Concomitant Swine Influenza A Virus Infection Alters PRRSV1 MLV Viremia in Piglets but Does Not Interfere with Vaccine Protection in Experimental Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Vaccines
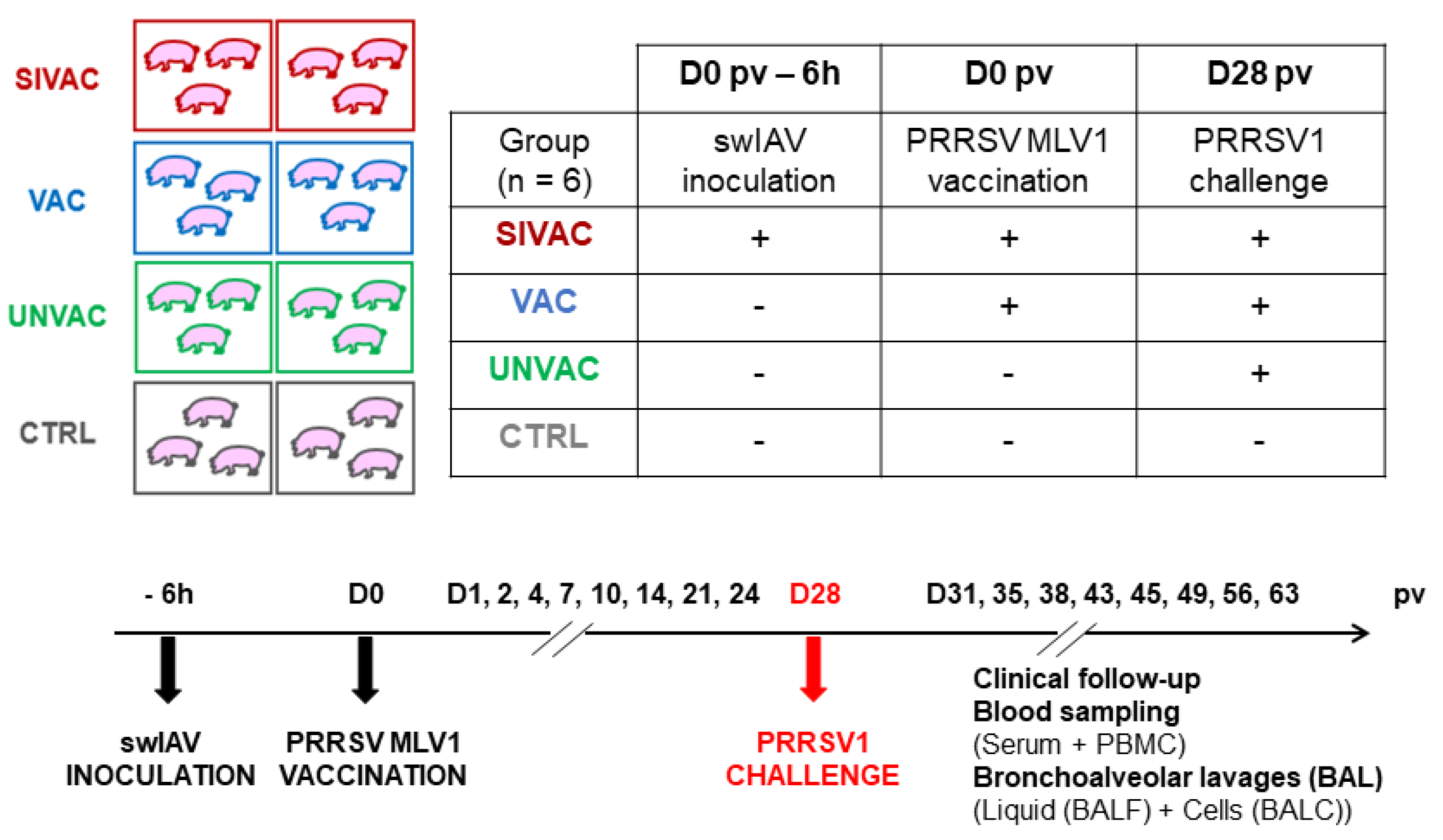
2.2. Animal Experiment
2.3. Viral Quantifications
2.4. Antibody Measurements
2.5. Cell-Mediated Immune Response (CMI) Assessment
2.6. Cytokine Measurements
2.7. Statistical Analyses
3. Results
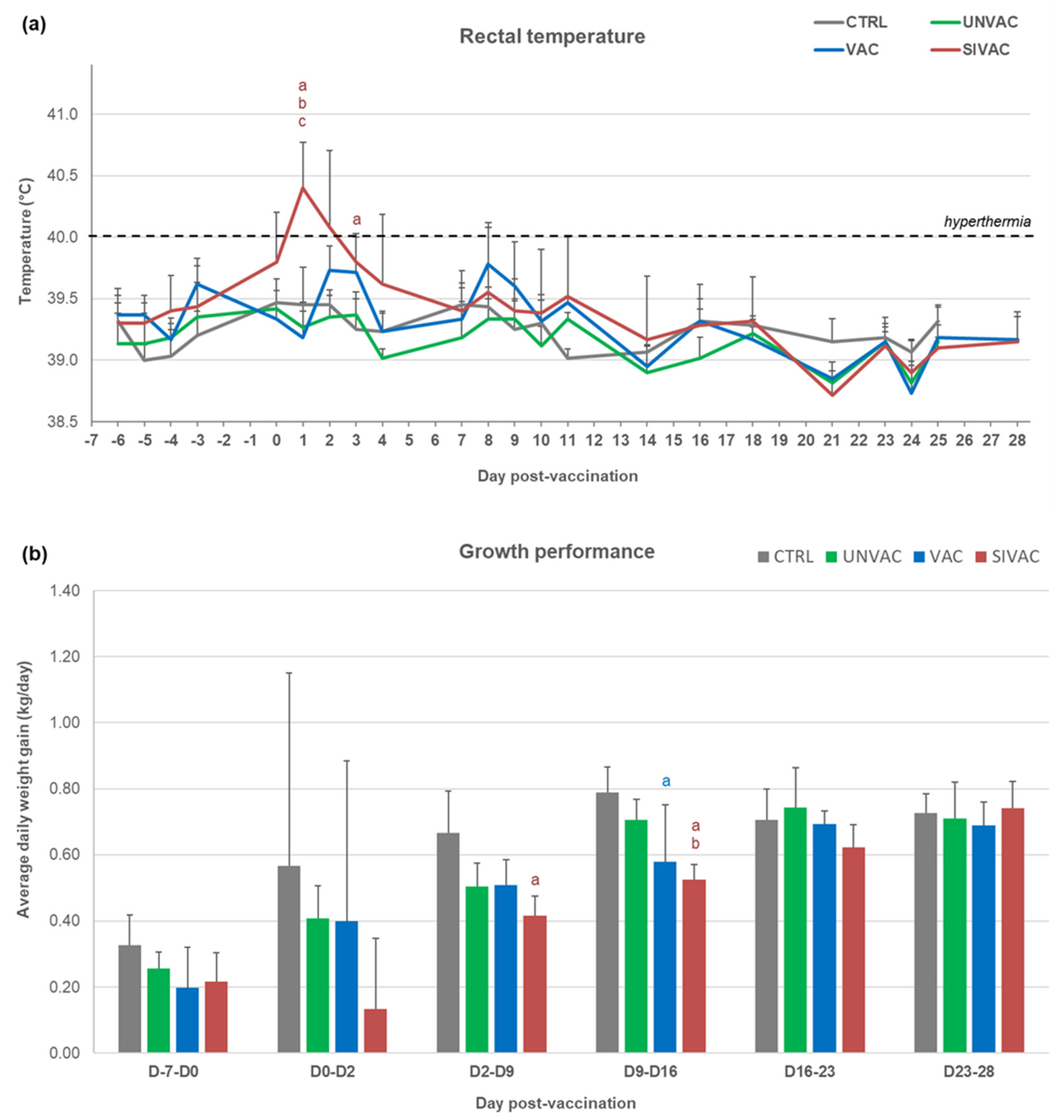
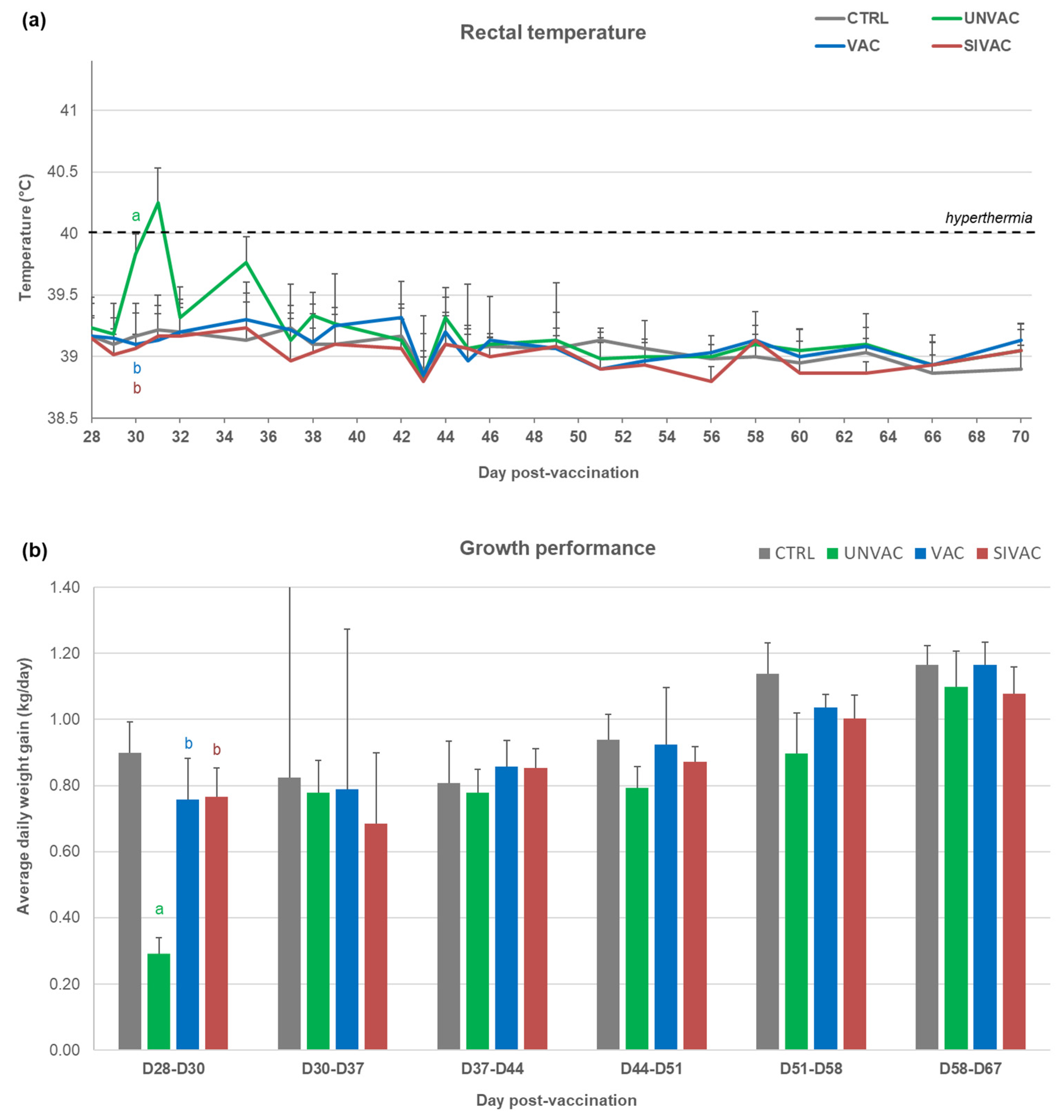
3.1. Clinical Follow-Up during the Post-Vaccination Phase
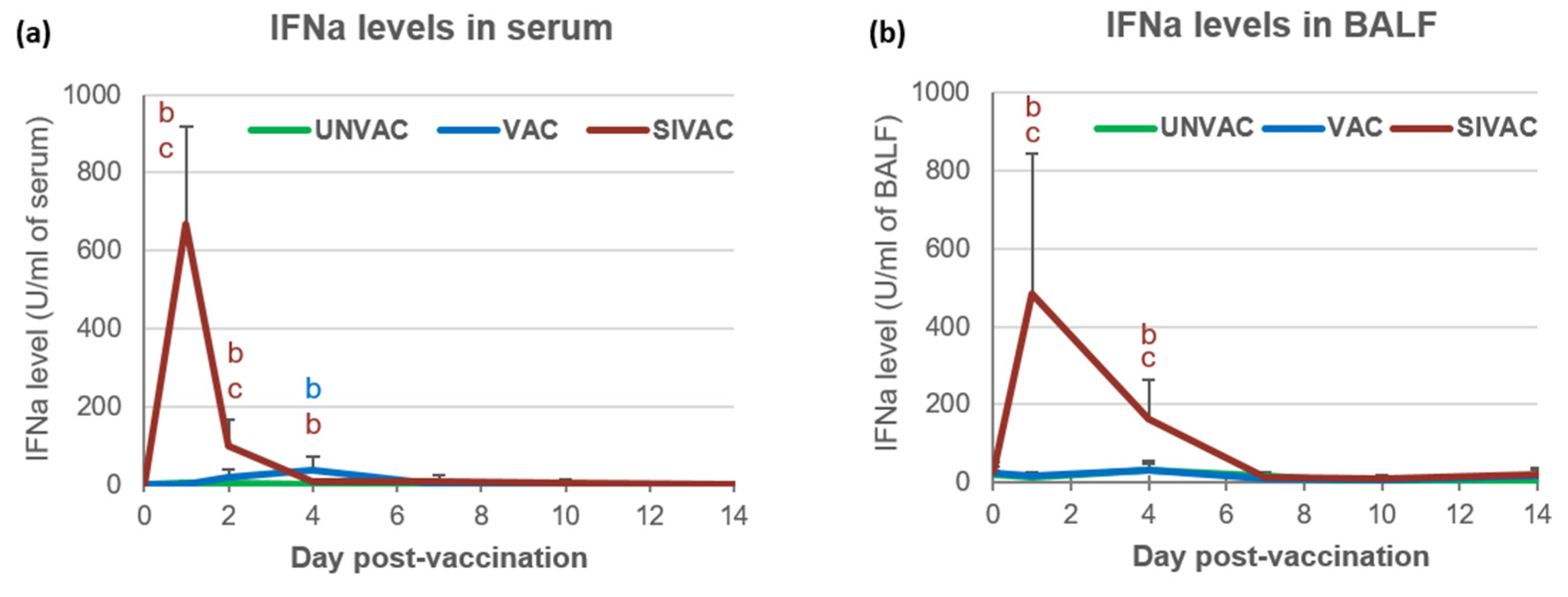
3.2. SwIAV Infection Assessment in the Post-Vaccination Period
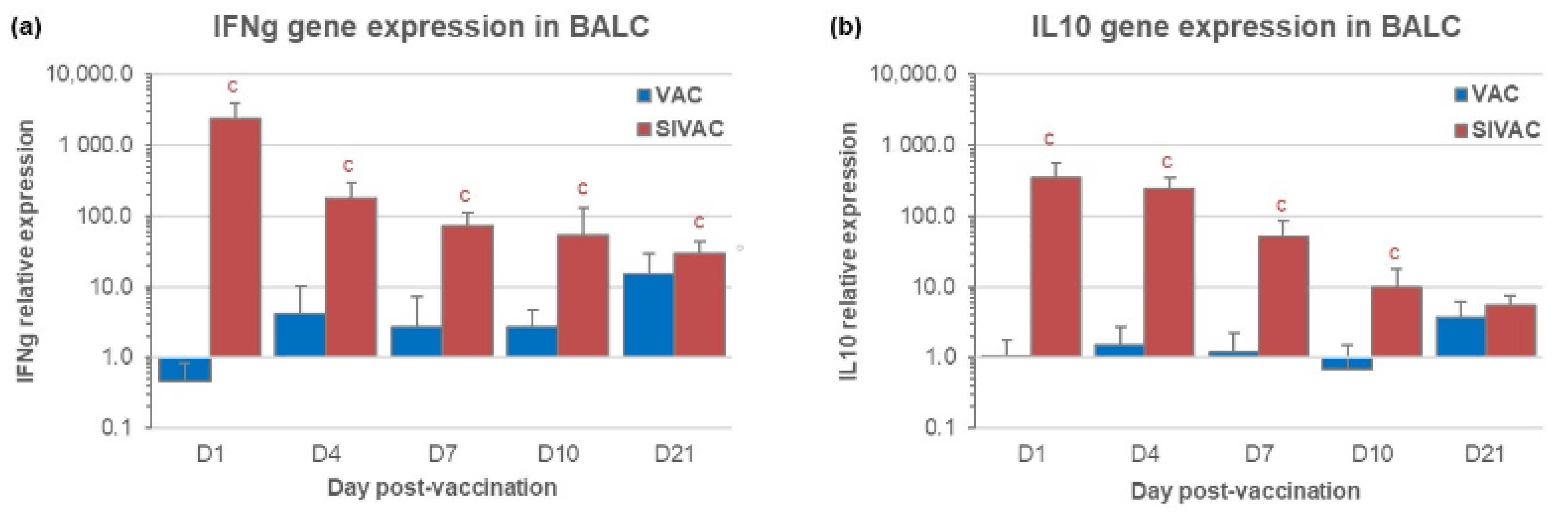
3.3. IFNg and IL10 Gene Expression in BALCs in the Post-Vaccination Period
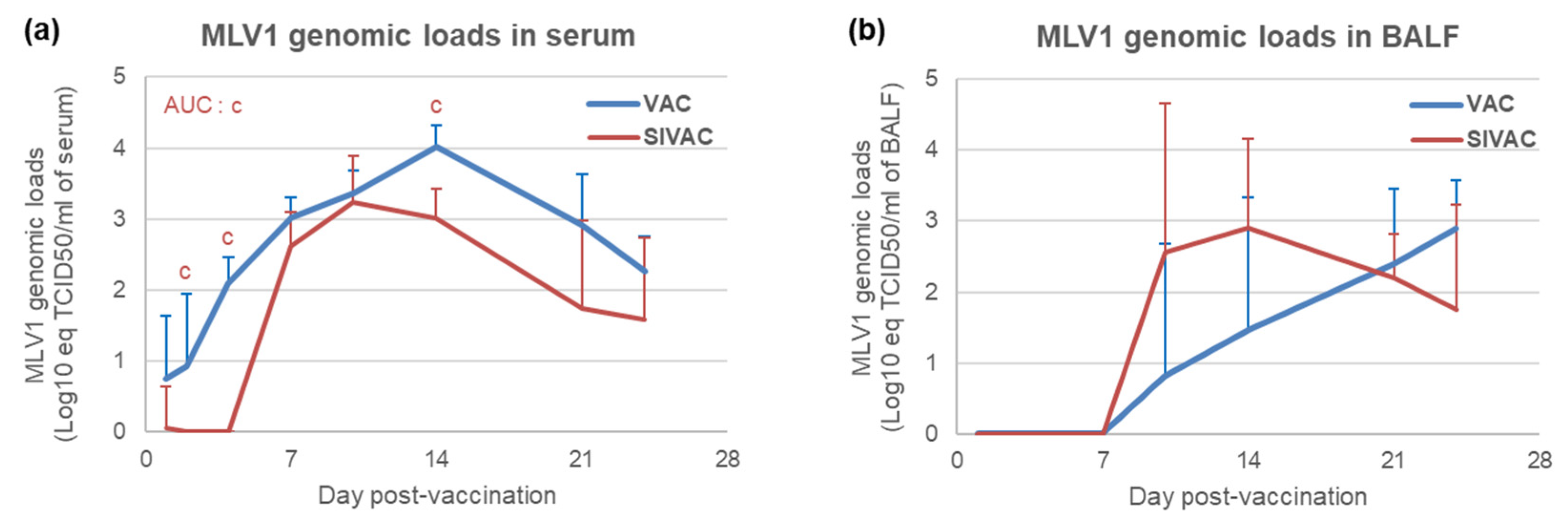
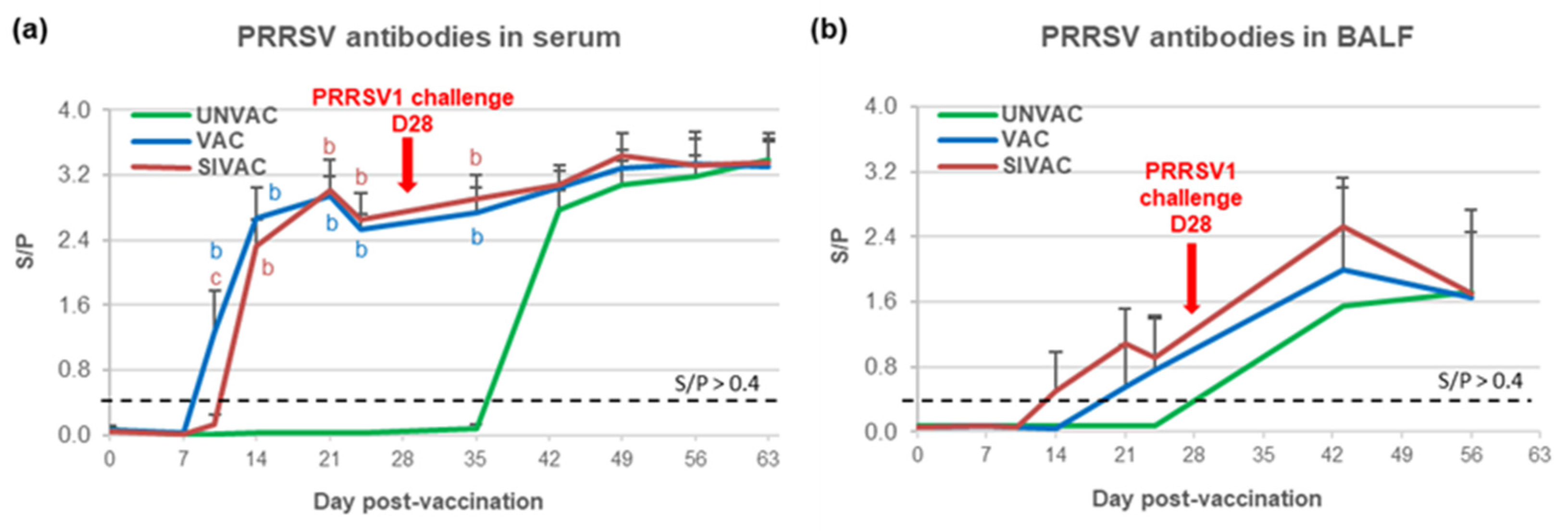
3.4. MLV1 Viremia and the PRRSV Immune Response Assessment during the Post-Vaccination Period
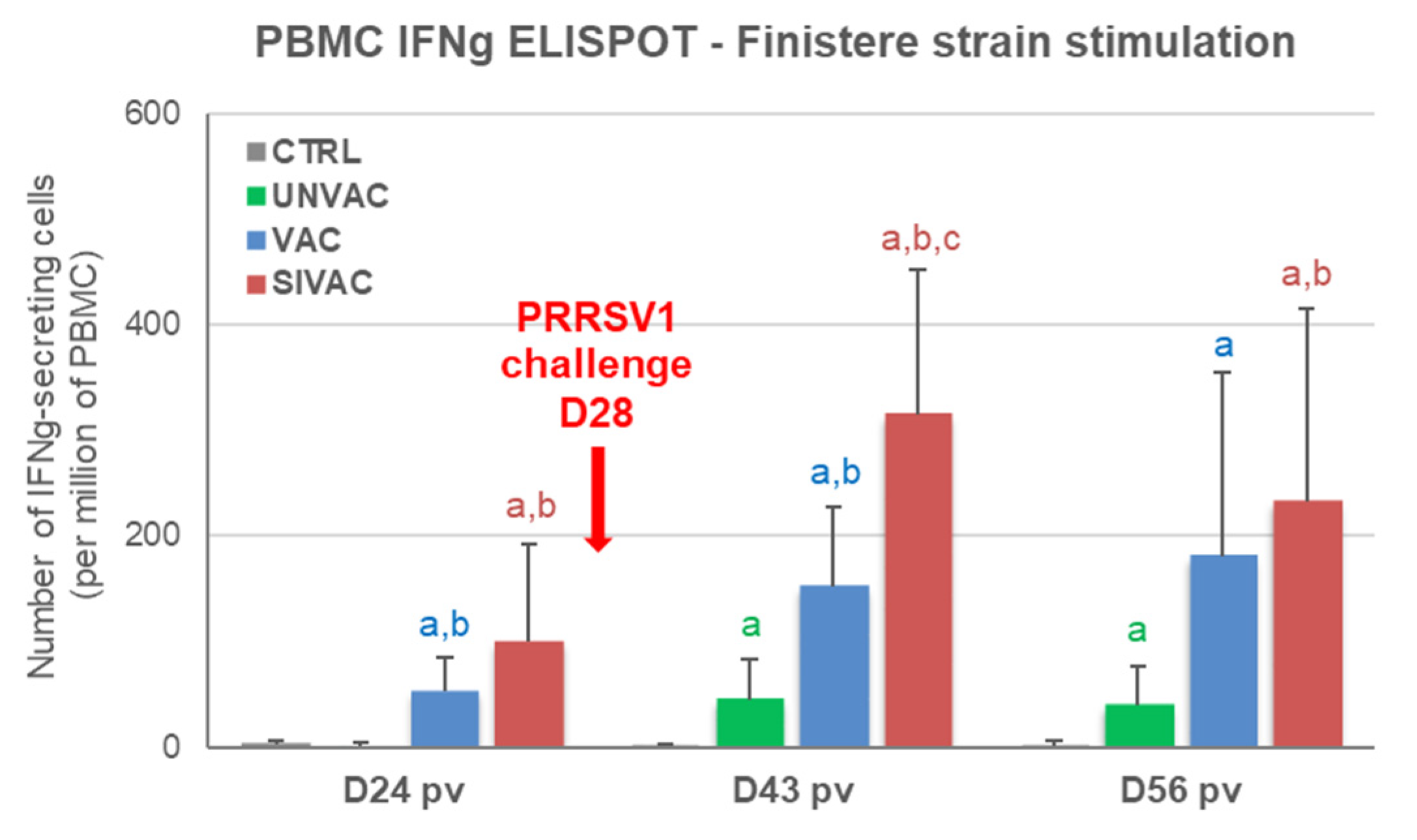
3.5. Evaluation of MLV1 Vaccine Efficacy after the PRRSV1 Challenge
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lunney, J.K.; Benfield, D.A.; Rowland, R.R. Porcine reproductive and respiratory syndrome virus: An update on an emerging and re-emerging viral disease of swine. Virus Res. 2010, 154, 1–6. [Google Scholar] [CrossRef]
- Nathues, H.; Alarcon, P.; Rushton, J.; Jolie, R.; Fiebig, K.; Jimenez, M.; Geurts, V.; Nathues, C. Cost of porcine reproductive and respiratory syndrome virus at individual farm level-An economic disease model. Prev. Vet. Med. 2017, 142, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Lauck, M.; Bailey, A.L.; Shchetinin, A.M.; Vishnevskaya, T.V.; Bao, Y.; Ng, T.F.; LeBreton, M.; Schneider, B.S.; Gillis, A.; et al. Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 2016, 161, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Siddell, S.G.; Walker, P.J.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; et al. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch. Virol. 2019, 164, 943–946. [Google Scholar] [CrossRef] [PubMed]
- Martelli, P.; Gozio, S.; Ferrari, L.; Rosina, S.; De Angelis, E.; Quintavalla, C.; Bottarelli, E.; Borghetti, P. Efficacy of a modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine in pigs naturally exposed to a heterologous European (Italian cluster) field strain: Clinical protection and cell-mediated immunity. Vaccine 2009, 27, 3788–3799. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.; Renson, P.; Andraud, M.; Paboeuf, F.; Le Potier, M.F.; Bourry, O. Porcine reproductive and respiratory syndrome virus (PRRSv) modified-live vaccine reduces virus transmission in experimental conditions. Vaccine 2015, 33, 2493–2499. [Google Scholar] [CrossRef]
- Fablet, C.; Renson, P.; Eono, F.; Mahe, S.; Eveno, E.; Le Dimna, M.; Normand, V.; Lebret, A.; Rose, N.; Bourry, O. Maternally-derived antibodies (MDAs) impair piglets’ humoral and cellular immune responses to vaccination against porcine reproductive and respiratory syndrome (PRRS). Vet. Microbiol. 2016, 192, 175–180. [Google Scholar] [CrossRef]
- Renson, P.; Fablet, C.; Andraud, M.; Normand, V.; Lebret, A.; Paboeuf, F.; Rose, N.; Bourry, O. Maternally-derived neutralizing antibodies reduce vaccine efficacy against porcine reproductive and respiratory syndrome virus infection. Vaccine 2019, 37, 4318–4324. [Google Scholar] [CrossRef] [PubMed]
- Charley, B.; Riffault, S.; Van Reeth, K. Porcine innate and adaptative immune responses to influenza and coronavirus infections. Ann. N. Y. Acad. Sci. 2006, 1081, 130–136. [Google Scholar] [CrossRef]
- Chepngeno, J.; Takanashi, S.; Diaz, A.; Michael, H.; Paim, F.C.; Rahe, M.C.; Hayes, J.R.; Baker, C.; Marthaler, D.; Saif, L.J.; et al. Comparative Sequence Analysis of Historic and Current Porcine Rotavirus C Strains and Their Pathogenesis in 3-Day-Old and 3-Week-Old Piglets. Front. Microbiol. 2020, 11, 780. [Google Scholar] [CrossRef]
- Barbe, F.; Atanasova, K.; Van Reeth, K. Cytokines and acute phase proteins associated with acute swine influenza infection in pigs. Vet. J. 2011, 187, 48–53. [Google Scholar] [CrossRef]
- Buddaert, W.; Van Reeth, K.; Pensaert, M. In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV). Adv. Exp. Med. Biol. 1998, 440, 461–467. [Google Scholar] [CrossRef]
- Luo, R.; Fang, L.; Jin, H.; Jiang, Y.; Wang, D.; Chen, H.; Xiao, S. Antiviral activity of type I and type III interferons against porcine reproductive and respiratory syndrome virus (PRRSV). Antivir. Res. 2011, 91, 99–101. [Google Scholar] [CrossRef]
- Liu, K.; Ma, G.; Liu, X.; Lu, Y.; Xi, S.; Ou, A.; Wei, J.; Li, B.; Shao, D.; Li, Y.; et al. Porcine reproductive and respiratory syndrome virus counteracts type I interferon-induced early antiviral state by interfering IRF7 activity. Vet. Microbiol. 2019, 229, 28–38. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y.J. Antagonizing interferon-mediated immune response by porcine reproductive and respiratory syndrome virus. BioMed Res. Int. 2014, 2014, 315470. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, S.L.; Loving, C.L.; Eberle, K.C.; Hau, S.J.; Buckley, A.; Van Geelen, A.; Montiel, N.A.; Nicholson, T.; Lager, K.M. Interferon alpha inhibits replication of a live-attenuated porcine reproductive and respiratory syndrome virus vaccine preventing development of an adaptive immune response in swine. Vet. Microbiol. 2017, 212, 48–51. [Google Scholar] [CrossRef] [PubMed]
- Saade, G.; Deblanc, C.; Bougon, J.; Marois-Créhan, C.; Fablet, C.; Auray, G.; Belloc, C.; Leblanc-Maridor, M.; Gagnon, C.A.; Zhu, J.; et al. Coinfections and their molecular consequences in the porcine respiratory tract. Vet. Res. 2020, 51, 80. [Google Scholar] [CrossRef]
- Fablet, C.; Marois-Crehan, C.; Grasland, B.; Simon, G.; Rose, N. Factors associated with herd-level PRRSV infection and age-time to seroconversion in farrow-to-finish herds. Vet. Microbiol. 2016, 192, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Hervé, S.; Gorin, S.; Quéguiner, S.; Barbier, N.; Eveno, E.; Dorenlor, V.; Eono, F.; Madec, F.; Rose, N.; Simon, G. Estimation of Influenza Seroprevalence in Slaughter Pigs in France in 2008–2009. In Proceedings of the Journées de la Recherche Porcine, Paris, France, 15–16 February 2011; pp. 281–282. [Google Scholar]
- Hervé, S.; Garin, E.; Calavas, D.; Lecarpentier, L.; Ngwa-Mbot, D.; Poliak, S.; Wendling, S.; Rose, N.; Simon, G. Virological and epidemiological patterns of swine influenza A virus infections in France: Cumulative data from the RESAVIP surveillance network, 2011–2018. Vet. Microbiol. 2019, 239, 108477. [Google Scholar] [CrossRef] [PubMed]
- Chamba Pardo, F.O.; Alba-Casals, A.; Nerem, J.; Morrison, R.B.; Puig, P.; Torremorell, M. Influenza Herd-Level Prevalence and Seasonality in Breed-to-Wean Pig Farms in the Midwestern United States. Front. Vet. Sci. 2017, 4, 167. [Google Scholar] [CrossRef]
- Diaz, A.; Marthaler, D.; Culhane, M.; Sreevatsan, S.; Alkhamis, M.; Torremorell, M. Complete Genome Sequencing of Influenza A Viruses within Swine Farrow-to-Wean Farms Reveals the Emergence, Persistence, and Subsidence of Diverse Viral Genotypes. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Deblanc, C.; Herve, S.; Gorin, S.; Cador, C.; Andraud, M.; Queguiner, S.; Barbier, N.; Paboeuf, F.; Rose, N.; Simon, G. Maternally-derived antibodies do not inhibit swine influenza virus replication in piglets but decrease excreted virus infectivity and impair post-infectious immune responses. Vet. Microbiol. 2018, 216, 142–152. [Google Scholar] [CrossRef]
- Cador, C.; Herve, S.; Andraud, M.; Gorin, S.; Paboeuf, F.; Barbier, N.; Queguiner, S.; Deblanc, C.; Simon, G.; Rose, N. Maternally-derived antibodies do not prevent transmission of swine influenza A virus between pigs. Vet. Res. 2016, 47, 86. [Google Scholar] [CrossRef]
- Deblanc, C.; Quéguiner, S.; Gorin, S.; Chastagner, A.; Hervé, S.; Paboeuf, F.; Simon, G. Evaluation of the Pathogenicity and the Escape from Vaccine Protection of a New Antigenic Variant Derived from the European Human-Like Reassortant Swine H1N2 Influenza Virus. Viruses 2020, 12, 1155. [Google Scholar] [CrossRef] [PubMed]
- Charpin, C.; Mahe, S.; Keranflec’h, A.; Belloc, C.; Cariolet, R.; Le Potier, M.F.; Rose, N. Infectiousness of pigs infected by the Porcine Reproductive and Respiratory Syndrome virus (PRRSV) is time-dependent. Vet. Res. 2012, 43, 69. [Google Scholar] [CrossRef] [PubMed]
- Renson, P.; Rose, N.; Le Dimna, M.; Mahe, S.; Keranflec’h, A.; Paboeuf, F.; Belloc, C.; Le Potier, M.F.; Bourry, O. Dynamic changes in bronchoalveolar macrophages and cytokines during infection of pigs with a highly or low pathogenic genotype 1 PRRSV strain. Vet. Res. 2017, 48, 15. [Google Scholar] [CrossRef] [PubMed]
- Jamin, A.; Gorin, S.; Le Potier, M.F.; Kuntz-Simon, G. Characterization of conventional and plasmacytoid dendritic cells in swine secondary lymphoid organs and blood. Vet. Immunol. Immunopathol. 2006, 114, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.; Beer, M.; Blome, S. Development and validation of a harmonized TaqMan-based triplex real-time RT-PCR protocol for the quantitative detection of normalized gene expression profiles of seven porcine cytokines. PLoS ONE 2014, 9, e108910. [Google Scholar] [CrossRef]
- Royaee, A.R.; Husmann, R.J.; Dawson, H.D.; Calzada-Nova, G.; Schnitzlein, W.M.; Zuckermann, F.A.; Lunney, J.K. Deciphering the involvement of innate immune factors in the development of the host response to PRRSV vaccination. Vet. Immunol. Immunopathol. 2004, 102, 199–216. [Google Scholar] [CrossRef]
- Brockmeier, S.L.; Loving, C.L.; Nelson, E.A.; Miller, L.C.; Nicholson, T.L.; Register, K.B.; Grubman, M.J.; Brough, D.E.; Kehrli, M.E., Jr. The presence of alpha interferon at the time of infection alters the innate and adaptive immune responses to porcine reproductive and respiratory syndrome virus. Clin. Vaccine Immunol. CVI 2012, 19, 508–514. [Google Scholar] [CrossRef]
- Brockmeier, S.L.; Lager, K.M.; Grubman, M.J.; Brough, D.E.; Ettyreddy, D.; Sacco, R.E.; Gauger, P.C.; Loving, C.L.; Vorwald, A.C.; Kehrli, M.E., Jr.; et al. Adenovirus-mediated expression of interferon-alpha delays viral replication and reduces disease signs in swine challenged with porcine reproductive and respiratory syndrome virus. Vir. Immunol. 2009, 22, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Lobo, F.J.; de Lome, L.C.; Díez-Fuertes, F.; Segalés, J.; García-Artiga, C.; Simarro, I.; Castro, J.M.; Prieto, C. Safety of Porcine Reproductive and Respiratory Syndrome Modified Live Virus (MLV) vaccine strains in a young pig infection model. Vet. Res. 2013, 44, 115. [Google Scholar] [CrossRef] [PubMed]
- Rowland, R.R.; Lawson, S.; Rossow, K.; Benfield, D.A. Lymphoid tissue tropism of porcine reproductive and respiratory syndrome virus replication during persistent infection of pigs originally exposed to virus in utero. Vet. Microbiol. 2003, 96, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Madapong, A.; Temeeyasen, G.; Saeng-Chuto, K.; Tripipat, T.; Navasakuljinda, W.; Boonsoongnern, A.; Tantituvanont, A.; Nilubol, D. Humoral immune responses and viral shedding following vaccination with modified live porcine reproductive and respiratory syndrome virus vaccines. Arch. Virol. 2017, 162, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Turlewicz-Podbielska, H.; Czyżewska-Dors, E.; Pomorska-Mól, M. Respiratory viral infections drive different lung cytokine profiles in pigs. BMC Vet. Res. 2021, 17, 5. [Google Scholar] [CrossRef]
- Pomorska-Mól, M.; Markowska-Daniel, I.; Kwit, K.; Czyżewska, E.; Dors, A.; Rachubik, J.; Pejsak, Z. Immune and inflammatory response in pigs during acute influenza caused by H1N1 swine influenza virus. Arch. Virol. 2014, 159, 2605–2614. [Google Scholar] [CrossRef]
- Ito, S.; Ansari, P.; Sakatsume, M.; Dickensheets, H.; Vazquez, N.; Donnelly, R.P.; Larner, A.C.; Finbloom, D.S. Interleukin-10 inhibits expression of both interferon alpha-and interferon gamma- induced genes by suppressing tyrosine phosphorylation of STAT1. Blood 1999, 93, 1456–1463. [Google Scholar] [CrossRef]
- Merolla, R.; Rebert, N.A.; Tsiviste, P.T.; Hoffmann, S.P.; Panuska, J.R. Respiratory syncytial virus replication in human lung epithelial cells: Inhibition by tumor necrosis factor alpha and interferon beta. Am. J. Respir. Crit. Care Med. 1995, 152, 1358–1366. [Google Scholar] [CrossRef]
- Van Reeth, K.; Ma, W. Swine influenza virus vaccines: To change or not to change-that’s the question. Curr. Top. Microbiol. Immunol. 2013, 370, 173–200. [Google Scholar] [CrossRef]
- Madapong, A.; Saeng-Chuto, K.; Boonsoongnern, A.; Tantituvanont, A.; Nilubol, D. Cell-mediated immune response and protective efficacy of porcine reproductive and respiratory syndrome virus modified-live vaccines against co-challenge with PRRSV-1 and PRRSV-2. Sci. Rep. 2020, 10, 1649. [Google Scholar] [CrossRef] [PubMed]
- Saade, G.; Ménard, D.; Hervet, C.; Renson, P.; Hue, E.; Zhu, J.; Dubreil, L.; Paillot, R.; Pronost, S.; Bourry, O.; et al. Porcine Reproductive and Respiratory Syndrome Virus Interferes with Swine Influenza A Virus Infection of Epithelial Cells. Vaccines 2020, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Boikos, C.; Papenburg, J.; Martineau, C.; Joseph, L.; Scheifele, D.; Chilvers, M.; Lands, L.C.; De Serres, G.; Quach, C. Viral interference and the live-attenuated intranasal influenza vaccine: Results from a pediatric cohort with cystic fibrosis. Hum. Vaccines Immunother. 2017, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Essaidi-Laziosi, M.; Geiser, J.; Huang, S.; Constant, S.; Kaiser, L.; Tapparel, C. Interferon-Dependent and Respiratory Virus-Specific Interference in Dual Infections of Airway Epithelia. Sci. Rep. 2020, 10, 10246. [Google Scholar] [CrossRef] [PubMed]
- Crisci, E.; Fraile, L.; Montoya, M. Cellular Innate Immunity against PRRSV and Swine Influenza Viruses. Vet. Sci. 2019, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Le Bon, A.; Tough, D.F. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 2002, 14, 432–436. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renson, P.; Deblanc, C.; Bougon, J.; Le Dimna, M.; Gorin, S.; Mahé, S.; Barbier, N.; Paboeuf, F.; Simon, G.; Bourry, O. Concomitant Swine Influenza A Virus Infection Alters PRRSV1 MLV Viremia in Piglets but Does Not Interfere with Vaccine Protection in Experimental Conditions. Vaccines 2021, 9, 356. https://doi.org/10.3390/vaccines9040356
Renson P, Deblanc C, Bougon J, Le Dimna M, Gorin S, Mahé S, Barbier N, Paboeuf F, Simon G, Bourry O. Concomitant Swine Influenza A Virus Infection Alters PRRSV1 MLV Viremia in Piglets but Does Not Interfere with Vaccine Protection in Experimental Conditions. Vaccines. 2021; 9(4):356. https://doi.org/10.3390/vaccines9040356
Chicago/Turabian StyleRenson, Patricia, Céline Deblanc, Juliette Bougon, Mireille Le Dimna, Stéphane Gorin, Sophie Mahé, Nicolas Barbier, Frédéric Paboeuf, Gaëlle Simon, and Olivier Bourry. 2021. "Concomitant Swine Influenza A Virus Infection Alters PRRSV1 MLV Viremia in Piglets but Does Not Interfere with Vaccine Protection in Experimental Conditions" Vaccines 9, no. 4: 356. https://doi.org/10.3390/vaccines9040356
APA StyleRenson, P., Deblanc, C., Bougon, J., Le Dimna, M., Gorin, S., Mahé, S., Barbier, N., Paboeuf, F., Simon, G., & Bourry, O. (2021). Concomitant Swine Influenza A Virus Infection Alters PRRSV1 MLV Viremia in Piglets but Does Not Interfere with Vaccine Protection in Experimental Conditions. Vaccines, 9(4), 356. https://doi.org/10.3390/vaccines9040356





