A Novel Effective and Safe Vaccine for Prevention of Marek’s Disease Caused by Infection with a Very Virulent Plus (vv+) Marek’s Disease Virus
Abstract
1. Introduction
2. Material and Methods
2.1. Cells and Viruses
2.2. Construction of meq and/or vIL8 Single and Double Deletion Viruses
2.3. Immunofluorescence (IFA) and Immunohistochemistry (IHC) Assays
2.4. In Vitro Growth Kinetics
2.5. MDV Genome Copy Numbers
2.6. Virus Reactivation Assay
2.7. Pathogenesis of meq or vIL8 Single and Double Deletion Mutant Viruses in SPF Ab– Chickens
- a
- Lymphoid organ atrophy: To evaluate the effect of virus replication in lymphoid organ atrophy, 5 randomly selected chickens from each experimental group were euthanatized at 14 dpi, and thymus and bursa were weighed. Results were presented as the average ratio of lymphoid organs weight to body weight of 5 chickens multiplied by one hundred.
- b
- Pathogenesis: To compare the pathogenic properties of parental 686BAC, 686BAC-∆Meq, 686BAC-∆vIL8, and 686BAC-∆Meq∆vIL8 viruses, the mortality of each experimental group was recorded daily for 65 days. All chickens that died during the experiment or were euthanized at the end of the experiment were necropsied and examined for MD associated gross tumors in visceral organs and nerves.
2.8. Vaccine Protection Experiments
2.9. Data and Statistical Analysis
3. Results
3.1. Construction and In Vitro Characterization of meq or vIL8 Single and Double Deletion Viruses
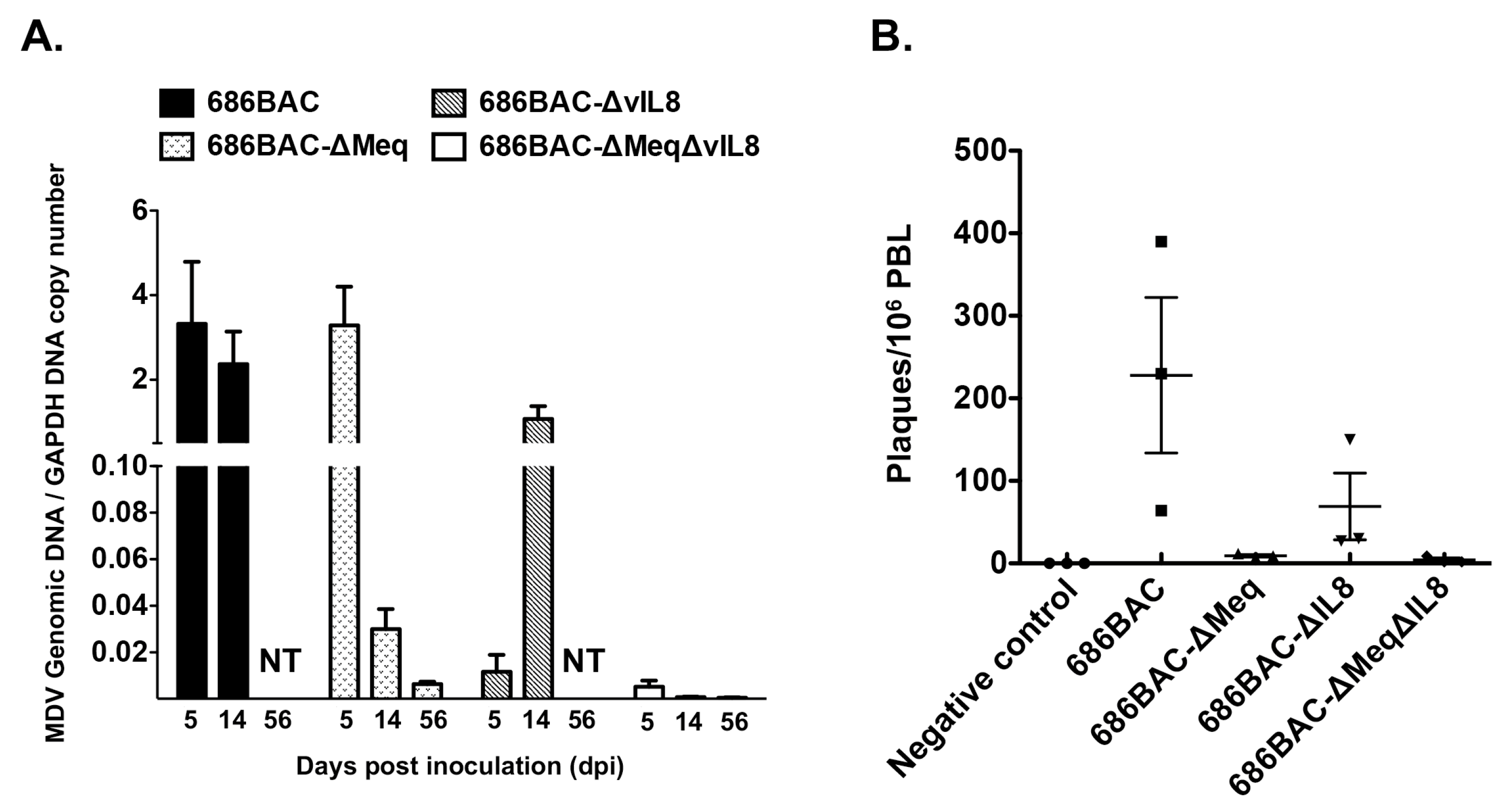
3.2. In Vivo Replication and Reactivation of meq or vIL8 Single and Double Deletion Viruses
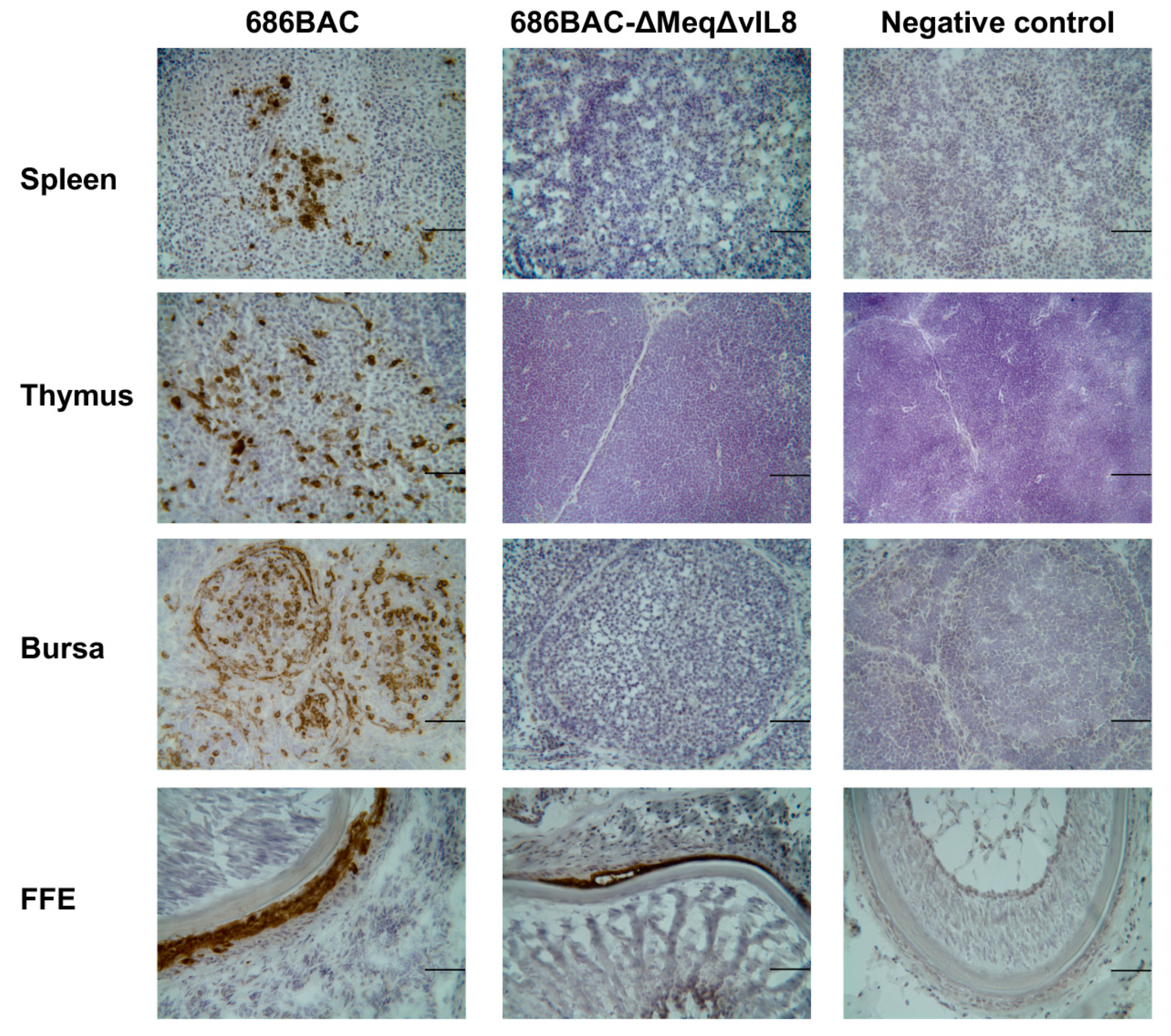
3.3. Evaluation of Lymphoid Organ Atrophy and Pathogenesis Induced by Inoculation of meq or vIL8 Single and Double Deletion Viruses
3.4. 686BAC-∆Meq∆vIL8 Provides Protection Comparable to CVI988 against Challenge with a vv+ MDV
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Churchill, A.E.; Biggs, P.M. Agent of Marek’s disease in tissue culture. Nature 1967, 215, 528–530. [Google Scholar] [CrossRef]
- Nazerian, K.; Solomon, J.J.; Witter, R.L.; Burmester, B.R. Studies on the etiology of Marek’s disease. II. Finding of a herpesvirus in cell culture. Proc. Soc. Exp. Biol. Med. 1968, 127, 177–182. [Google Scholar] [CrossRef]
- Calnek, B.W. Pathogenesis of Marek’s disease virus infection. Curr Top. Microbiol. Immunol. 2001, 255, 25–55. [Google Scholar]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Witter, R.L. Increased virulence of Marek’s disease virus field isolates. Avian Dis. 1997, 41, 149–163. [Google Scholar] [CrossRef]
- Nair, V. Successful control of Marek’s disease by vaccination. Dev. Biol. 2004, 119, 147–154. [Google Scholar]
- Okazaki, W.; Purchase, H.G.; Burmester, B.R. Protection against Marek’s disease by vaccination with a herpesvirus of turkeys. Avian Dis. 1970, 14, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef]
- Churchill, A.E.; Chubb, R.C.; Baxendale, W. The attenuation, with loss of oncogenicity, of the herpes-type virus of Marek’s disease (strain HPRS-16) on passage in cell culture. J. Gen. Virol. 1969, 4, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Witter, R.L.; Nazerian, K.; Purchase, H.G.; Burgoyne, G.H. Isolation from turkeys of a cell-associated herpesvirus antigenically related to Marek’s disease virus. Am. J. Vet. Res. 1970, 31, 525–538. [Google Scholar] [PubMed]
- Witter, R.L.; Lee, L.F. Polyvalent Marek’s disease vaccines: Safety, efficacy and protective synergism in chickens with maternal antibodies. Avian Pathol. 1984, 13, 75–92. [Google Scholar] [CrossRef]
- Witter, R.L. New serotype 2 and attenuated serotype 1 Marek’s disease vaccine viruses: Comparative efficacy. Avian Dis. 1987, 31, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Rispens, B.H.; van Vloten, H.; Mastenbroek, N.; Maas, H.J.; Schat, K.A. Control of Marek’s disease in the Netherlands. I. Isolation of an avirulent Marek’s disease virus (strain CVI 988) and its use in laboratory vaccination trials. Avian Dis. 1972, 16, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Rispens, B.H.; van Vloten, H.; Mastenbroek, N.; Maas, J.L.; Schat, K.A. Control of Marek’s disease in the Netherlands. II. Field trials on vaccination with an avirulent strain (CVI 988) of Marek’s disease virus. Avian Dis. 1972, 16, 126–138. [Google Scholar] [CrossRef]
- Lupiani, B.; Lee, L.F.; Kreager, K.S.; Witter, R.L.; Reddy, S.M. Insertion of reticuloendotheliosis virus long terminal repeat into the genome of CVI988 strain of Marek’s disease virus results in enhanced growth and protection. Avian Dis. 2013, 57 (Suppl. 2), 427–431. [Google Scholar] [CrossRef]
- Lee, L.F.; Wu, P.; Sui, D.; Ren, D.; Kamil, J.; Kung, H.J.; Witter, R.L. The complete unique long sequence and the overall genomic organization of the GA strain of Marek’s disease virus. Proc. Natl. Acad. Sci. USA 2000, 97, 6091–6096. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Rock, D.L.; Kutish, G.F. The genome of a very virulent Marek’s disease virus. J. Virol. 2000, 74, 7980–7988. [Google Scholar] [CrossRef]
- Cui, Z.Z.; Lee, L.F.; Liu, J.L.; Kung, H.J. Structural analysis and transcriptional mapping of the Marek’s disease virus gene encoding pp38, an antigen associated with transformed cells. J. Virol. 1991, 65, 6509–6515. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.S.; Francesconi, A.; Morgan, R.W. Complete nucleotide sequence of the Marek’s disease virus ICP4 gene. Virology 1992, 189, 657–667. [Google Scholar] [CrossRef]
- Parcells, M.S.; Lin, S.F.; Dienglewicz, R.L.; Majerciak, V.; Robinson, D.R.; Chen, H.C.; Wu, Z.; Dubyak, G.R.; Brunovskis, P.; Hunt, H.D.; et al. Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): Characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 2001, 75, 5159–5173. [Google Scholar] [CrossRef]
- Lupiani, B.; Lee, L.F.; Cui, X.; Gimeno, I.; Anderson, A.; Morgan, R.W.; Silva, R.F.; Witter, R.L.; Kung, H.J.; Reddy, S.M. Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 2004, 101, 11815–11820. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.; Lee, L.; Liu, J.L.; Kung, H.J.; Tillotson, J.K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc. Natl. Acad. Sci. USA 1992, 89, 4042–4046. [Google Scholar] [CrossRef]
- Brown, A.C.; Smith, L.P.; Kgosana, L.; Baigent, S.J.; Nair, V.; Allday, M.J. Homodimerization of the Meq viral oncoprotein is necessary for induction of T-cell lymphoma by Marek’s disease virus. J. Virol. 2009, 83, 11142–11151. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, P.F.; Izumiya, Y.; Lupiani, B.; Ajithdoss, D.K.; Gilad, O.; Lee, L.F.; Kung, H.J.; Reddy, S.M. Homodimerization of Marek’s disease virus-encoded Meq protein is not sufficient for transformation of lymphocytes in chickens. J. Virol. 2009, 83, 859–869. [Google Scholar] [CrossRef]
- Suchodolski, P.F.; Izumiya, Y.; Lupiani, B.; Ajithdoss, D.K.; Lee, L.F.; Kung, H.J.; Reddy, S.M. Both homo and heterodimers of Marek’s disease virus encoded Meq protein contribute to transformation of lymphocytes in chickens. Virology 2010, 399, 312–321. [Google Scholar] [CrossRef]
- Liu, J.L.; Ye, Y.; Lee, L.F.; Kung, H.J. Transforming potential of the herpesvirus oncoprotein MEQ: Morphological transformation, serum-independent growth, and inhibition of apoptosis. J. Virol. 1998, 72, 388–395. [Google Scholar] [CrossRef]
- Levy, A.M.; Gilad, O.; Xia, L.; Izumiya, Y.; Choi, J.; Tsalenko, A.; Yakhini, Z.; Witter, R.; Lee, L.; Cardona, C.J.; et al. Marek’s disease virus Meq transforms chicken cells via the v-Jun transcriptional cascade: A converging transforming pathway for avian oncoviruses. Proc. Natl. Acad. Sci. USA 2005, 102, 14831–14836. [Google Scholar] [CrossRef]
- Ajithdoss, D.K.; Reddy, S.M.; Suchodolski, P.F.; Lee, L.F.; Kung, H.J.; Lupiani, B. In vitro characterization of the Meq proteins of Marek’s disease virus vaccine strain CVI988. Virus Res. 2009, 142, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.F.; Lupiani, B.; Silva, R.F.; Kung, H.J.; Reddy, S.M. Recombinant Marek’s disease virus (MDV) lacking the Meq oncogene confers protection against challenge with a very virulent plus strain of MDV. Vaccine 2008, 26, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.F.; Kreager, K.S.; Arango, J.; Paraguassu, A.; Beckman, B.; Zhang, H.; Fadly, A.; Lupiani, B.; Reddy, S.M. Comparative evaluation of vaccine efficacy of recombinant Marek’s disease virus vaccine lacking Meq oncogene in commercial chickens. Vaccine 2010, 28, 1294–1299. [Google Scholar] [CrossRef]
- Dunn, J.R.; Silva, R.F. Ability of MEQ-deleted MDV vaccine candidates to adversely affect lymphoid organs and chicken weight gain. Avian Dis. 2012, 56, 494–500. [Google Scholar] [CrossRef]
- Lee, L.F.; Heidari, M.; Zhang, H.; Lupiani, B.; Reddy, S.M.; Fadly, A. Cell culture attenuation eliminates rMd5DeltaMeq-induced bursal and thymic atrophy and renders the mutant virus as an effective and safe vaccine against Marek’s disease. Vaccine 2012, 30, 5151–5158. [Google Scholar] [CrossRef]
- Cui, X.; Lee, L.F.; Reed, W.M.; Kung, H.J.; Reddy, S.M. Marek’s disease virus-encoded vIL-8 gene is involved in early cytolytic infection but dispensable for establishment of latency. J. Virol. 2004, 78, 4753–4760. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lee, L.F.; Hunt, H.D.; Reed, W.M.; Lupiani, B.; Reddy, S.M. A Marek’s disease virus vIL-8 deletion mutant has attenuated virulence and confers protection against challenge with a very virulent plus strain. Avian Dis. 2005, 49, 199–206. [Google Scholar] [CrossRef]
- Reddy, S.M.; Sun, A.; Khan, O.A.; Lee, L.F.; Lupiani, B. Cloning of a very virulent plus, 686 strain of Marek’s disease virus as a bacterial artificial chromosome. Avian Dis. 2013, 57 (Suppl. 2), 469–473. [Google Scholar] [CrossRef]
- Schumacher, D.; Tischer, B.K.; Fuchs, W.; Osterrieder, N. Reconstitution of Marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 2000, 74, 11088–11098. [Google Scholar] [CrossRef] [PubMed]
- Tischer, B.K.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 2006, 40, 191–197. [Google Scholar] [PubMed]
- Lee, L.F.; Liu, X.; Witter, R.L. Monoclonal antibodies with specificity for three different serotypes of Marek’s disease viruses in chickens. J. Immunol. 1983, 130, 1003–1006. [Google Scholar]
- Jarosinski, K.W.; Osterrieder, N.; Nair, V.K.; Schat, K.A. Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J. Virol. 2005, 79, 11647–11659. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Lupiani, B.; Bajwa, K.; Khan, O.A.; Izumiya, Y.; Reddy, S.M. Role of Marek’s disease virus encoded US3 serine/threonine protein kinase in regulating MDV Meq and cellular CREB phosphorylation. J. Virol. 2020. [Google Scholar] [CrossRef]
- Davison, F.; Nair, V. Use of Marek’s disease vaccines: Could they be driving the virus to increasing virulence? Expert Rev. Vaccines 2005, 4, 77–88. [Google Scholar] [CrossRef]
- Gandon, S.; Mackinnon, M.J.; Nee, S.; Read, A.F. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001, 414, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, I.M. Marek’s disease vaccines: A solution for today but a worry for tomorrow? Vaccine 2008, 26 (Suppl. 3), C31–C41. [Google Scholar] [CrossRef]
- Reddy, S.M.; Izumiya, Y.; Lupiani, B. Marek’s disease vaccines: Current status, and strategies for improvement and development of vector vaccines. Vet. Microbiol. 2016. [Google Scholar] [CrossRef]
- Bulow, V.V. Further characterisation of the CVI 988 strain of Marek’s disease virus. Avian Pathol. 1977, 6, 395–403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Witter, R. Safety and comparative efficacy of the CVI988/Rispens vaccine strain. In 4th International Symposium on Marek’s Disease, 19th World’s Poultry Congress; World Poultry Science Association: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Witter, R.L.; Lee, L.F.; Fadly, A.M. Characteristics of CVI988/Rispens and R2/23, two prototype vaccine strains of serotype 1 Marek’s disease virus. Avian Dis. 1995, 39, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Witter, R.L.; Kreager, K.S. Serotype 1 viruses modified by backpassage or insertional mutagenesis: Approaching the threshold of vaccine efficacy in Marek’s disease. Avian Dis. 2004, 48, 768–782. [Google Scholar] [CrossRef]
- Witter, R.L. Induction of strong protection by vaccination with partially attenuated serotype 1 Marek’s disease viruses. Avian Dis. 2002, 46, 925–937. [Google Scholar] [CrossRef]
- Gimeno, I.M.; Witter, R.L.; Hunt, H.D.; Reddy, S.M.; Reed, W.M. Biocharacteristics shared by highly protective vaccines against Marek’s disease. Avian Pathol. 2004, 33, 59–68. [Google Scholar] [CrossRef]
- Hildebrandt, E.; Dunn, J.R.; Cheng, H.H. The Mut UL5-I682R Marek’s Disease Virus with a Single Nucleotide Mutation Within the Helicase-Primase Subunit Gene not only Reduces Virulence but also Provides Partial Vaccinal Protection against Marek’s Disease. Avian Dis. 2015, 59, 94–97. [Google Scholar] [CrossRef]
- Hildebrandt, E.; Dunn, J.R.; Cheng, H.H. Addition of a UL5 helicase-primase subunit point mutation eliminates bursal-thymic atrophy of Marek’s disease virus Meq recombinant virus but reduces vaccinal protection. Avian Pathol. 2015, 44, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.F.; Zhang, H.; Heidari, M.; Lupiani, B.; Reddy, S.M. Evaluation of factors affecting vaccine efficacy of recombinant Marek’s disease virus lacking the Meq oncogene in chickens. Avian Dis. 2011, 55, 172–179. [Google Scholar] [CrossRef]
- Gimeno, I.M.; Schat, K.A. Virus-Induced Immunosuppression in Chickens. Avian Dis. 2018, 62, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Faiz, N.M.; Cortes, A.L.; Guy, J.S.; Fletcher, O.J.; West, M.; Montiel, E.; Gimeno, I.M. Early infection with Marek’s disease virus can jeopardize protection conferred by laryngotracheitis vaccines: A method to study MDV-induced immunosuppression. Avian Pathol. 2016, 45, 606–615. [Google Scholar] [CrossRef] [PubMed][Green Version]



| Primers | Sequences (5′ to 3′) | Purposes |
|---|---|---|
| vIL8Kan-F | aaaatcggaaaaaaaagtgccttcttttaattacaggaggtagcaattaaaggatgacgacgataagtaggg | Amplification of KanR cassette with MDV sequences flanking vIL8 gene |
| vIL8Kan-R | gatatataatgcagggggtgtgggtttgatgagcagttggggcggcaaaattaattgctacctcctgtaattaaaagaaggcactttttttttccgattttcaaccaattaaccaattctgattag | |
| MeqKan-F | cttgcaggtgtataccagggagaaggcgggcacggtacaggtgtaaagagaggatgacgacgataagtaggg | Amplification of KanR cassette with MDV sequences flanking meq gene |
| MeqKan-R | aacatggggcatagacgatgtgctgctgagagtcacaatgcggatcatcactctttacacctgtaccgtgcccgccttctccctggtatacacctgcaagcaaccaattaaccaattctgattag | |
| vIL8-F | gccaagcttcgaggagtcaaaatcgg | Amplification of vIL8 gene |
| vIL8-R | gccgaattcggtggagacccaataac | |
| Meq-F | ccgcacactgattcctag | Amplification of meq gene |
| Meq-R | ccttatgttgatcttccca | |
| RR-F | ccgcgatcgttaggttgggtatta | Amplification of RR gene |
| RR-R | ccttatgttgatcttccca | |
| ICP4-F | ttattgccccgtactcaccg | Viral genomic copy number detection |
| ICP4-R | catttaaagtctttccatgccaaac | |
| GAPDH-F | gtcaacggatttggccgtat | Viral genomic copy number detection |
| GAPDH-R | ccacttggactttgccagaga |
| Vaccination | Challenge | Ab– | Ab+ | ||
|---|---|---|---|---|---|
| Tumors (%) 1 | PI | Tumors (%) | PI 3 | ||
| CVI988/Rispens | 686 | 3/15 (20) | 80 a | 4/14 (29) | 71 a |
| 686BAC-∆Meq | 686 | 0/13 (0) | 100 a | 1/14 (7) | 93 a |
| 686BAC-∆Meq∆vIL8 | 686 | 0/13 (0) | 100 a | 2/15 (13) | 87 a |
| None | 686 | 12/12 (100) | NA 2 | 13/13 (100) | NA |
| None | None | 0/10 (0) | NA | 0/15 (0) | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, Y.; Reddy, S.M.; Khan, O.A.; Sun, A.; Lupiani, B. A Novel Effective and Safe Vaccine for Prevention of Marek’s Disease Caused by Infection with a Very Virulent Plus (vv+) Marek’s Disease Virus. Vaccines 2021, 9, 159. https://doi.org/10.3390/vaccines9020159
Liao Y, Reddy SM, Khan OA, Sun A, Lupiani B. A Novel Effective and Safe Vaccine for Prevention of Marek’s Disease Caused by Infection with a Very Virulent Plus (vv+) Marek’s Disease Virus. Vaccines. 2021; 9(2):159. https://doi.org/10.3390/vaccines9020159
Chicago/Turabian StyleLiao, Yifei, Sanjay M. Reddy, Owais A. Khan, Aijun Sun, and Blanca Lupiani. 2021. "A Novel Effective and Safe Vaccine for Prevention of Marek’s Disease Caused by Infection with a Very Virulent Plus (vv+) Marek’s Disease Virus" Vaccines 9, no. 2: 159. https://doi.org/10.3390/vaccines9020159
APA StyleLiao, Y., Reddy, S. M., Khan, O. A., Sun, A., & Lupiani, B. (2021). A Novel Effective and Safe Vaccine for Prevention of Marek’s Disease Caused by Infection with a Very Virulent Plus (vv+) Marek’s Disease Virus. Vaccines, 9(2), 159. https://doi.org/10.3390/vaccines9020159






