Delivery of mRNA Vaccine against SARS-CoV-2 Using a Polyglucin:Spermidine Conjugate
Abstract
1. Introduction
2. Materials and Methods
2.1. mRNA Synthesis
2.2. Preparation of the Polyglucin:Spermidine Conjugate
2.3. Dynamic and Electrophoretic Light Scattering
2.4. Electron Microscopy
2.5. Cells Transfection with mRNA-GFP
2.6. Mice Immunization
2.7. Serum ELISA
2.8. Neutralizing Activity of Immune Sera
2.9. Cell Viability Assay (MTT)
3. Results
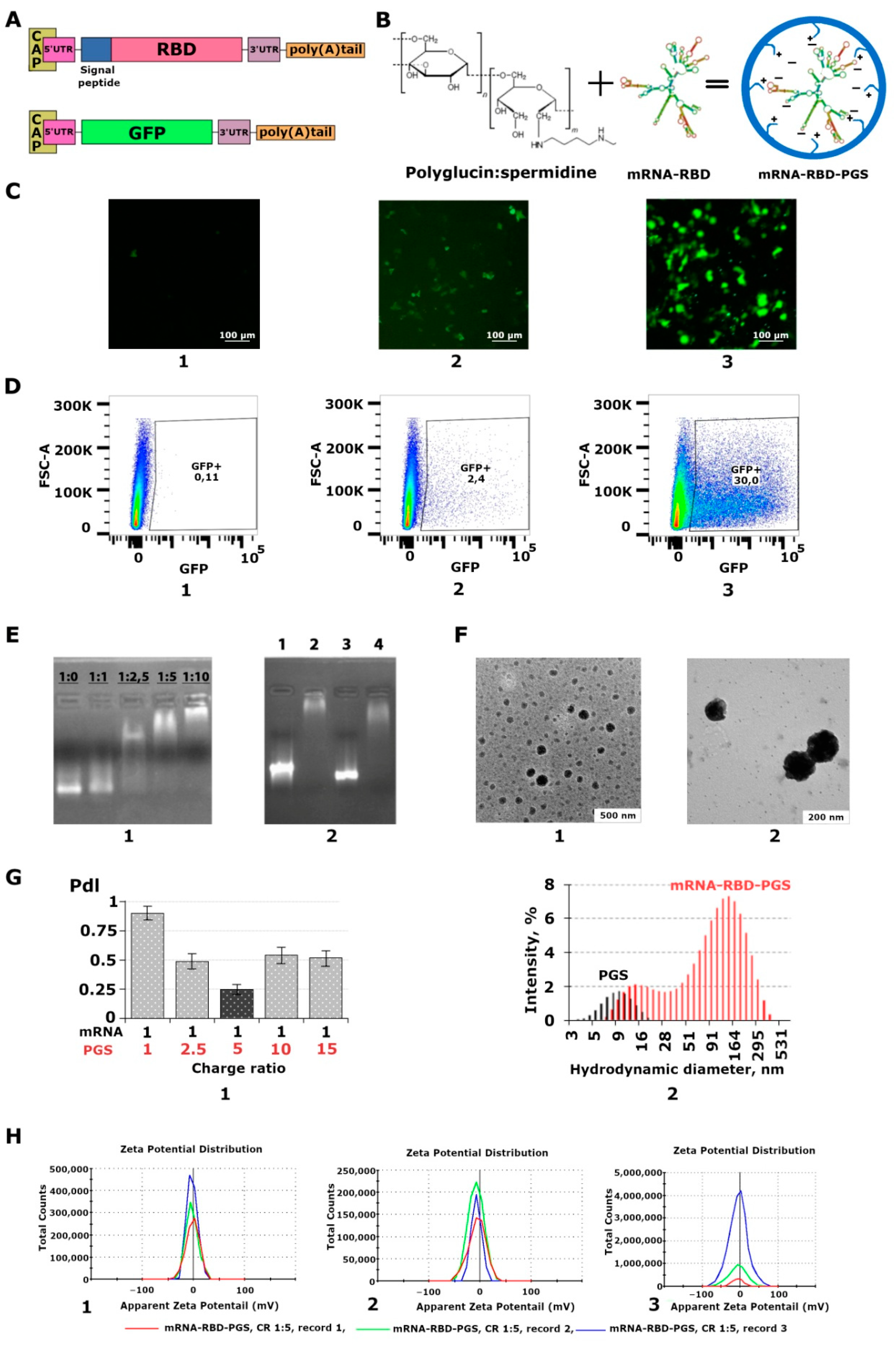
3.1. Characterization of the Synthesized mRNA
3.2. Characterization of the mRNA-PGS Complex
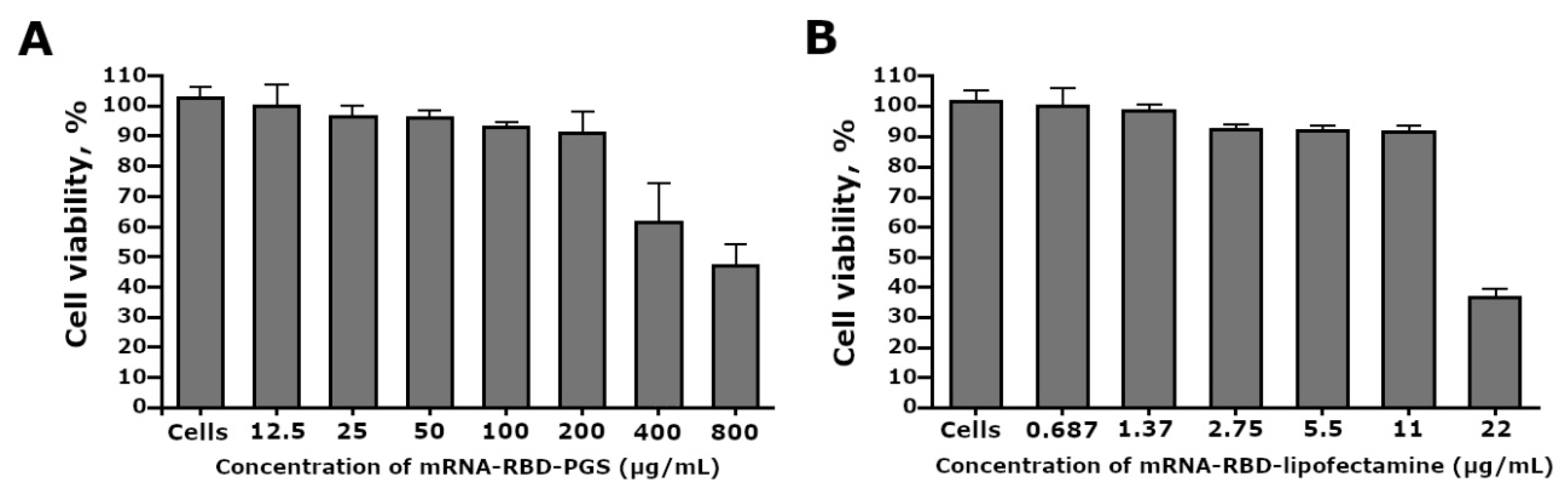
3.3. Cytotoxicity of the mRNA-RBD-PGS Complexes
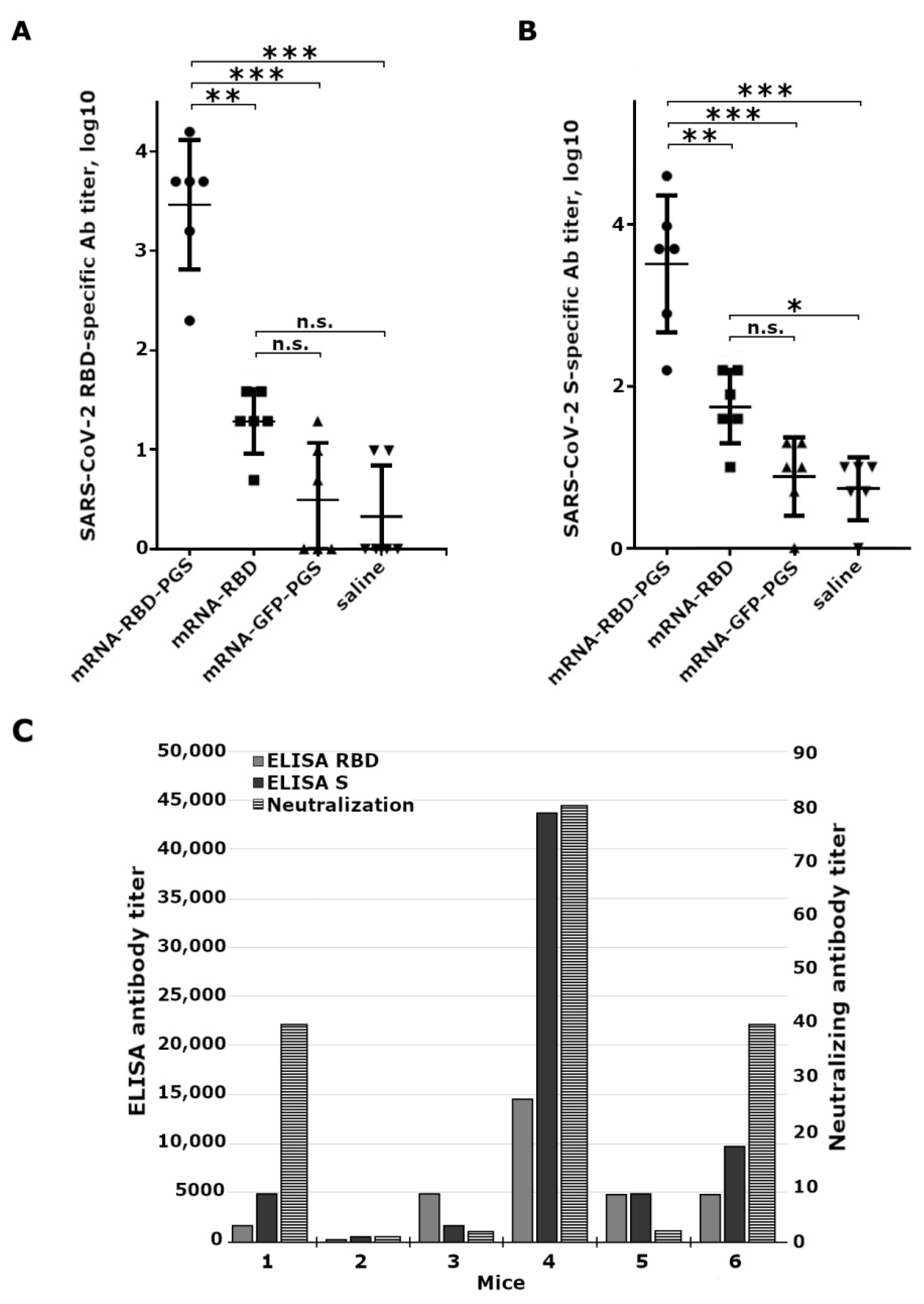
3.4. Mice Immune Response to Vaccine
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Volchkov, V.E.; Volchkova, V.A.; Vorac, J.; Repiquet-Paire, L.; Lawrence, P. Ebola Virus GP Gene Polyadenylation Versus RNA Editing. J. Infect. Dis. 2015, 212, S191–S198. [Google Scholar] [CrossRef] [PubMed]
- Schnee, M.; Vogel, A.B.; Voss, D.; Petsch, B.; Baumhof, P.; Kramps, T.; Stitz, L. An mRNA Vaccine Encoding Rabies Virus Glycoprotein Induces Protection against Lethal Infection in Mice and Correlates of Protection in Adult and Newborn Pigs. PLoS Negl. Trop. Dis. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Liu, M.A. A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Qi, Y.; Wang, M.; Yu, N.; Nan, F.; Zhang, H.; Tian, M.; Li, C.; Lu, H.; Jin, N. MRNA vaccines encoding the HA protein of influenza a H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines 2020, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Chahal, J.S.; Fang, T.; Woodham, A.W.; Khan, O.F.; Ling, J.J.; Anderson, D.G.; Ploegh, H.L. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+T cell responses in a mouse model. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Zhang, N.N.; Li, X.F.; Deng, Y.Q.; Zhao, H.; Huang, Y.J.; Yang, G.; Huang, W.J.; Gao, P.; Zhou, C.; Zhang, R.R.; et al. A Thermostable mRNA Vaccine against COVID-19. Cell 2020, 182, 1271–1283.e1216. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Tai, W.; Zhang, X.; Drelich, A.; Shi, J.; Hsu, J.C.; Luchsinger, L.; Hillyer, C.D.; Tseng, C.T.K.; Jiang, S.; Du, L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020, 30, 932–935. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2-Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Aguado, I.; Rodriguez-Castejon, J.; Vicente-Pascual, M.; Rodriguez-Gascon, A.; Solinis, M.A.; del Pozo-Rodriguez, A. Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials 2020, 10, 364. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and challenges in the delivery of mrna-based vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef]
- WHO. Novel Coronavirus (2019-nCoV). Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 12 December 2020).
- Fischer, D.; Bieber, T.; Li, Y.; Elsässer, H.P.; Kissel, T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: Effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 1999, 16, 1273–1279. [Google Scholar] [CrossRef] [PubMed]
- Pun, S.H.; Hoffman, A.S. 8—nucleic acid delivery. In Biomaterial Science, 3rd ed.; Ratner, B.D., Hoffman, A.S., Schoen, F.J., Lemons, J.E., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 1047–1054. [Google Scholar]
- Xue, H.Y.; Guo, P.; Wen, W.C.; Wong, H.L. Lipid-based nanocarriers for RNA delivery. Curr. Pharm. Des. 2015, 21, 3140–3147. [Google Scholar] [CrossRef]
- Sedic, M.; Senn, J.J.; Lynn, A.; Laska, M.; Smith, M.; Platz, S.J.; Bolen, J.; Hoge, S.; Bulychev, A.; Jacquinet, E.; et al. Safety Evaluation of Lipid Nanoparticle–Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2018, 55, 341–354. [Google Scholar] [CrossRef]
- Chang, H.I.; Yeh, M.K. Clinical development of liposome-based drugs: Formulation, characterization, and therapeutic efficacy. Int. J. Nanomed. 2012, 7, 49–60. [Google Scholar] [CrossRef]
- Lball, R.; Bajaj, P.; Whitehead, K.A. Achieving long-term stability of lipid nanoparticles: Examining the effect of pH, temperature, and lyophilization. Int. J. Nanomed. 2017, 12, 305–315. [Google Scholar] [CrossRef]
- Lebedev, L.R.; Karpenko, L.I.; Poryvaeva, V.A.; Azaev, M.S.; Ryabchikova, E.I.; Gileva, I.P.; Il’ichev, A.A. Construction of virus-like particles exposing HIV-1 epitopes. Mol. Biol. 2000, 34, 413–417. [Google Scholar] [CrossRef]
- Karpenko, L.I.; Lebedev, L.R.; Ignatyev, G.M.; Agafonov, A.P.; Poryvaeva, V.A.; Pronyaeva, T.R.; Ryabchikova, E.I.; Pokrovsky, A.G.; Ilyichev, A.A. Construction of artificial virus-like particles exposing HIV epitopes, and the study of their immunogenic properties. Vaccine 2003, 21, 386–392. [Google Scholar] [CrossRef]
- Karpenko, L.I.; Apartsin, E.K.; Dudko, S.G.; Starostina, E.V.; Kaplina, O.N.; Antonets, D.V.; Volosnikova, E.A.; Zaitsev, B.N.; Bakulina, A.Y.; Venyaminova, A.G.; et al. Cationic polymers for the delivery of the ebola dna vaccine encoding artificial t-cell immunogen. Vaccines 2020, 8, 718. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- van Meerloo, J.; Kaspers, G.J.L.; Cloos, J. Cell Sensitivity Assays: The MTT Assay. In Cancer Cell Culture: Methods and Protocols; Cree, I.A., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 237–245. [Google Scholar] [CrossRef]
- Ihnatsyeu-Kachan, A.; Dzmitruk, V.; Apartsin, E.; Krasheninina, O.; Ionov, M.; Loznikova, S.; Venyaminova, A.; Milowska, K.; Shcharbin, D.; Mignani, S.; et al. Multi-Target Inhibition of Cancer Cell Growth by SiRNA Cocktails and 5-Fluorouracil Using Effective Piperidine-Terminated Phosphorus Dendrimers. Colloids Interfaces 2017, 1, 6. [Google Scholar] [CrossRef]
- Filion, M.C.; Phillips, N.C. Toxicity and immunomodulatory activity of liposomal vectors formulated with cationic lipids toward immune effector cells. Biochim. Biophys. Acta Biomembr. 1997, 1329, 345–356. [Google Scholar] [CrossRef]
- Zhao, Y.N.; Piao, Y.Z.; Zhang, C.M.; Jiang, Y.M.; Liu, A.; Cui, S.H.; Zhi, D.F.; Zhen, Y.H.; Zhang, S.B. Replacement of quaternary ammonium headgroups by tri-ornithine in cationic lipids for the improvement of gene delivery: In vitro and in vivo. J. Mater. Chem. B 2017, 5, 7963–7973. [Google Scholar] [CrossRef] [PubMed]
- Landesman-Milo, D.; Peer, D. Toxicity profiling of several common RNAi-based nanomedicines: A comparative study. Drug Deliv. Transl. Res. 2014, 4, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L.; Drane, D.; Gowans, E.J. Long-term storage of DNA-free RNA for use in vaccine studies. BioTechniques 2007, 43, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. How COVID unlocked the power of RNA vaccines. Nature 2021, 589, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e899. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.D.; Zhao, Y.G.; Ren, J.S.; Zhou, D.M.; Duyvesteyn, H.M.E.; Ginn, H.M.; Carrique, L.; Malinauskas, T.; Ruza, R.R.; Shah, P.N.M.; et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe 2020, 28, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.F.; Zhao, F.; Huang, D.; Beutler, N.; Burns, A.; He, W.T.; Limbo, O.; Smith, C.; Song, G.; Woehl, J.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 369, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Karpenko, L.I.; Ilyichev, A.A.; Eroshkin, A.M.; Lebedev, L.R.; Uzhachenko, R.V.; Nekrasova, N.A.; Plyasunova, O.A.; Belavin, P.A.; Seregin, S.V.; Danilyuk, N.K.; et al. Combined virus-like particle-based polyepitope DNA/protein HIV-1 vaccine. Design, immunogenicity and toxicity studies. Vaccine 2007, 25, 4312–4323. [Google Scholar] [CrossRef]
- Karpenko, L.I.; Bazhan, S.I.; Bogryantseva, M.P.; Ryndyuk, N.N.; Ginko, Z.I.; Kuzubov, V.I.; Lebedev, L.R.; Kaplina, O.N.; Reguzova, A.Y.; Ryzhikov, A.B.; et al. Results of phase i clinical trials of a combined vaccine against HIV-1 based on synthetic polyepitope immunogens. Russ. J. Bioorganic Chem. 2016, 42, 170–182. [Google Scholar] [CrossRef]
- Karpenko, L.I.; Bazhan, S.I.; Eroshkin, A.M.; Antonets, D.V.; Chikaev, A.N.; Ilyichev, A.A. Artificial Epitope-Based Immunogens in HIV-Vaccine Design. In Advances in HIV and AIDS Control; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Singh, D.V.; Singh, R.; Sodhi, S.P.S. Effect of blood transfusion in combination with Dextran-40 and hypertonic saline solution on cardiopulmonary haemodynamics of endotoxin (Lipopolysaccharide) shock in buffalo calves. Vet. Res. Commun. 2005, 29, 421–430. [Google Scholar] [CrossRef]
- Cohen, S.S. A Guide to Polyamines; Oxford University Press: New York, NY, USA, 1998. [Google Scholar]
- Tabor, C.W.; Tabor, H. Polyamines. Annu. Rev. Biochem. 1984, 53, 749–790. [Google Scholar] [CrossRef]
- Perepelytsya, S.; Ulicny, J.; Laaksonen, A.; Mocci, F. Pattern preferences of DNA nucleotide motifs by polyamines putrescine(2+), spermidine(3+)and spermine(4+). Nucleic Acids Res. 2019, 47, 6084–6097. [Google Scholar] [CrossRef]
- Lightfoot, H.L.; Hall, J. Endogenous polyamine function-the RNA perspective. Nucleic Acids Res. 2014, 42, 11275–11290. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [PubMed]
- Farris, E.; Brown, D.M.; Ramer-Tait, A.E.; Pannier, A.K. Micro- and nanoparticulates for DNA vaccine delivery. Exp. Biol. Med. 2016, 241, 919–929. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karpenko, L.I.; Rudometov, A.P.; Sharabrin, S.V.; Shcherbakov, D.N.; Borgoyakova, M.B.; Bazhan, S.I.; Volosnikova, E.A.; Rudometova, N.B.; Orlova, L.A.; Pyshnaya, I.A.; et al. Delivery of mRNA Vaccine against SARS-CoV-2 Using a Polyglucin:Spermidine Conjugate. Vaccines 2021, 9, 76. https://doi.org/10.3390/vaccines9020076
Karpenko LI, Rudometov AP, Sharabrin SV, Shcherbakov DN, Borgoyakova MB, Bazhan SI, Volosnikova EA, Rudometova NB, Orlova LA, Pyshnaya IA, et al. Delivery of mRNA Vaccine against SARS-CoV-2 Using a Polyglucin:Spermidine Conjugate. Vaccines. 2021; 9(2):76. https://doi.org/10.3390/vaccines9020076
Chicago/Turabian StyleKarpenko, Larisa I., Andrey P. Rudometov, Sergei V. Sharabrin, Dmitry N. Shcherbakov, Mariya B. Borgoyakova, Sergei I. Bazhan, Ekaterina A. Volosnikova, Nadezhda B. Rudometova, Lyubov A. Orlova, Inna A. Pyshnaya, and et al. 2021. "Delivery of mRNA Vaccine against SARS-CoV-2 Using a Polyglucin:Spermidine Conjugate" Vaccines 9, no. 2: 76. https://doi.org/10.3390/vaccines9020076
APA StyleKarpenko, L. I., Rudometov, A. P., Sharabrin, S. V., Shcherbakov, D. N., Borgoyakova, M. B., Bazhan, S. I., Volosnikova, E. A., Rudometova, N. B., Orlova, L. A., Pyshnaya, I. A., Zaitsev, B. N., Volkova, N. V., Azaev, M. S., Zaykovskaya, A. V., Pyankov, O. V., & Ilyichev, A. A. (2021). Delivery of mRNA Vaccine against SARS-CoV-2 Using a Polyglucin:Spermidine Conjugate. Vaccines, 9(2), 76. https://doi.org/10.3390/vaccines9020076






