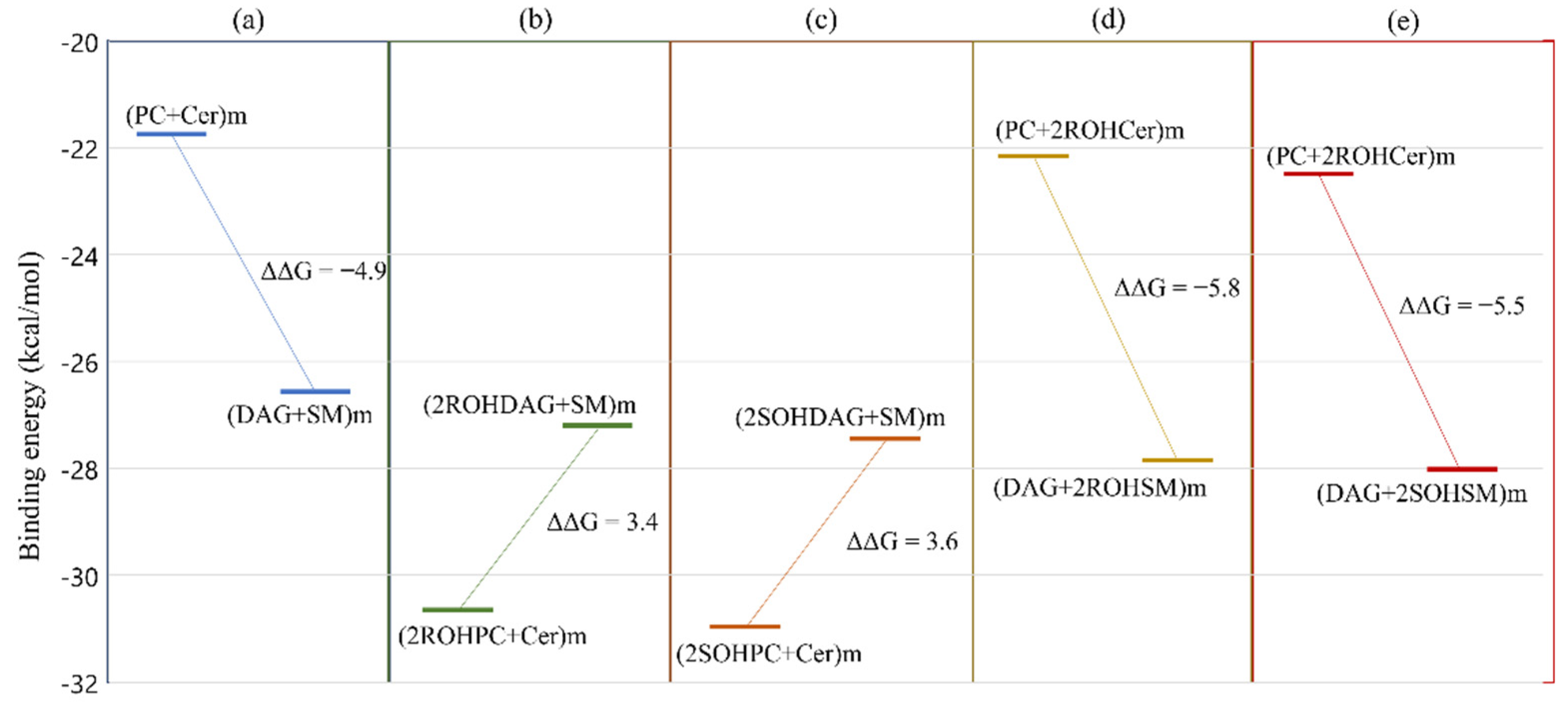
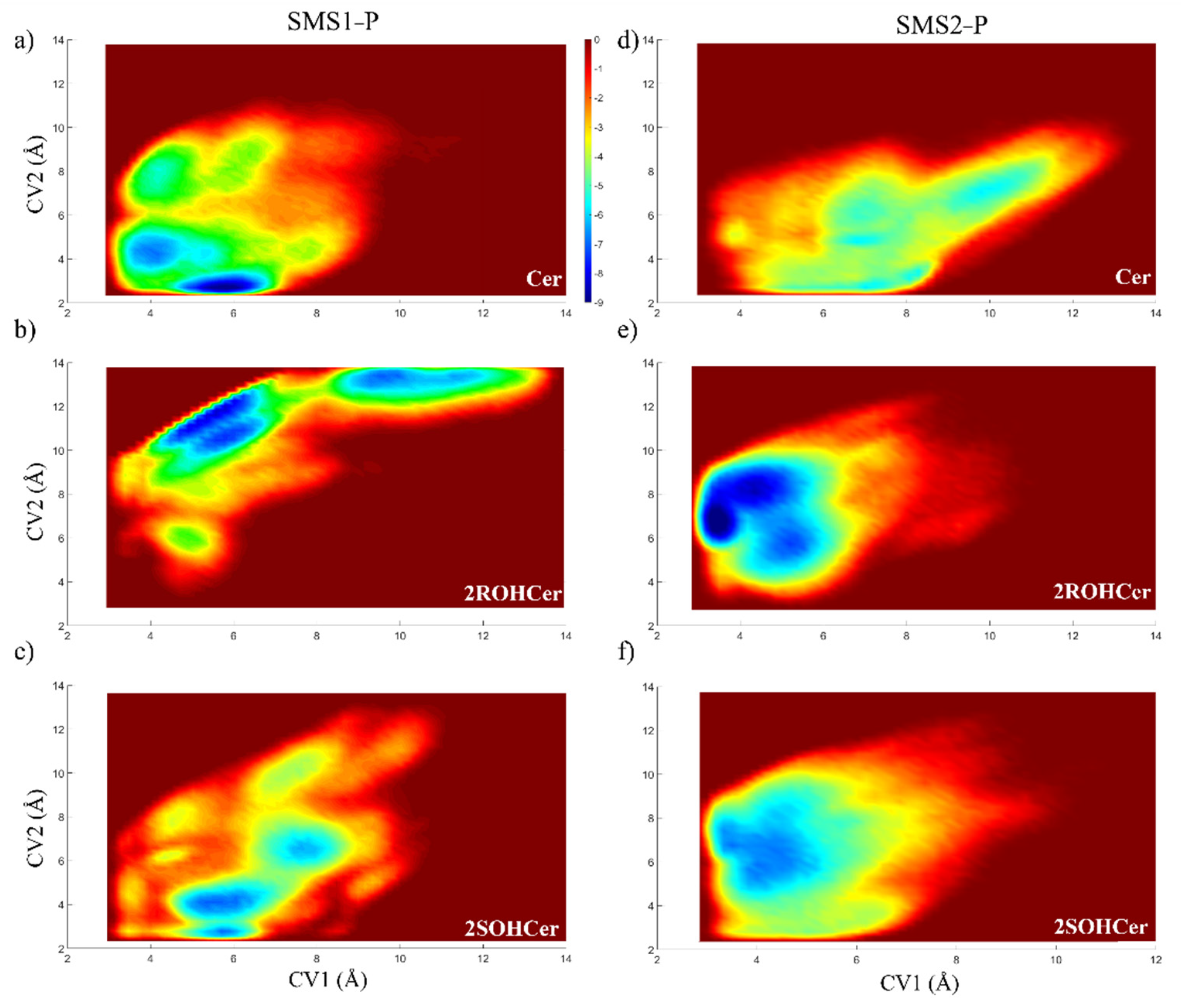
3.4. Free Energy Profile: Hydroxylated Ceramide Is the Better Substrate of SMSs
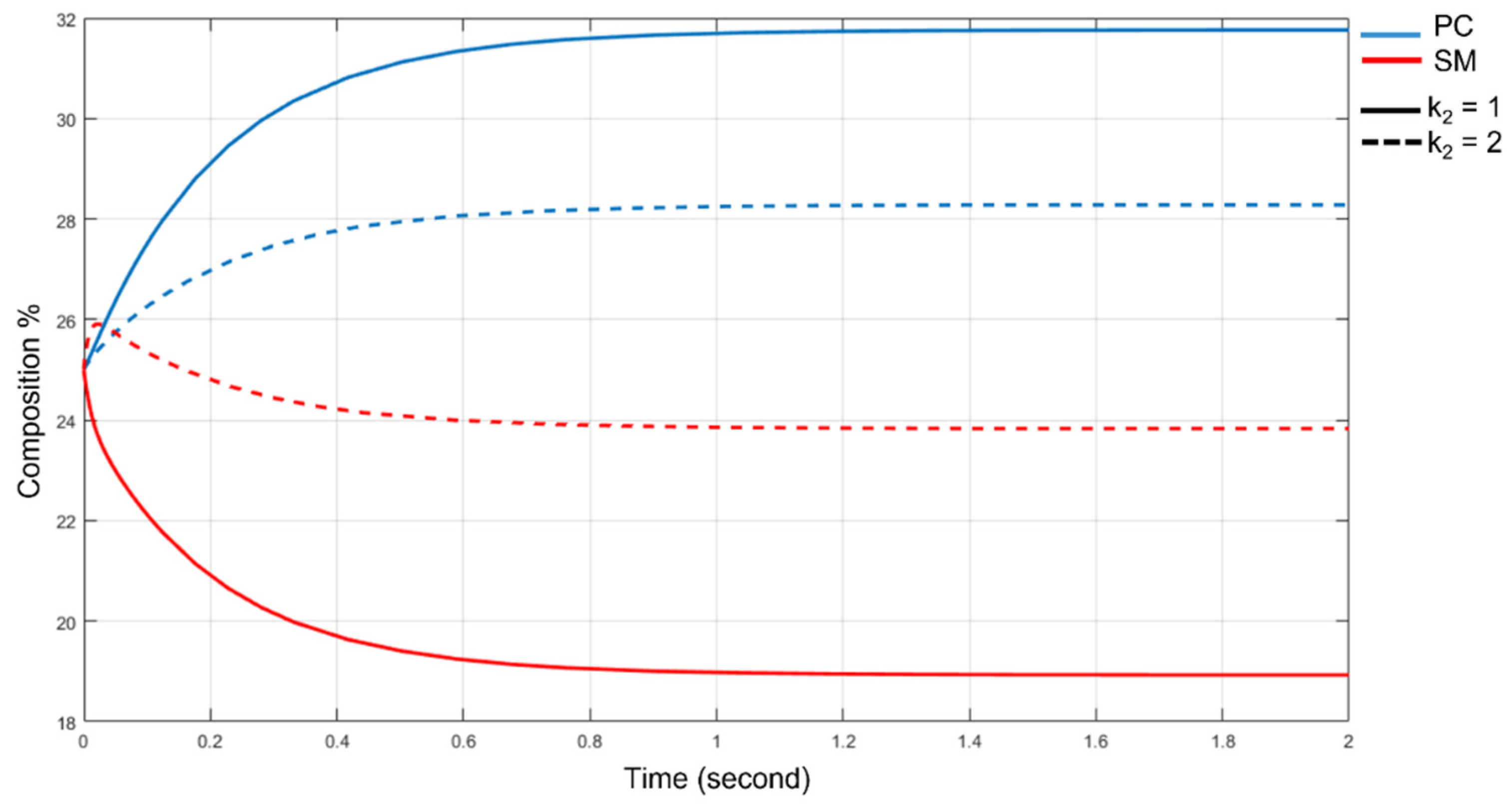
3.5. The SMS Pathway
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Merrill, A.H. Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef] [PubMed]
- Deevska, G.M.; Rozenova, K.A.; Giltiay, N.V.; Chambers, M.A.; White, J.; Boyanovsky, B.B.; Wei, J.; Daugherty, A.; Smart, E.J.; Reid, M.B. Acid sphingomyelinase deficiency prevents diet-induced hepatic triacylglycerol accumulation and hyperglycemia in mice. J. Biol. Chem. 2009, 284, 8359–8368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, N.W.; Gellings, N.M.; Guo, M.; Barlow, S.B.; Glembotski, C.C.; Sabbadini, R.A. Factor associated with neutral sphingomyelinase activation and its role in cardiac cell death. Circ. Res. 2003, 92, 589–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrödinger LLC. Schrödinger Release 2021-3: Maestro; Schrödinger LLC: New York, NY, USA, 2021. [Google Scholar]
- Goñi, F.M.; Alonso, A. Biophysics of sphingolipids I. Membrane properties of sphingosine, ceramides and other simple sphingolipids. Biochim. Biophys. Acta 2006, 1758, 1902–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Török, Z.; Crul, T.; Maresca, B.; Schütz, G.J.; Viana, F.; Dindia, L.; Piotto, S.; Brameshuber, M.; Balogh, G.; Péter, M.; et al. Plasma membranes as heat stress sensors: From lipid-controlled molecular switches to therapeutic applications. Biochim. Biophys. Acta 2014, 1838, 1594–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vígh, L.; Török, Z.; Balogh, G.; Glatz, A.; Piotto, S.; Horváth, I. Membrane-regulated stress response: A theoretical and practical approach. Adv. Exp. Med. Biol. 2007, 594, 114–131. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.; Khalid, S.; Aponte-Santamaría, C.; Arutyunova, E.; Becker, M.; Boyd, K.J.; Christensen, M.; Coimbra, J.T.S.; Concilio, S.; Daday, C.; et al. Understanding Conformational Dynamics of Complex Lipid Mixtures Relevant to Biology. J. Membr. Biol. 2018, 251, 609–631. [Google Scholar] [CrossRef] [Green Version]
- Concilio, S.; Ferrentino, I.; Sessa, L.; Massa, A.; Iannelli, P.; Diana, R.; Panunzi, B.; Rella, A.; Piotto, S. A novel fluorescent solvatochromic probe for lipid bilayers. Supramol. Chem. 2017, 29, 887–895. [Google Scholar] [CrossRef]
- Holthuis, J.C.; Luberto, C. Tales and mysteries of the enigmatic sphingomyelin synthase family. Adv. Exp. Med. Biol. 2010, 688, 72–85. [Google Scholar] [CrossRef] [Green Version]
- Lingwood, D.; Kaiser, H.J.; Levental, I.; Simons, K. Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 2009, 37, 955–960. [Google Scholar] [CrossRef] [Green Version]
- Engberg, O.; Lin, K.-L.; Hautala, V.; Slotte, J.P.; Nyholm, T.K. Sphingomyelin acyl chains influence the formation of sphingomyelin-and cholesterol-enriched domains. Biophys. J. 2020, 119, 913–923. [Google Scholar] [CrossRef]
- Balleza, D.; Mescola, A.; Marín–Medina, N.; Ragazzini, G.; Pieruccini, M.; Facci, P.; Alessandrini, A. Complex phase behavior of GUVs containing different sphingomyelins. Biophys. J. 2019, 116, 503–517. [Google Scholar] [CrossRef] [Green Version]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Carreira, A.C.; Ventura, A.E.; Varela, A.R.; Silva, L.C. Tackling the biophysical properties of sphingolipids to decipher their biological roles. Biol. Chem. 2015, 396, 597–609. [Google Scholar] [CrossRef]
- Piotto, S.; Di Biasi, L.; Sessa, L.; Concilio, S. Transmembrane peptides as sensors of the membrane physical state. Front. Phys. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Adada, M.; Luberto, C.; Canals, D. Inhibitors of the sphingomyelin cycle: Sphingomyelin synthases and sphingomyelinases. Chem. Phys. Lipids 2016, 197, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.S.; Goldschmidt-Arzi, M.; Sabanay, H.; Storch, J.; Levade, T.; Ribeiro, M.G.; Addadi, L.; Futerman, A.H. Accumulation of ordered ceramide-cholesterol domains in farber disease fibroblasts. In JIMD Reports-Volume 12; Springer: Cham, Switzerland, 2013; pp. 71–77. [Google Scholar] [CrossRef] [Green Version]
- Kirkegaard, T.; Roth, A.G.; Petersen, N.H.; Mahalka, A.K.; Olsen, O.D.; Moilanen, I.; Zylicz, A.; Knudsen, J.; Sandhoff, K.; Arenz, C. Hsp70 stabilizes lysosomes and reverts Niemann–Pick disease-associated lysosomal pathology. Nature 2010, 463, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Pralhada Rao, R.; Vaidyanathan, N.; Rengasamy, M.; Mammen Oommen, A.; Somaiya, N.; Jagannath, M. Sphingolipid metabolic pathway: An overview of major roles played in human diseases. J. Lipids 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, N.; Keep, R.F.; Hua, Y.; Xi, G. Critical role of the sphingolipid pathway in stroke: A review of current utility and potential therapeutic targets. Transl. Stroke Res. 2016, 7, 420–438. [Google Scholar] [CrossRef] [Green Version]
- Van Helvoort, A.; Van’t Hof, W.; Ritsema, T.; Sandra, A.; Van Meer, G. Conversion of diacylglycerol to phosphatidylcholine on the basolateral surface of epithelial (Madin-Darby canine kidney) cells. Evidence for the reverse action of a sphingomyelin synthase. J. Biol. Chem. 1994, 269, 1763–1769. [Google Scholar] [CrossRef]
- Huitema, K.; van den Dikkenberg, J.; Brouwers, J.F.; Holthuis, J.C. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004, 23, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Piotto, S.; Sessa, L.; Iannelli, P.; Concilio, S. Computational study on human sphingomyelin synthase 1 (hSMS1). Biochim. Biophys. Acta. Biomembr. 2017, 1859, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Villani, M.; Subathra, M.; Im, Y.-B.; Choi, Y.; Signorelli, P.; Del Poeta, M.; Luberto, C. Sphingomyelin synthases regulate production of diacylglycerol at the Golgi. Biochem. J. 2008, 414, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Hailemariam, T.K.; Zhou, H.; Li, Y.; Duckworth, D.C.; Peake, D.A.; Zhang, Y.; Kuo, M.-S.; Cao, G.; Jiang, X.-C. Inhibition of sphingomyelin synthase (SMS) affects intracellular sphingomyelin accumulation and plasma membrane lipid organization. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 1186–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, D.H.; Fiol-deRoque, M.A.; Noguera-Salvà, M.A.; Terés, S.; Campana, F.; Piotto, S.; Castro, J.A.; Mohaibes, R.J.; Escribá, P.V.; Busquets, X. 2-Hydroxy Arachidonic Acid: A New Non-Steroidal Anti-Inflammatory Drug. PLoS ONE 2013, 8, e72052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slotte, P.J. Molecular properties of various structurally defined sphingomyelins–correlation of structure with function. Prog. Lipid Res. 2013, 52, 206–219. [Google Scholar] [CrossRef]
- Markham, J.E.; Lynch, D.V.; Napier, J.A.; Dunn, T.M.; Cahoon, E.B. Plant sphingolipids: Function follows form. Curr. Opin. Plant Biol. 2013, 16, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Tani, M. Structure–Function Relationship of Complex Sphingolipids in Yeast. Trends Glycosci. Glycotechnol. 2016, 28, E109–E116. [Google Scholar] [CrossRef] [Green Version]
- Casas, J.; Ibarguren, M.; Álvarez, R.; Terés, S.; Lladó, V.; Piotto, S.P.; Concilio, S.; Busquets, X.; López, D.J.; Escribá, P.V. G protein-membrane interactions II: Effect of G protein-linked lipids on membrane structure and G protein-membrane interactions. Biochim. Biophys. Acta. Biomembr. 2017, 1859, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Marquês, J.T.; Marinho, H.S.; de Almeida, R.F.M. Sphingolipid hydroxylation in mammals, yeast and plants—An integrated view. Prog. Lipid Res. 2018, 71, 18–42. [Google Scholar] [CrossRef]
- Jaikishan, S.; Slotte, J.P. Stabilization of sphingomyelin interactions by interfacial hydroxyls—A study of phytosphingomyelin properties. Biochim. Biophys. Acta Biomembr. 2013, 1828, 391–397. [Google Scholar] [CrossRef] [Green Version]
- Piotto, S.; Trapani, A.; Bianchino, E.; Ibarguren, M.; López, D.J.; Busquets, X.; Concilio, S. The effect of hydroxylated fatty acid-containing phospholipids in the remodeling of lipid membranes. Biochim. Et Biophys. Acta 2014, 1838, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Hama, H. Fatty acid 2-Hydroxylation in mammalian sphingolipid biology. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2010, 1801, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Alderson, N.L.; Rembiesa Bm Fau—Walla, M.D.; Walla Md Fau—Bielawska, A.; Bielawska, A. Fau—Bielawski, J.; Bielawski, J. Fau—Hama, H.; Hama, H. The human FA2H gene encodes a fatty acid 2-hydroxylase. J. Biol. Chem. 2004, 279, 48562–48568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, A.B.; Astudillo, A.M.; Balboa, M.A.; Cuevas, C.; Balsinde, J.; Moreno, S. Levels of SCS7/FA2H-mediated fatty acid 2-hydroxylation determine the sensitivity of cells to antitumor PM02734. Cancer Res. 2008, 68, 9779–9787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Zhang, X.; Zhou, D.; Okunade, A.L.; Su, X. Stereospecificity of fatty acid 2-hydroxylase and differential functions of 2-hydroxy fatty acid enantiomers. J. Lipid Res. 2012, 53, 1327–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizutani, Y.; Kihara, A.; Chiba, H.; Tojo, H.; Igarashi, Y. 2-Hydroxy-ceramide synthesis by ceramide synthase family: Enzymatic basis for the preference of FA chain length. J. Lipid Res. 2008, 49, 2356–2364. [Google Scholar] [CrossRef] [Green Version]
- Schaeren-Wiemers, N.; van der Bijl, P.; Schwab, M.E. The UDP-galactose:ceramide galactosyltransferase: Expression pattern in oligodendrocytes and Schwann cells during myelination and substrate preference for hydroxyceramide. J. Neurochem. 1995, 65, 2267–2278. [Google Scholar] [CrossRef]
- Piotto, S.; Concilio, S.; Bianchino, E.; Iannelli, P.; López, D.J.; Terés, S.; Ibarguren, M.; Barceló-Coblijn, G.; Martin, M.L.; Guardiola-Serrano, F.; et al. Differential effect of 2-hydroxyoleic acid enantiomers on protein (sphingomyelin synthase) and lipid (membrane) targets. Biochim. Biophys. Acta 2014, 1838, 1628–1637. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- SM Platform 1.1 Powered by SoftMining srl. Available online: https://smp.softmining.it (accessed on 1 January 2021).
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Vriend, G. WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 1190, 8, 52–56. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotto, S.; Di Biasi, L.; Fino, R.; Parisi, R.; Sessa, L.; Concilio, S. Yada: A novel tool for molecular docking calculations. J. Comp.-Aided Mol. Des. 2016, 30, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Debiec, K.T.; Cerutti, D.S.; Baker, L.R.; Gronenborn, A.M.; Case, D.A.; Chong, L.T. Further along the Road Less Traveled: AMBER ff15ipq, an Original Protein Force Field Built on a Self-Consistent Physical Model. J. Chem. Theory Comput. 2016, 12, 3926–3947. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Sessa, L.; Biasi, L.D.; Concilio, S.; Cattaneo, G.; De Santis, A.; Iannelli, P.; Piotto, S. A new flexible protocol for docking studies; Stano, P., Rossi, F., Mavelli, F., Caivano, D., Eds.; Springer: Cham, Switzerland, 2016; Volume 587, pp. 117–126. [Google Scholar]
- Sessa, L.; Concilio, S.; Piotto, S. Molecular Dynamics and Morphing Protocols for High Accuracy Molecular Docking. In Advances in Bionanomaterials; Lecture Notes in Bioengineering; Springer: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the SC’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L. OPLS3e: Extending force field coverage for drug-like small molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef] [PubMed]
- SymBiology. Available online: https://it.mathworks.com/products/simbiology.html (accessed on 1 February 2021).
- Mavelli, F.; Piotto, S. Stochastic simulations of homogeneous chemically reacting systems. J. Mol. Struct. THEOCHEM 2006, 771, 55–64. [Google Scholar] [CrossRef]
- Celniker, G.; Nimrod, G.; Ashkenazy, H.; Glaser, F.; Martz, E.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013, 53, 199–206. [Google Scholar] [CrossRef]
- Yeang, C.; Varshney, S.; Wang, R.; Zhang, Y.; Ye, D.; Jiang, X.-C. The domain responsible for sphingomyelin synthase (SMS) activity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2008, 1781, 610–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, B.; Liu, Q.; Hou, J.; Kabir, I.; Liu, P.; Ding, T.; Dong, J.; Mo, M.; Ye, D.; Chen, Y.; et al. 2-Hydroxy-oleic acid does not activate sphingomyelin synthase activity. J. Biol. Chem. 2018, 293, 18328–18336. [Google Scholar] [CrossRef] [Green Version]
- Barceló-Coblijn, G.; Martin, M.L.; de Almeida, R.F.M.; Noguera-Salvà, M.A.; Marcilla-Etxenike, A.; Guardiola-Serrano, F.; Lüth, A.; Kleuser, B.; Halver, J.E.; Escribá, P.V. Sphingomyelin and sphingomyelin synthase (SMS) in the malignant transformation of glioma cells and in 2-hydroxyoleic acid therapy. Proc. Natl. Acad. Sci. USA 2011, 108, 19569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigh, L.; Escribá, P.V.; Sonnleitner, A.; Sonnleitner, M.; Piotto, S.; Maresca, B.; Horváth, I.; Harwood, J.L. The significance of lipid composition for membrane activity: New concepts and ways of assessing function. Prog. Lipid Res. 2005, 44, 303–344. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.A. The drug–target residence time model: A 10-year retrospective. Nat. Rev. Drug Discov. 2016, 15, 87–95. [Google Scholar] [CrossRef]
- Tanaka, K.; Tamiya-Koizumi, K.; Yamada, M.; Murate, T.; Kannagi, R.; Kyogashima, M. Individual profiles of free ceramide species and the constituent ceramide species of sphingomyelin and neutral glycosphingolipid and their alteration according to the sequential changes of environmental oxygen content in human colorectal cancer Caco-2 cells. Glycoconj. J. 2014, 31, 209–219. [Google Scholar] [CrossRef]
- Shaner, R.L.; Allegood, J.C.; Park, H.; Wang, E.; Kelly, S.; Haynes, C.A.; Sullards, M.C.; Merrill, A.H., Jr. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 2009, 50, 1692–1707. [Google Scholar] [CrossRef] [Green Version]
- Ou, P.; Stanek, A.; Huan, Z.; Roman, C.A.; Huan, C. SMS2 deficiency impairs PKCδ-regulated B cell tolerance in the germinal center. Cell Rep. 2021, 36, 109624. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sessa, L.; Nardiello, A.M.; Santoro, J.; Concilio, S.; Piotto, S. Hydroxylated Fatty Acids: The Role of the Sphingomyelin Synthase and the Origin of Selectivity. Membranes 2021, 11, 787. https://doi.org/10.3390/membranes11100787
Sessa L, Nardiello AM, Santoro J, Concilio S, Piotto S. Hydroxylated Fatty Acids: The Role of the Sphingomyelin Synthase and the Origin of Selectivity. Membranes. 2021; 11(10):787. https://doi.org/10.3390/membranes11100787
Chicago/Turabian StyleSessa, Lucia, Anna Maria Nardiello, Jacopo Santoro, Simona Concilio, and Stefano Piotto. 2021. "Hydroxylated Fatty Acids: The Role of the Sphingomyelin Synthase and the Origin of Selectivity" Membranes 11, no. 10: 787. https://doi.org/10.3390/membranes11100787
APA StyleSessa, L., Nardiello, A. M., Santoro, J., Concilio, S., & Piotto, S. (2021). Hydroxylated Fatty Acids: The Role of the Sphingomyelin Synthase and the Origin of Selectivity. Membranes, 11(10), 787. https://doi.org/10.3390/membranes11100787








