Influenza Virus-like Particle (VLP) Vaccines Expressing the SARS-CoV-2 S Glycoprotein, S1, or S2 Domains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Animals
2.2. Codon Optimization, Gene Cloning, Recombinant Baculovirus, and VLP Production
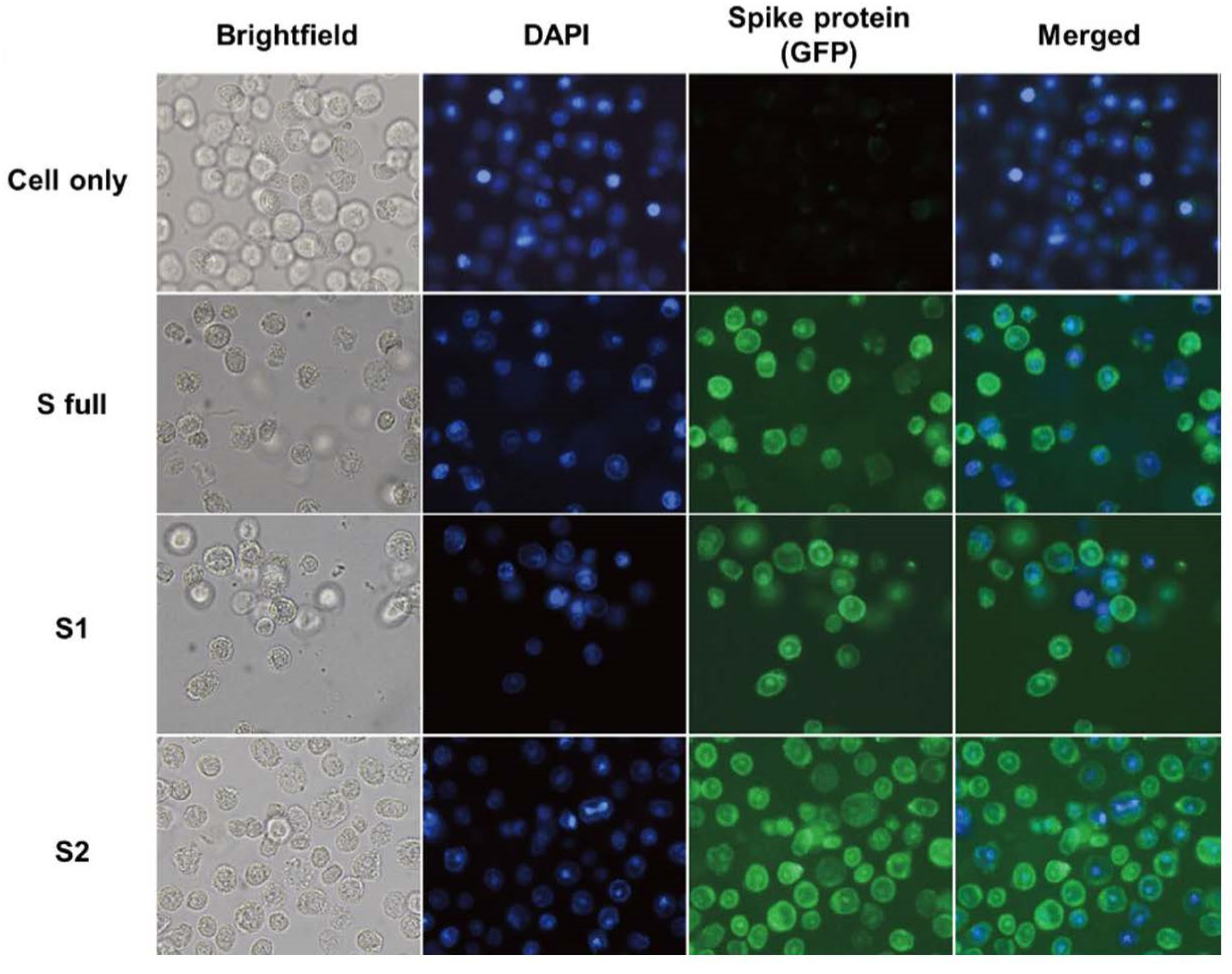
2.3. Immunocytochemistry Using the Transfected rBVs
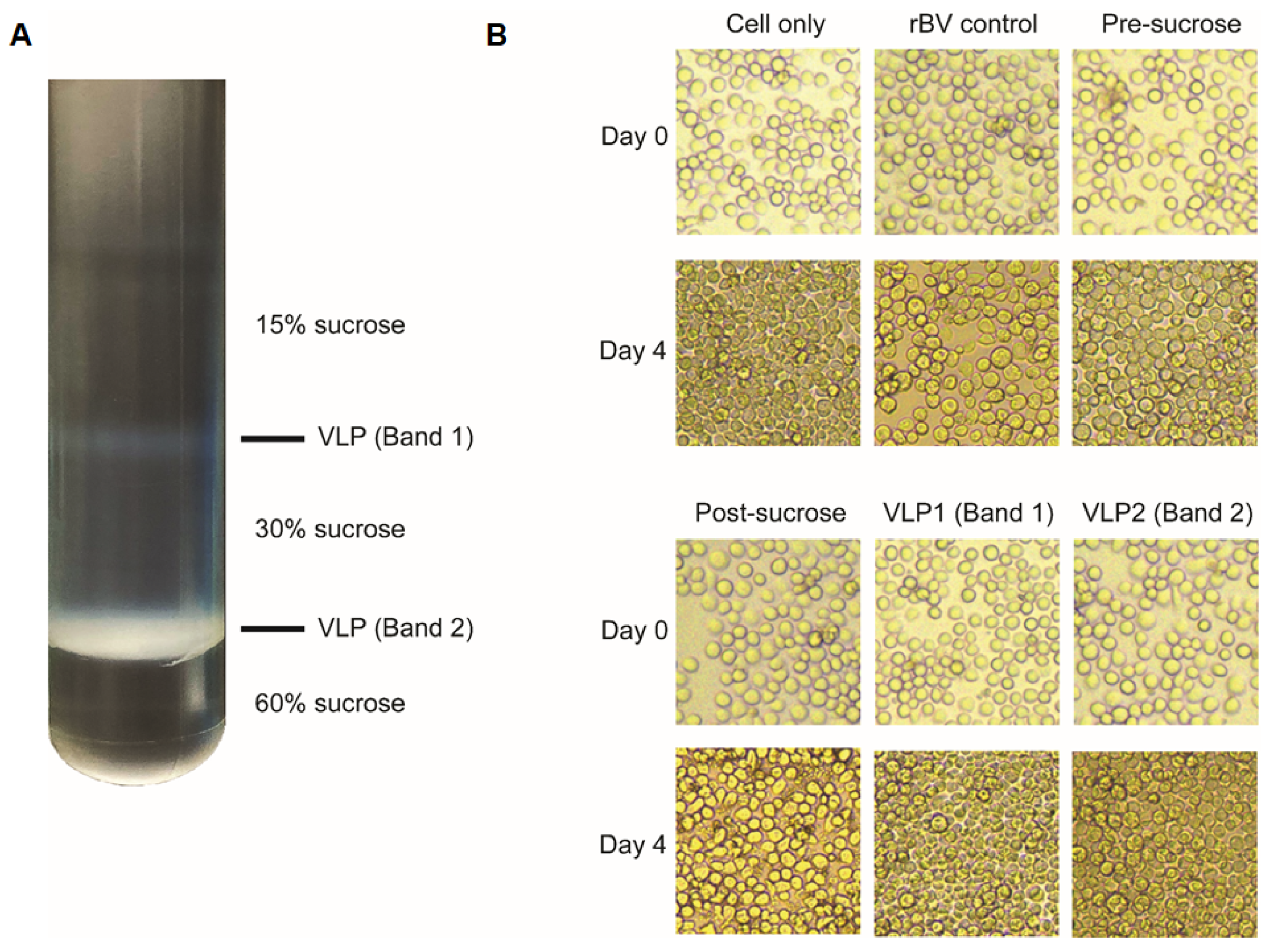
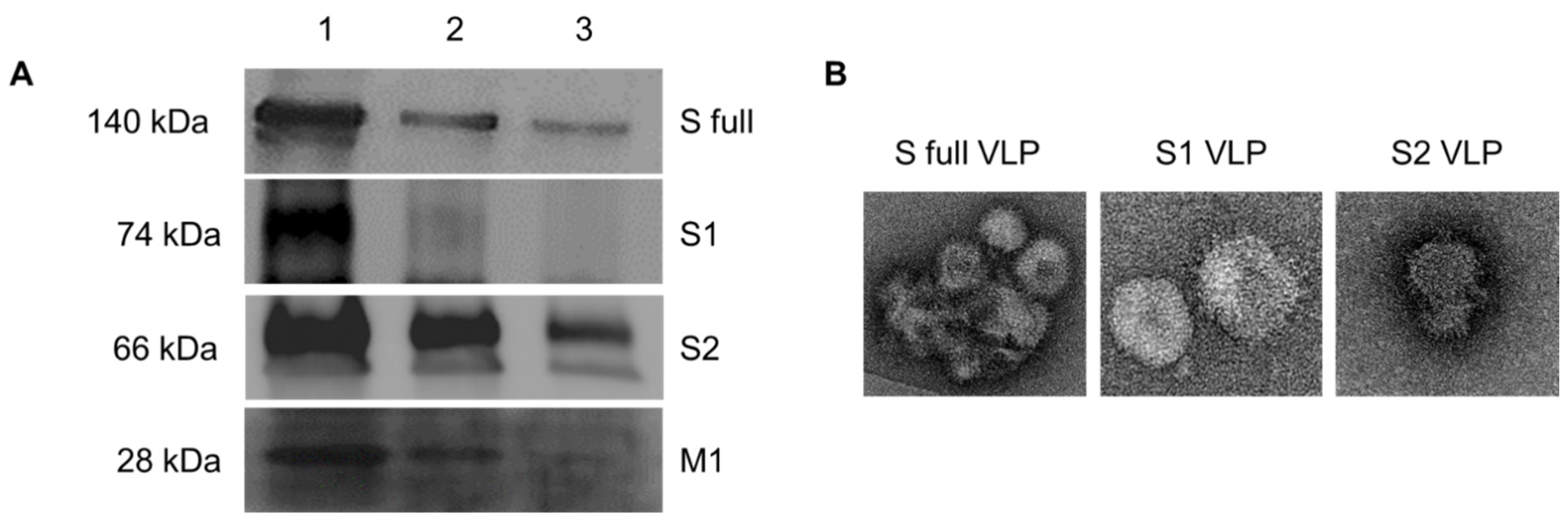
2.4. Characterization of VLPs
2.5. Animals, Immunization, and Sample Collection
2.6. Antibody Responses against S1 and S2 Proteins of SARS-CoV-2
2.7. Surrogate Virus Neutralization Assay (sVNT)
2.8. Statistical Analysis
3. Results
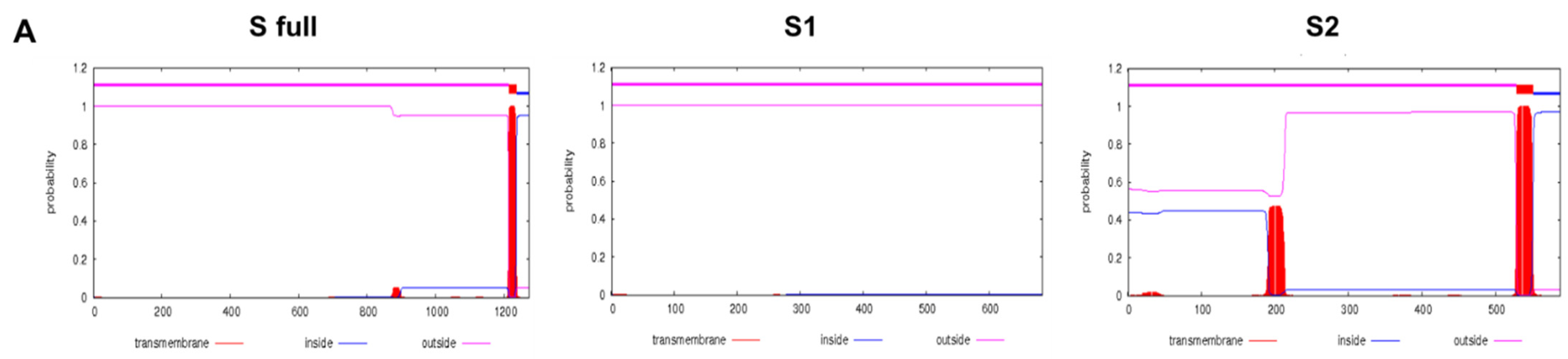
3.1. Transmembrane Protein Topology and Construct Schematic
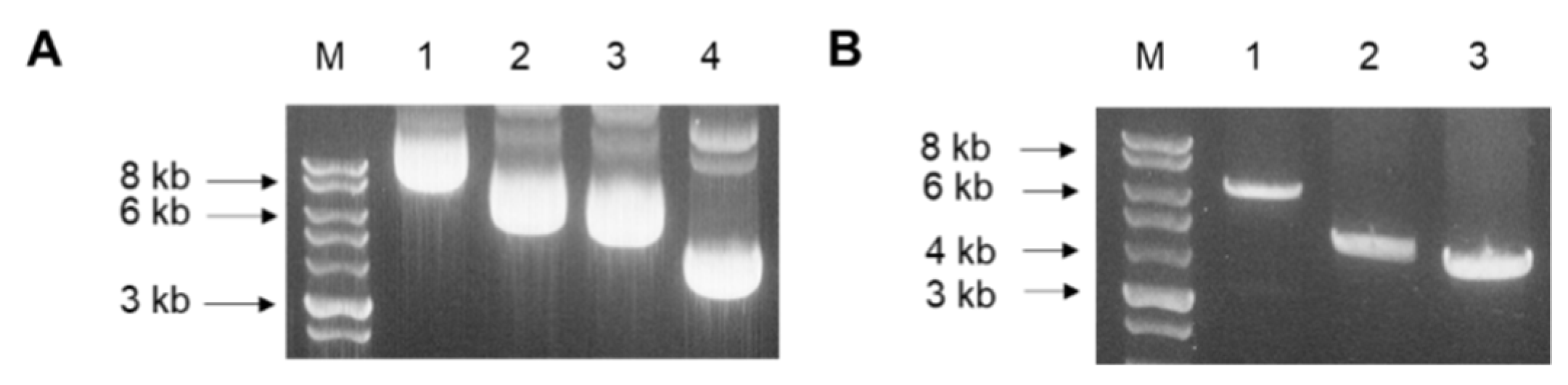
3.2. Codon-Optimized Gene Cloning into pFastBac Vectors and Colony PCR
3.3. Construction of Spike Protein-Expressing Recombinant Baculoviruses
3.4. Characterization of VLPs by Western blot and TEM
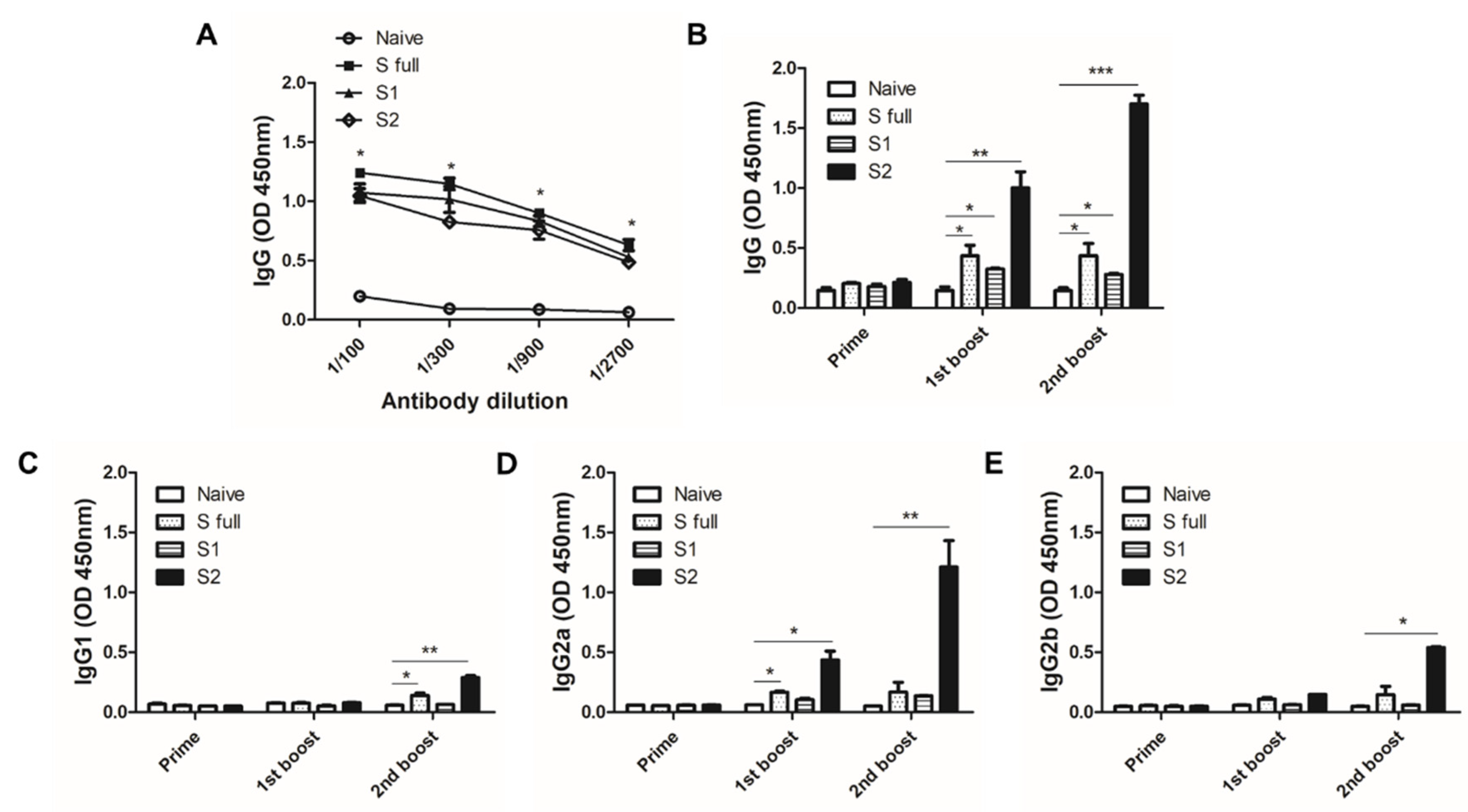
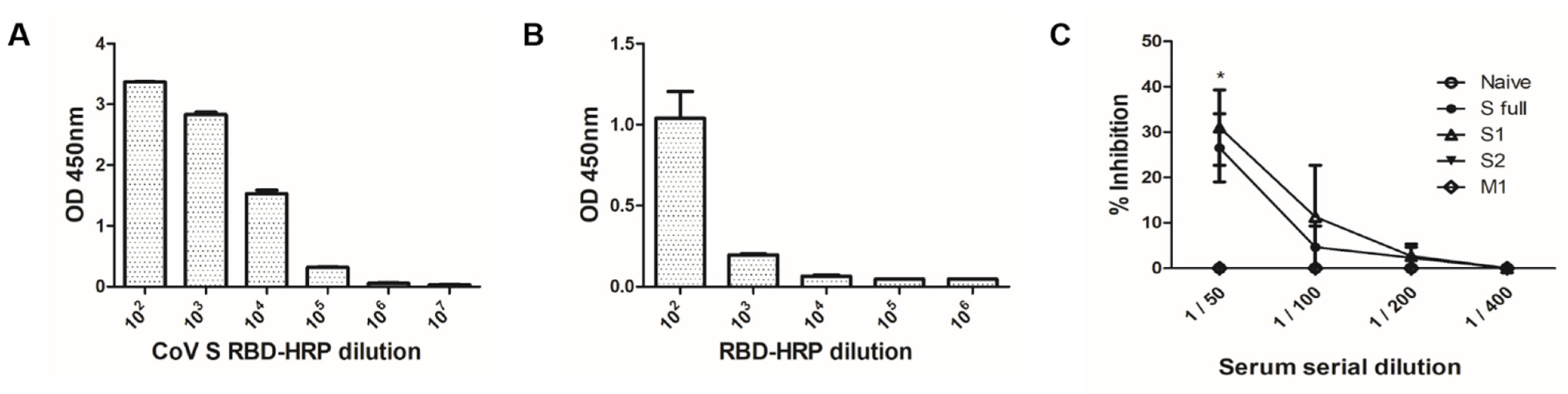
3.5. VLP Immunization Induced Antibody Response in Sera
3.6. Effective Inhibition Requires Antibodies Raised against the S1 Domain
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Hoek, L. Human coronaviruses: What do they cause? Antivir. Ther. 2007, 12, 651–658. [Google Scholar]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Karthik, K.; Senthilkumar, T.M.A.; Udhayavel, S.; Raj, G.D. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum. Vaccin. Immunother. 2020, 16, 3055–3060. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 21 April 2021).
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Le, T.T.; Cramer, J.P.; Chen, R.; Mayhew, S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 667–668. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 COVID-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef]
- Chen, Q.; Lai, H. Plant-derived virus-like particles as vaccines. Hum. Vaccin. Immunother. 2013, 9, 26–49. [Google Scholar] [CrossRef]
- Kumar, A.; Dowling, W.E.; Román, R.G.; Chaudhari, A.; Gurry, C.; Le, T.T.; Tollefson, S.; Clark, C.E.; Bernasconi, V.; Kristiansen, P.A. Status Report on COVID-19 Vaccines Development. Curr. Infect. Dis. Rep. 2021, 23, 9. [Google Scholar] [CrossRef]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.K.; Du, P.; Ignacio, C.; Mehta, S.; Newton, I.G.; Steinmetz, N.F. Biomimetic Virus-Like Particles as Severe Acute Respiratory Syndrome Coronavirus 2 Diagnostic Tools. ACS Nano 2021, 15, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ghani, K.; de Campos-Lima, P.O.; Caruso, M. A stable platform for the production of virus-like particles pseudotyped with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike protein. Virus Res. 2021, 295, 198305. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.K.; Rijal, P.; Rahikainen, R.; Keeble, A.H.; Schimanski, L.; Hussain, S.; Harvey, R.; Hayes, J.W.P.; Edwards, J.C.; McLean, R.K.; et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 2021, 12, 542. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, W.; Abiona, O.M.; Nazzari, A.; Olia, A.S.; Ou, L.; Phung, E.; Stephens, T.; Tsybovsky, Y.; Verardi, R.; et al. Newcastle Disease Virus-Like Particles Displaying Prefusion-Stabilized SARS-CoV-2 Spikes Elicit Potent Neutralizing Responses. Vaccines 2021, 9, 73. [Google Scholar] [CrossRef]
- Plotkin, S.; Robinson, J.M.; Cunningham, G.; Iqbal, R.; Larsen, S. The complexity and cost of vaccine manufacturing—An overview. Vaccine 2017, 35, 4064–4071. [Google Scholar] [CrossRef]
- Chu, K.B.; Quan, F.S. Virus-Like Particle Vaccines Against Respiratory Viruses and Protozoan Parasites. Curr. Top. Microbiol Immunol. 2021, 1–30. [Google Scholar] [CrossRef]
- Kang, H.J.; Chu, K.B.; Lee, D.H.; Lee, S.H.; Park, B.R.; Kim, M.C.; Kang, S.M.; Quan, F.S. Influenza M2 virus-like particle vaccination enhances protection in combination with avian influenza HA VLPs. PLoS ONE 2019, 14, e0216871. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.Z.; Quan, F.S.; Kang, S.M.; Bozja, J.; Skountzou, I.; Compans, R.W. Incorporation of membrane-anchored flagellin into influenza virus-like particles enhances the breadth of immune responses. J. Virol. 2008, 82, 11813–11823. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Kang, H.J.; Lee, D.H.; Kang, S.M.; Quan, F.S. Virus-like particle vaccines expressing Toxoplasma gondii rhoptry protein 18 and microneme protein 8 provide enhanced protection. Vaccine 2018, 36, 5692–5700. [Google Scholar] [CrossRef]
- Moon, E.K.; Kang, H.J.; Chu, K.B.; Lee, S.H.; Lee, D.H.; Soh, Y.; Quan, F.S. Immune Correlates of Protection Induced by Virus-Like Particles Containing 2009 H1N1 Pandemic Influenza HA, NA or M1 Proteins. Immunol. Invest. 2019, 48, 355–366. [Google Scholar] [CrossRef]
- Mäkelä, A.R.; Ernst, W.; Grabherr, R.; Oker-Blom, C. Immunofluorescence analysis of baculovirus-displayed viral proteins on infected insect cells. Cold Spring Harb. Protoc. 2010, 2010, pdb-prot5395. [Google Scholar] [CrossRef]
- Chu, K.B.; Lee, H.A.; Kang, H.J.; Moon, E.K.; Quan, F.S. Preliminary Trichinella spiralis Infection Ameliorates Subsequent RSV Infection-Induced Inflammatory Response. Cells 2020, 9, 1314. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef]
- Tessier, D.C.; Thomas, D.Y.; Khouri, H.E.; Laliberté, F.; Vernet, T. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene 1991, 98, 177–183. [Google Scholar] [CrossRef]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef]
- Tang, J.W.; Tambyah, P.A.; Hui, D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2021, 82, e27–e28. [Google Scholar] [CrossRef]
- Krammer, F.; Hai, R.; Yondola, M.; Tan, G.S.; Leyva-Grado, V.H.; Ryder, A.B.; Miller, M.S.; Rose, J.K.; Palese, P.; García-Sastre, A.; et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J. Virol. 2014, 88, 3432–3442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steel, J.; Lowen, A.C.; Wang, T.T.; Yondola, M.; Gao, Q.; Haye, K.; García-Sastre, A.; Palese, P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 2010, 1, e00018-10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margine, I.; Krammer, F.; Hai, R.; Heaton, N.S.; Tan, G.S.; Andrews, S.A.; Runstadler, J.A.; Wilson, P.C.; Albrecht, R.A.; García-Sastre, A.; et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 2013, 87, 10435–10446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roubidoux, E.K.; Carreño, J.M.; McMahon, M.; Jiang, K.; van Bakel, H.; Wilson, P.; Krammer, F. Mutations in the Hemagglutinin Stalk Domain Do Not Permit Escape from a Protective, Stalk-Based Vaccine-Induced Immune Response in the Mouse Model. MBio 2021, 12, e03617-20. [Google Scholar] [CrossRef]
- Gao, T.; Ren, Y.; Li, S.; Lu, X.; Lei, H. Immune response induced by oral administration with a Saccharomyces cerevisiae-based SARS-CoV-2 vaccine in mice. Microb. Cell Fact. 2021, 20, 95. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Mantus, G.; Nyhoff, L.E.; Kauffman, R.C.; Edara, V.V.; Lai, L.; Floyd, K.; Shi, P.Y.; Menachery, V.D.; Edupuganti, S.; Scherer, E.M.; et al. Evaluation of Cellular and Serological Responses to Acute SARS-CoV-2 Infection Demonstrates the Functional Importance of the Receptor-Binding Domain. J. Immunol. 2021, 206, 2605–2613. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.D.; Hay, K.; Rini, J.M.; Yu, M.; Wang, L.; Plummer, F.A.; Corbett, C.R.; Andonov, A. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs 2010, 2, 53–66. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Liu, M.; Wang, A.; Lu, L.; Wang, Q.; Gu, C.; Chen, J.; Wu, Y.; Xia, S.; Ling, Y.; et al. Evaluating the Association of Clinical Characteristics With Neutralizing Antibody Levels in Patients Who Have Recovered From Mild COVID-19 in Shanghai, China. JAMA Intern. Med. 2020, 180, 1356–1362. [Google Scholar] [CrossRef]
- Ding, S.; Laumaea, A.; Benlarbi, M.; Beaudoin-Bussières, G.; Gasser, R.; Medjahed, H.; Pancera, M.; Stamatatos, L.; McGuire, A.T.; Bazin, R.; et al. Antibody Binding to SARS-CoV-2 S Glycoprotein Correlates with but Does Not Predict Neutralization. Viruses 2020, 12, 1214. [Google Scholar] [CrossRef]
- Anderson, E.M.; Goodwin, E.C.; Verma, A.; Arevalo, C.P.; Bolton, M.J.; Weirick, M.E.; Gouma, S.; McAllister, C.M.; Christensen, S.R.; Weaver, J.; et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell 2021, 184, 1858–1864. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Haynes, L.; Struble, E.B.; Tamin, A.; Virata-Theimer, M.L.; Zhang, P. Antibody-mediated synergy and interference in the neutralization of SARS-CoV at an epitope cluster on the spike protein. Biochem. Biophys. Res. Commun. 2009, 390, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xiao, Y.; Lu, R.; Chen, B.; Liu, F.; Wang, L.; Yao, H.; Wu, N.; Wu, H. Generation of neutralizing and non-neutralizing monoclonal antibodies against H7N9 influenza virus. Emerg. Microbes Infect. 2020, 9, 664–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asthagiri Arunkumar, G.; Ioannou, A.; Wohlbold, T.J.; Meade, P.; Aslam, S.; Amanat, F.; Ayllon, J.; García-Sastre, A.; Krammer, F. Broadly Cross-Reactive, Nonneutralizing Antibodies against Influenza B Virus Hemagglutinin Demonstrate Effector Function-Dependent Protection against Lethal Viral Challenge in Mice. J. Virol. 2019, 93, e01696-18. [Google Scholar] [CrossRef] [Green Version]
- Martinez, M.E.; Capella Gonzalez, C.; Huey, D.; Peeples, M.E.; McCarty, D.; Niewiesk, S. Immunogenicity and inflammatory properties of respiratory syncytial virus attachment G protein in cotton rats. PLoS ONE 2021, 16, e0246770. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.A.; Gramespacher, J.A.; Pance, K.; Rettko, N.J.; Solomon, P.; Jin, J.; Lui, I.; Elledge, S.K.; Liu, J.; Bracken, C.J.; et al. Bispecific VH/Fab antibodies targeting neutralizing and non-neutralizing Spike epitopes demonstrate enhanced potency against SARS-CoV-2. MAbs 2021, 13, 1893426. [Google Scholar] [CrossRef]
- Fang, J.; Hu, B.; Li, S.; Zhang, C.; Liu, Y.; Wang, P. A multi-antigen vaccine in combination with an immunotoxin targeting tumor-associated fibroblast for treating murine melanoma. Mol. Ther. Oncolytics 2016, 3, 16007. [Google Scholar] [CrossRef] [Green Version]
- Anderson, A.M.; Baranowska-Hustad, M.; Braathen, R.; Grodeland, G.; Bogen, B. Simultaneous Targeting of Multiple Hemagglutinins to APCs for Induction of Broad Immunity against Influenza. J. Immunol. 2018, 200, 2057–2066. [Google Scholar] [CrossRef] [Green Version]
- Mansour, A.A.; Banik, S.; Suresh, R.V.; Kaur, H.; Malik, M.; McCormick, A.A.; Bakshi, C.S. An Improved Tobacco Mosaic Virus (TMV)-Conjugated Multiantigen Subunit Vaccine Against Respiratory Tularemia. Front. Microbiol. 2018, 9, 1195. [Google Scholar] [CrossRef]
- Johansen, M.D.; Irving, A.; Montagutelli, X.; Tate, M.D.; Rudloff, I.; Nold, M.F.; Hansbro, N.G.; Kim, R.Y.; Donovan, C.; Liu, G.; et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020, 13, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.I.; Kim, S.G.; Kim, S.M.; Kim, E.H.; Park, S.J.; Yu, K.M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709. [Google Scholar] [CrossRef] [PubMed]

| Optimized 1 | ATGTTCGTTTTCCTCGTGCTCCTCCCCCTCGTTTCCTCCCAATGCGTCAACCTCACTACC |
| Original 1 | ATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTTAATCTTACAACC |
| Optimized 61 | CGTACCCAGCTCCCACCAGCCTACACCAACAGCTTCACTCGCGGTGTGTACTACCCCGAC |
| Original 61 | AGAACTCAATTACCCCCTGCATACACTAATTCTTTCACACGTGGTGTTTATTACCCTGAC |
| Optimized 121 | AAGGTCTTCCGTTCCAGCGTGCTGCACAGCACTCAGGACCTGTTCCTGCCATTCTTCTCT |
| Original 121 | AAAGTTTTCAGATCCTCAGTTTTACATTCAACTCAGGACTTGTTCTTACCTTTCTTTTCC |
| Optimized 181 | AACGTGACCTGGTTCCACGCCATCCACGTGAGCGGCACCAACGGCACTAAGCGCTTCGAC |
| Original 181 | AATGTTACTTGGTTCCATGCTATACATGTCTCTGGGACCAATGGTACTAAGAGGTTTGAT |
| Optimized 241 | AACCCTGTGCTGCCCTTCAACGACGGTGTCTACTTCGCTTCCACCGAGAAGTCTAACATC |
| Original 241 | AACCCTGTCCTACCATTTAATGATGGTGTTTATTTTGCTTCCACTGAGAAGTCTAACATA |
| Optimized 301 | ATCCGTGGATGGATCTTCGGCACCACTCTGGACTCAAAGACTCAGTCCCTGCTGATCGTC |
| Original 301 | ATAAGAGGCTGGATTTTTGGTACTACTTTAGATTCGAAGACCCAGTCCCTACTTATTGTT |
| Optimized 361 | AACAACGCCACCAACGTGGTCATCAAGGTGTGCGAGTTCCAGTTCTGCAACGACCCTTTC |
| Original 361 | AATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGTAATGATCCATTT |
| Optimized 421 | CTGGGCGTCTACTACCACAAGAACAACAAGAGCTGGATGGAGTCTGAGTTCCGCGTCTAC |
| Original 421 | TTGGGTGTTTATTACCACAAAAACAACAAAAGTTGGATGGAAAGTGAGTTCAGAGTTTAT |
| Optimized 481 | TCTTCAGCTAACAACTGCACTTTCGAGTACGTGAGCCAGCCCTTCCTGATGGACCTGGAA |
| Original 481 | TCTAGTGCGAATAATTGCACTTTTGAATATGTCTCTCAGCCTTTTCTTATGGACCTTGAA |
| Optimized 541 | GGAAAGCAGGGTAACTTCAAGAACCTGAGGGAGTTCGTGTTCAAGAACATCGACGGATAC |
| Original 541 | GGAAAACAGGGTAATTTCAAAAATCTTAGGGAATTTGTGTTTAAGAATATTGATGGTTAT |
| Optimized 601 | TTCAAGATTTACTCAAAGCACACCCCTATCAACCTGGTGCGCGACCTGCCACAGGGTTTC |
| Original 601 | TTTAAAATATATTCTAAGCACACGCCTATTAATTTAGTGCGTGATCTCCCTCAGGGTTTT |
| Optimized 661 | TCCGCTCTGGAGCCTCTGGTGGACCTGCCCATCGGCATCAACATCACCCGCTTCCAGACT |
| Original 661 | TCGGCTTTAGAACCATTGGTAGATTTGCCAATAGGTATTAACATCACTAGGTTTCAAACT |
| Optimized 721 | CTGCTGGCTCTGCACCGTTCCTACCTGACTCCTGGCGACTCCAGCTCTGGATGGACCGCC |
| Original 721 | TTACTTGCTTTACATAGAAGTTATTTGACTCCTGGTGATTCTTCTTCAGGTTGGACAGCT |
| Optimized 781 | GGAGCTGCCGCTTACTACGTGGGTTACCTGCAACCCAGGACCTTCCTGCTGAAGTACAAC |
| Original 781 | GGTGCTGCAGCTTATTATGTGGGTTATCTTCAACCTAGGACTTTTCTATTAAAATATAAT |
| Optimized 841 | GAAAACGGAACCATCACAGACGCTGTGGACTGCGCTCTGGACCCCCTGAGCGAAACCAAG |
| Original 841 | GAAAATGGAACCATTACAGATGCTGTAGACTGTGCACTTGACCCTCTCTCAGAAACAAAG |
| Optimized 901 | TGCACTCTGAAGTCTTTCACCGTGGAGAAGGGCATCTACCAGACTAGCAACTTCAGGGTG |
| Original 901 | TGTACGTTGAAATCCTTCACTGTAGAAAAAGGAATCTATCAAACTTCTAACTTTAGAGTC |
| Optimized 961 | CAGCCAACCGAATCTATCGTCAGATTCCCCAACATCACTAACCTGTGCCCATTCGGAGAG |
| Original 961 | CAACCAACAGAATCTATTGTTAGATTTCCTAATATTACAAACTTGTGCCCTTTTGGTGAA |
| Optimized 1021 | GTCTTCAACGCCACCAGATTCGCTTCCGTGTACGCCTGGAACAGGAAGAGAATCAGCAAC |
| Original 1021 | GTTTTTAACGCCACCAGATTTGCATCTGTTTATGCTTGGAACAGGAAGAGAATCAGCAAC |
| Optimized 1081 | TGCGTCGCTGACTACTCTGTGCTGTACAACAGCGCCTCTTTCTCAACCTTCAAGTGCTAC |
| Original 1081 | TGTGTTGCTGATTATTCTGTCCTATATAATTCCGCATCATTTTCCACTTTTAAGTGTTAT |
| Optimized 1141 | GGCGTGAGCCCTACTAAGCTGAACGACCTGTGCTTCACCAACGTCTACGCCGACTCTTTC |
| Original 1141 | GGAGTGTCTCCTACTAAATTAAATGATCTCTGCTTTACTAATGTCTATGCAGATTCATTT |
| Optimized 1201 | GTGATCAGGGGAGACGAGGTCAGACAGATCGCTCCCGGCCAGACTGGAAAGATCGCCGAC |
| Original 1201 | GGAGTGTCTCCTACTAAATTAAATGATCTCTGCTTTACTAATGTCTATGCAGATTCATTT |
| Optimized 1261 | TACAACTACAAGCTGCCAGACGACTTCACCGGCTGCGTCATCGCTTGGAACTCAAACAAC |
| Original 1261 | TATAATTATAAATTACCAGATGATTTTACAGGCTGCGTTATAGCTTGGAATTCTAACAAT |
| Optimized 1321 | CTGGACTCCAAAGTGGGTGGCAACTACAACTACCTGTACCGCCTGTTCCGTAAGAGCAAC |
| Original 1321 | CTTGATTCTAAGGTTGGTGGTAATTATAATTACCTGTATAGATTGTTTAGGAAGTCTAAT |
| Optimized 1381 | CTGAAGCCTTTCGAGAGGGACATCTCAACTGAAATCTACCAGGCTGGTTCCACCCCCTGC |
| Original 1381 | CTCAAACCTTTTGAGAGAGATATTTCAACTGAAATCTATCAGGCCGGTAGCACACCTTGT |
| Optimized 1441 | AACGGTGTCGAGGGCTTCAACTGCTACTTCCCACTGCAATCTTACGGTTTCCAGCCTACT |
| Original 1441 | AATGGTGTTGAAGGTTTTAATTGTTACTTTCCTTTACAATCATATGGTTTCCAACCCACT |
| Optimized 1501 | AACGGTGTGGGCTACCAGCCCTACAGAGTGGTCGTGCTGTCATTCGAACTGCTGCACGCC |
| Original 1501 | AATGGTGTTGGTTACCAACCATACAGAGTAGTAGTACTTTCTTTTGAACTTCTACATGCA |
| Optimized 1561 | CCAGCTACTGTGTGCGGTCCTAAGAAGTCCACCAACCTGGTCAAGAACAAGTGCGTGAAC |
| Original 1561 | CCAGCAACTGTTTGTGGACCTAAAAAGTCTACTAATTTGGTTAAAAACAAATGTGTCAAT |
| Optimized 1621 | TTCAACTTCAACGGCCTGACCGGAACTGGTGTCCTGACCGAGTCAAACAAGAAGTTCCTG |
| Original 1621 | TTCAACTTCAATGGTTTAACAGGCACAGGTGTTCTTACTGAGTCTAACAAAAAGTTTCTG |
| Optimized 1681 | CCATTCCAGCAGTTCGGAAGGGACATCGCTGACACCACTGACGCTGTGCGCGACCCTCAG |
| Original 1681 | CCTTTCCAACAATTTGGCAGAGACATTGCTGACACTACTGATGCTGTCCGTGATCCACAG |
| Optimized 1741 | ACCCTGGAAATCCTGGACATCACTCCTTGCAGCTTCGGAGGTGTCTCTGTGATCACCCCT |
| Original 1741 | ACACTTGAGATTCTTGACATTACACCATGTTCTTTTGGTGGTGTCAGTGTTATAACACCA |
| Optimized 1801 | GGCACCAACACTTCCAACCAGGTCGCTGTGCTGTACCAGGACGTCAACTGCACCGAGGTG |
| Original 1801 | GGAACAAATACTTCTAACCAGGTTGCTGTTCTTTATCAGGATGTTAACTGCACAGAAGTC |
| Optimized 1861 | CCTGTGGCTATCCACGCTGACCAGCTGACCCCAACTTGGCGCGTGTACTCCACCGGCTCC |
| Original 1861 | CCTGTTGCTATTCATGCAGATCAACTTACTCCTACTTGGCGTGTTTATTCTACAGGTTCT |
| Optimized 1921 | AACGTCTTCCAGACTCGTGCTGGTTGCCTGATCGGCGCCGAGCACGTGAACAACTCATAC |
| Original 1921 | AATGTTTTTCAAACACGTGCAGGCTGTTTAATAGGGGCTGAACATGTCAACAACTCATAT |
| Optimized 1981 | GAATGCGACATCCCAATCGGCGCTGGAATCTGCGCCTCCTACCAGACCCAGACTAACTCA |
| Original 1981 | GAGTGTGACATACCCATTGGTGCAGGTATATGCGCTAGTTATCAGACTCAGACTAATTCT |
| Optimized 2041 | CCTCGCCGTGCTCGCTCCGTCGCCTCCCAGAGCATCATCGCTTACACCATGAGCCTGGGC |
| Original 2041 | CCTCGGCGGGCACGTAGTGTAGCTAGTCAATCCATCATTGCCTACACTATGTCACTTGGT |
| Optimized 2101 | GCTGAAAACTCTGTGGCCTACTCCAACAACAGCATCGCCATCCCAACCAACTTCACTATC |
| Original 2101 | GCAGAAAATTCAGTTGCTTACTCTAATAACTCTATTGCCATACCCACAAATTTTACTATT |
| Optimized 2161 | TCAGTGACCACTGAGATCCTGCCTGTCTCAATGACCAAGACTTCCGTGGACTGCACTATG |
| Original 2161 | AGTGTTACCACAGAAATTCTACCAGTGTCTATGACCAAGACATCAGTAGATTGTACAATG |
| Optimized 2221 | TACATCTGCGGAGACTCAACCGAATGCTCCAACCTGCTGCTGCAATACGGCTCCTTCTGC |
| Original 2221 | TACATTTGTGGTGATTCAACTGAATGCAGCAATCTTTTGTTGCAATATGGCAGTTTTTGT |
| Optimized 2281 | ACCCAGCTGAACCGTGCTCTGACTGGAATCGCCGTGGAGCAGGACAAGAACACTCAGGAA |
| Original 2281 | ACACAATTAAACCGTGCTTTAACTGGAATAGCTGTTGAACAAGACAAAAACACCCAAGAA |
| Optimized 2341 | GTCTTCGCTCAGGTGAAGCAAATCTACAAGACCCCTCCCATCAAGGACTTCGGCGGATTC |
| Original 2341 | GTTTTTGCACAAGTCAAACAAATTTACAAAACACCACCAATTAAAGATTTTGGTGGTTTT |
| Optimized 2401 | AACTTCTCCCAGATCCTGCCCGACCCATCTAAGCCTTCAAAGCGCTCCTTCATCGAGGAC |
| Original 2401 | AATTTTTCACAAATATTACCAGATCCATCAAAACCAAGCAAGAGGTCATTTATTGAAGAT |
| Optimized 2461 | CTGCTGTTCAACAAGGTCACCCTGGCCGACGCTGGATTCATCAAGCAGTACGGAGACTGC |
| Original 2461 | CTACTTTTCAACAAAGTGACACTTGCAGATGCTGGCTTCATCAAACAATATGGTGATTGC |
| Optimized 2521 | CTGGGTGACATCGCCGCTCGTGACCTGATCTGCGCTCAGAAGTTCAACGGTCTGACCGTG |
| Original 2521 | CTTGGTGATATTGCTGCTAGAGACCTCATTTGTGCACAAAAGTTTAACGGCCTTACTGTT |
| Optimized 2581 | CTGCCACCTCTGCTGACTGACGAAATGATCGCCCAGTACACTTCAGCCCTGCTGGCTGGA |
| Original 2581 | TTGCCACCTTTGCTCACAGATGAAATGATTGCTCAATACACTTCTGCACTGTTAGCGGGT |
| Optimized 2641 | ACCATCACTTCCGGTTGGACCTTCGGTGCTGGTGCTGCTCTGCAAATCCCCTTCGCTATG |
| Original 2641 | ACAATCACTTCTGGTTGGACCTTTGGTGCAGGTGCTGCATTACAAATACCATTTGCTATG |
| Optimized 2701 | CAGATGGCCTACAGGTTCAACGGAATCGGTGTCACCCAGAACGTGCTGTACGAGAACCAG |
| Original 2701 | |
| Optimized 2761 | CAAATGGCTTATAGGTTTAATGGTATTGGAGTTACACAGAATGTTCTCTATGAGAACCAA |
| Original 2761 | AAGCTGATCGCTAACCAGTTCAACAGCGCCATCGGAAAGATCCAGGACTCACTGTCATCC |
| Optimized 2821 | AAATTGATTGCCAACCAATTTAATAGTGCTATTGGCAAAATTCAAGACTCACTTTCTTCC |
| Original 2821 | ACTGCCTCCGCTCTGGGCAAGCTGCAAGACGTCGTGAACCAGAACGCCCAGGCTCTGAAC |
| Optimized 2881 | ACAGCAAGTGCACTTGGAAAACTTCAAGATGTGGTCAACCAAAATGCACAAGCTTTAAAC |
| Original 2881 | ACCCTGGTCAAGCAGCTGTCCTCCAACTTCGGTGCTATCTCATCCGTGCTGAACGACATC |
| Optimized 2941 | ACGCTTGTTAAACAACTTAGCTCCAATTTTGGTGCAATTTCAAGTGTTTTAAATGATATC |
| Original 2941 | CTGTCTCGCCTGGACAAGGTCGAGGCCGAAGTGCAGATCGACCGCCTGATCACCGGCCGC |
| Optimized 3001 | CTTTCACGTCTTGACAAAGTTGAGGCTGAAGTGCAAATTGATAGGTTGATCACAGGCAGA |
| Original 3001 | CTGCAATCCCTGCAAACCTACGTGACTCAGCAGCTGATCAGGGCCGCTGAAATCAGAGCC |
| Optimized 3061 | CTTCAAAGTTTGCAGACATATGTGACTCAACAATTAATTAGAGCTGCAGAAATCAGAGCT |
| Original 3061 | TCTGCTAACCTGGCCGCTACCAAGATGTCAGAGTGCGTCCTGGGTCAGTCCAAGCGTGTG |
| Optimized 3121 | TCTGCTAATCTTGCTGCTACTAAAATGTCAGAGTGTGTACTTGGACAATCAAAAAGAGTT |
| Original 3121 | GACTTCTGCGGCAAGGGATACCACCTGATGAGCTTCCCCCAGTCTGCTCCACACGGCGTC |
| Optimized 3181 | GATTTTTGTGGAAAGGGCTATCATCTTATGTCCTTCCCTCAGTCAGCACCTCATGGTGTA |
| Original 3181 | GTGTTCCTGCACGTCACCTACGTGCCTGCCCAGGAGAAGAACTTCACCACTGCCCCCGCT |
| Optimized 3241 | GTCTTCTTGCATGTGACTTATGTCCCTGCACAAGAAAAGAACTTCACAACTGCTCCTGCC |
| Original 3241 | ATCTGCCACGACGGCAAGGCTCACTTCCCAAGGGAAGGTGTCTTCGTGTCAAACGGCACC |
| Optimized 3301 | ATTTGTCATGATGGAAAAGCACACTTTCCTCGTGAAGGTGTCTTTGTTTCAAATGGCACA |
| Original 3301 | CACTGGTTCGTCACTCAGAGAAACTTCTACGAGCCTCAGATCATCACCACTGACAACACT |
| Optimized 3361 | CACTGGTTTGTAACACAAAGGAATTTTTATGAACCACAAATCATTACTACAGACAACACA |
| Original 3361 | TTCGTGTCCGGAAACTGCGACGTCGTGATCGGTATCGTCAACAACACCGTGTACGACCCA |
| Optimized 3421 | TTTGTGTCTGGTAACTGTGATGTTGTAATAGGAATTGTCAACAACACAGTTTATGATCCT |
| Original 3421 | CTGCAACCTGAGCTGGACAGCTTCAAGGAGGAACTGGACAAATACTTCAAGAACCACACC |
| Optimized 3481 | TTGCAACCTGAATTAGACTCATTCAAGGAGGAGTTAGATAAATATTTTAAGAATCATACA |
| Original 3481 | TCTCCCGACGTGGACCTGGGTGACATCAGCGGAATCAACGCTTCTGTCGTGAACATCCAG |
| Optimized 3541 | TCACCAGATGTTGATTTAGGTGACATCTCTGGCATTAATGCTTCAGTTGTAAACATTCAA |
| Original 3541 | AAGGAGATCGACCGTCTGAACGAAGTGGCTAAGAACCTGAACGAATCCCTGATCGACCTG |
| Optimized 3601 | AAAGAAATTGACCGCCTCAATGAGGTTGCCAAGAATTTAAATGAATCTCTCATCGATCTC |
| Original 3601 | CAAGAGCTGGGCAAGTACGAACAGTACATCAAGTGGCCTTGGTACATCTGGCTGGGTTTC |
| Optimized 3661 | CAAGAACTTGGAAAGTATGAGCAGTATATAAAATGGCCATGGTACATTTGGCTAGGTTTT |
| Original 3661 | ATCGCTGGCCTGATCGCCATCGTCATGGTGACCATCATGCTGTGCTGCATGACTAGCTGC |
| Optimized 3721 | ATAGCTGGCTTGATTGCCATAGTAATGGTGACAATTATGCTTTGCTGTATGACCAGTTGC |
| Original 3721 | TGCTCTTGCCTGAAGGGCTGCTGCTCATGCGGTTCCTGCTGCAAGTTCGATGAAGACGAT |
| Optimized 3781 | TGTAGTTGTCTCAAGGGCTGTTGTTCTTGTGGATCCTGCTGCAAATTTGATGAAGACGAC |
| Original 3781 | TCCGAGCCCGTTCTCAAAGGAGTGAAGTTGCATTACACATAA |
| TCTGAGCCAGTGCTCAAAGGAGTCAAATTACATTACACATAA | |
| S1 HA-TM-CT | CAGATCCTGGCTATCTACTCTACTGTGGCCTCCAGCCTGGTGCTGCTGGTCTCCCTGGGTGCTATCTCTTTCTGGATGTGCTCTAACGGCTCACTGCAGTGCCGCATCTGCATCTAA |
| S2 SP | ATGAAGTTCCTGGTGAACGTCGCCCTGGTGTTCATGGTGGTCTACATCTCCTACATCTACGCTGCCGCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, K.-B.; Kang, H.-J.; Yoon, K.-W.; Lee, H.-A.; Moon, E.-K.; Han, B.-K.; Quan, F.-S. Influenza Virus-like Particle (VLP) Vaccines Expressing the SARS-CoV-2 S Glycoprotein, S1, or S2 Domains. Vaccines 2021, 9, 920. https://doi.org/10.3390/vaccines9080920
Chu K-B, Kang H-J, Yoon K-W, Lee H-A, Moon E-K, Han B-K, Quan F-S. Influenza Virus-like Particle (VLP) Vaccines Expressing the SARS-CoV-2 S Glycoprotein, S1, or S2 Domains. Vaccines. 2021; 9(8):920. https://doi.org/10.3390/vaccines9080920
Chicago/Turabian StyleChu, Ki-Back, Hae-Ji Kang, Keon-Woong Yoon, Hae-Ahm Lee, Eun-Kyung Moon, Beom-Ku Han, and Fu-Shi Quan. 2021. "Influenza Virus-like Particle (VLP) Vaccines Expressing the SARS-CoV-2 S Glycoprotein, S1, or S2 Domains" Vaccines 9, no. 8: 920. https://doi.org/10.3390/vaccines9080920
APA StyleChu, K.-B., Kang, H.-J., Yoon, K.-W., Lee, H.-A., Moon, E.-K., Han, B.-K., & Quan, F.-S. (2021). Influenza Virus-like Particle (VLP) Vaccines Expressing the SARS-CoV-2 S Glycoprotein, S1, or S2 Domains. Vaccines, 9(8), 920. https://doi.org/10.3390/vaccines9080920






