Virus-like Particle Vaccines: A Prospective Panacea Against an Avian Influenza Panzootic
Abstract
1. Introduction
2. Avian Influenza Viruses (AIVs)
Pathogenicity of AIVs
3. Surveillance of Avian Influenza Viruses (AIVs)
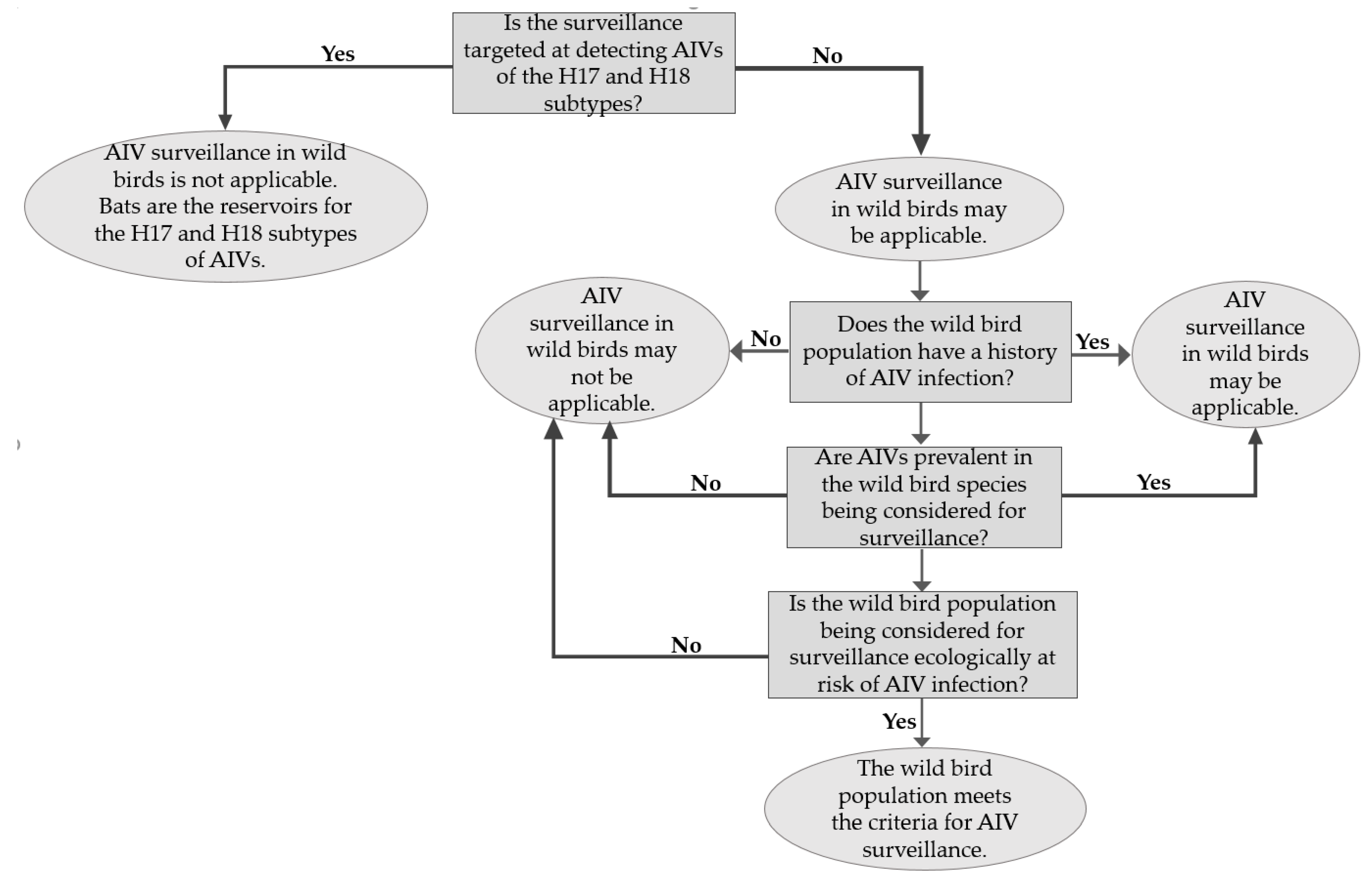
3.1. AIV Surveillance in Wild Birds
3.2. AIV Surveillance in Domestic Birds
3.3. Antiviral Treatment Regimens for Avian Influenza
4. Prevention of a Future Avian Influenza Panzootic
4.1. AI Vaccines
4.2. VLP Vaccine Development Technology
4.2.1. AIV Antigens for VLP Vaccine Development
4.2.2. AI VLP Vaccines
4.3. VLP Vaccines as Universal Avian Influenza Vaccines
4.4. Limitations and Challenges Associated with VLP Vaccines
4.5. Current Trends in AIV VLP Vaccine Development
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lupiani, B.; Reddy, S.M. The history of avian influenza. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Lycett, S.J.; Duchatel, F.; Digard, P. A brief history of bird flu. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180257. [Google Scholar] [CrossRef] [PubMed]
- Joannis, T.M.; Meseko, A.C.; Oladokun, A.T.; Ularamu, H.; Hussaini, G.; Egbuji, A.N.; Solomon, P.; Nyam, D.C.; Gado, A.D.; Luka, P.; et al. Serologic and virologic surveillance of avian influenza in Nigeria, 2006–2007. Eurosurveillance 2008, 42, 1–5. [Google Scholar]
- Biswas, P.K.; Christensen, J.P.; Ahmed, S.S.; Barua, H.; Das, A.; Rahman, M.H.; Giasuddin, M.; Hannan, A.S.; Habib, M.A.; Ahad, A.; et al. Avian influenza outbreaks in chickens, Bangladesh. Emerg. Infect. Dis. 2008, 14, 1909–1912. [Google Scholar] [CrossRef]
- Sockett, P. Avian influenza. Cmaj 2016, 158, 369–407. [Google Scholar]
- Kishida, N.; Sakoda, Y.; Eto, M.; Sunaga, Y.; Kida, H. Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Arch. Virol. 2004, 149, 2095–2104. [Google Scholar] [CrossRef]
- Bano, S.; Naeem, K.; Malik, S.A. Evaluation of pathogenic potential of avian influenza virus serotype H9N2 in chickens. Avian Dis. 2003, 47, 817–822. [Google Scholar] [CrossRef]
- Akanbi, O.B.; Taiwo, V.O. Mortality and pathology associated with highly pathogenic avian influenza H5N1 outbreaks in commercial poultry production systems in Nigeria. Int. Sch. Res. Not. 2014, 1–7. [Google Scholar] [CrossRef]
- Rushton, J.; Viscarra, R.; Guerne Bleich, E.; Mcleod, A. Impact of avian influenza outbreaks in the poultry sectors of five South East Asian countries (Cambodia, Indonesia, Lao PDR, Thailand, Viet Nam) outbreak costs, responses and potential long term control. World’s Poult. Sci. J. 2005, 61, 491–514. [Google Scholar] [CrossRef]
- Fan, X.; Hu, Y.; Zhang, G.; Wang, M. Veterinary influenza vaccines against avian influenza in China. Future Virol. 2015, 10, 585–595. [Google Scholar] [CrossRef]
- Iqbal, M.; Yaqub, T.; Reddy, K.; McCauley, J.W. Novel genotypes of H9N2 influenza a viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS ONE 2009, 4, e5788. [Google Scholar] [CrossRef]
- Lee, D.H.; Torchetti, M.K.; Winker, K.; Ip, H.S.; Song, C.S.; Swayne, D.E. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J. Virol. 2015, 89, 6521–6524. [Google Scholar] [CrossRef] [PubMed]
- Kida, H.; Ito, T.; Yasuda, J.; Shimizu, Y.; Itakura, C.; Shortridge, K.F.; Kawaoka, Y.; Webster, R.G. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 1994, 75, 2183–2188. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Kang, B.; Lee, C.; Jung, K.; Ha, G.; Kang, D.; Park, S.; Park, B.; Oh, J. Transmission of avian influenza virus (H3N2) to dogs. Emerg. Infect. Dis. 2008, 14, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Borkenhagen, L.K.; Salman, M.D.; Ma, M.J.; Gray, G.C. Animal influenza virus infections in humans: A commentary. Int. J. Infect. Dis. 2019, 88, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Yasué, M.; Feare, C.J.; Bennun, L.; Fiedler, W. The epidemiology of H5N1 avian influenza in wild birds: Why we need better ecological data. BioScience 2006, 56, 923. [Google Scholar] [CrossRef]
- Li, K.S.; Guan, Y.; Wang, J.; Smith, G.J.D.; Xu, K.M.; Duan, L.; Rahardjo, A.P.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 2004, 430, 209–213. [Google Scholar] [CrossRef]
- Venkatesh, D.; Poen, M.J.; Bestebroer, T.M.; Scheuer, R.D.; Vuong, O.; Chkhaidze, M.; Machablishvili, A.; Mamuchadze, J.; Ninua, L.; Fedorova, N.B.; et al. Avian Influenza Viruses in Wild Birds: Virus Evolution in a Multihost Ecosystem. J. Virol. 2018, 92, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Munster, V.J.; Wallensten, A.; Baas, C.; Rimmelzwaan, G.F.; Schutten, M.; Olsen, B.; Osterhaus, A.D.; Fouchier, R.A.M. Mallards and highly pathogenic avian influenza ancestral viruses, Northern Europe. Emerg. Infect. Dis. 2005, 11, 1545–1551. [Google Scholar] [CrossRef]
- Machalaba, C.C.; Elwood, S.E.; Forcella, S.; Smith, K.M.; Hamilton, K.; Jebara, K.B.; Swayne, D.E.; Webby, R.J.; Mumford, E.; Mazet, J.A.; et al. Global avian influenza surveillance in wild birds: A strategy to capture viral diversity. Emerg. Infect. Dis. 2015, 21. [Google Scholar] [CrossRef]
- Yong, C.Y.; Yeap, S.K.; Ho, K.L.; Omar, A.R.; Tan, W.S. Potential recombinant vaccine against influenza: A virus based on M2e displayed on nodaviral capsid nanoparticles. Int. J. Nanomed. 2015, 10, 2751–2763. [Google Scholar] [CrossRef]
- Elaish, M.; Xia, M.; Ngunjiri, J.M.; Ghorbani, A.; Jang, H.; Kc, M.; Abundo, M.C.; Dhakal, S.; Renukaradhya, G.J.; Jiang, X.; et al. Protective immunity against influenza virus challenge by norovirus P particle-M2e and HA2-AtCYN vaccines in chickens. Vaccine 2019, 37, 6454–6462. [Google Scholar] [CrossRef] [PubMed]
- Bertran, K.; Criado, M.F.; Lee, D.-H.; Killmaster, L.; Silva, E.M.S.; Lucio, E.; Widener, J.; Pritchard, N.; Atkins, E.; Mebatsion, T.; et al. Protection of white leghorn chickens by recombinant fowlpox vector vaccine with an updated H5 insert against Mexican H5N2 avian influenza viruses. Vaccine 2020, 38, 1526–1534. [Google Scholar] [CrossRef]
- Pu, J.; Wang, S.; Yin, Y.; Zhang, G.; Carter, R.A.; Wang, J.; Xu, G.; Sun, H.; Wang, M.; Wen, C.; et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc. Natl. Acad. Sci. USA 2015, 112, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.H.; Seong, B.L. The quest for a truly universal influenza vaccine. Front. Cell. Infect. Microbiol. 2019, 9, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.; O’Kennedy, M.M.; Wandrag, D.B.R.; Adeyemi, M.; Abolnik, C. Efficacy of a plant-produced virus-like particle vaccine in chickens challenged with Influenza A H6N2 virus. Plant Biotech. J. 2020, 18, 502–512. [Google Scholar] [CrossRef]
- Steel, J.; Lowen, A.C.; Wang, T.T.; Yondola, M.; Gao, Q.; Haye, K.; García-Sastre, A.; Palesea, P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 2010, 1, 1–9. [Google Scholar] [CrossRef]
- Ong, H.K.; Yong, C.Y.; Tan, W.S.; Yeap, S.K.; Omar, A.R.; Razak, M.A.; Ho, K.L. An influenza A vaccine based on the extracellular domain of matrix 2 protein protects BALB/c mice against H1N1 and H3N2. Vaccines 2019, 7, 91. [Google Scholar] [CrossRef]
- Mostafa, A.; Abdelwhab, E.M.; Mettenleiter, T.C.; Pleschka, S. Zoonotic potential of influenza A viruses: A comprehensive overview. Viruses 2018, 10, 497. [Google Scholar] [CrossRef]
- Vandegrift, K.J.; Sokolow, S.H.; Daszak, P.; Kilpatrick, A.M. Ecology of avian influenza viruses in a changing world. Anny. N. Y. Acad. Sci. 2010, 1195, 113–128. [Google Scholar] [CrossRef]
- Kosik, I.; Yewdell, J.W. Influenza hemagglutinin and neuraminidase: Yin–yang proteins coevolving to thwart immunity. Viruses 2019, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Holmes, E.C. Avian influenza virus exhibits rapid evolutionary dynamics. Mol. Biol. Evol. 2006, 23, 2336–2341. [Google Scholar] [CrossRef]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef]
- Killingley, B.; Nguyen-Van-Tam, J. Routes of influenza transmission. Influenza Other Respi. Viruses 2013, 7, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, Y.; Shinya, K.; Deng, G.; Jiang, Y.; Li, Z.; Guan, Y.; Tian, G.; Li, Y.; Shi, J.; et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Qu, N.; Guo, Y.; Cao, L.; Wu, S.; Mei, K.; Sun, H.; Lu, Y.; Qin, Z.; Liao, M.; et al. Phylogeny, pathogenicity, and transmission of H5N1 avian influenza viruses in chickens. Front. Cell. Infect. Microbiol. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Wu, Z.-Q.; Zhang, Y.; Zhao, N.; Yu, Z.; Pan, H.; Chan, T.-C.; Zhang, Z.; Liu, S. Comparative epidemiology of human fatal infections with novel, high (H5N6 and H5N1) and low (H7N9 and H9N2) pathogenicity avian influenza a viruses. Int. J. Environ. Res. Public Health 2017, 14, 263. [Google Scholar] [CrossRef] [PubMed]
- Chatziprodromidou, I.P.; Arvanitidou, M.; Guitian, J.; Apostolou, T.; Vantarakis, G.; Vantarakis, A. Global avian influenza outbreaks 2010–2016: A systematic review of their distribution, avian species and virus subtype. Syst. Rev. 2018, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- OIE World Organisation for Animal Health. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. In OIE Terrestial Manual; OIE World Organisation for Animal Health: Paris, France, 2018; pp. 821–843. [Google Scholar]
- Rimi, N.A.; Hassan, Z.; Chowdhury, S.; Rahman, M.; Sultana, R.; Biswas, P.K.; Debnath, N.C.; Islam, S.S.; Ross, A.G. A decade of avian influenza in Bangladesh: Where are we now? Trop. Med. Infect. Dis. 2019, 4, 119. [Google Scholar] [CrossRef] [PubMed]
- Wan Norulhuda, W.A.W.; Tariq, J. An overview of highly pathogenic avian influenza (H5N1) outbreak cases in Kelantan, West Malaysia in year 2017. MJVR 2018, 9, 102–108. [Google Scholar]
- Sutton, T.C. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses 2018, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.P.; Shu, L.L.; Wright, S.; Bean, W.J.; Sharp, G.B.; Shortridge, K.F.; Webster, R.G. Analysis of the influenza virus gene pool of avian species from Southern China. Virology 1994, 198, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, Y.; Chambers, T.M.; Sladen, W.L.; Gwebster, R. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology 1988, 163, 247–250. [Google Scholar] [CrossRef]
- Okazaki, K.; Takada, A.; Ito, T.; Imai, M.; Takakuwa, H.; Hatta, M.; Ozaki, H.; Tanizaki, T.; Nagano, T.; Ninomiya, A.; et al. Precursor genes of future pandemic influenza viruses are perpetuated in ducks nesting in Siberia. Arch. Virol. 2000, 145, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Belser, J.A.; Tumpey, T.M. Mammalian models for the study of H7 virus pathogenesis and transmission. Curr. Top. Microbiol. Immunol. 2014, 385, 275–305. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Deng, Y.M.; Su, Y.C.F.; Fourment, M.; Iannello, P.; Arzey, G.G.; Hansbro, P.M.; Arzey, K.E.; Kirkland, P.D.; Warner, S. The recent establishment of north American H10 lineage influenza viruses in Australian wild waterfowl and the evolution of Australian avian influenza viruses. J. Virol. 2013, 87, 10182–10189. [Google Scholar] [CrossRef]
- Suguitan, A.L.; Matsuoka, Y.; Lau, Y.F.; Santos, C.P.; Vogel, L.; Cheng, L.I.; Orandle, M.; Subbarao, K. The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J. Virol. 2012, 86, 2706–2714. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza a viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- De Filette, M.; Jou, W.M.; Birkett, A.; Lyons, K.; Schultz, B.; Tonkyro, A.; Resch, S.; Fiers, W. Universal influenza A vaccine: Optimization of M2-based constructs. Virology 2005, 337, 149–161. [Google Scholar] [CrossRef]
- Stallknecht, D.E.; Brown, J.D. Wild birds and the epidemiology of avian influenza. J. Wildl. Dis. 2007, 43, 15–20. [Google Scholar]
- Gomaa, M.R.; Khalil, A.A.; Kandeil, A.; Sabir, J.S.M.; Kayed, A.; Moatasim, Y.; El saied, M.F.; El-safty, M.M.; Kayali, G.; Ali, M.A. Development of an effective contemporary trivalent avian influenza vaccine against circulating H5N1, H5N8, and H9N2 in Egypt. Poult. Sci. 2019, 98, 6289–6295. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Shi, W.; Shi, Y.; Wang, D.; Xiao, H.; Li, W.; Bi, Y.; Wu, Y.; Li, X.; Yan, J.; et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: Phylogenetic, structural, and coalescent analyses. Lancet 2013, 381, 1926–1932. [Google Scholar] [CrossRef]
- Yu, D.; Xiang, G.; Zhu, W.; Lei, X.; Li, B.; Meng, Y.; Yang, L.; Jiao, H.; Li, X.; Huang, W.; et al. The re-emergence of highly pathogenic avian influenza H7N9 viruses in humans in Mainland China, 2019. Eurosurveillance 2019, 24, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Van Gils, J.A.; Munster, V.J.; Radersma, R.; Liefhebber, D.; Fouchier, R.A.M.; Klaassen, M. Hampered foraging and migratory performance in swans infected with low-pathogenic avian influenza A virus. PLoS ONE 2007, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Osterhaus, A.; Fouchier, R.; Rimmelzwaan, G. Towards universal influenza vaccines? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 2766–2773. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Vries, R.D.; Herfst, S.; Richard, M. Avian influenza: A virus pandemic preparedness and vaccine development. Vaccines 2018, 6, 46. [Google Scholar] [CrossRef]
- Wang, G.; Zhan, D.; Li, L.; Lei, F.; Liu, B.; Liu, D.; Xiao, H.; Feng, Y.; Li, J.; Yang, B.; et al. H5N1 avian influenza re-emergence of Lake Qinghai: Phylogenetic and antigenic analyses of the newly isolated viruses and roles of migratory birds in virus circulation. J. Gen. Virol. 2008, 89, 697–702. [Google Scholar] [CrossRef]
- Hoye, B.J.; Munster, V.J.; Nishiura, H.; Klaassen, M. Surveillance of Wild Birds for Avian Influenza Virus. Emerg. Infect. Dis. 2010, 16, 1–18. [Google Scholar] [CrossRef]
- Bergervoet, S.A.; Pritz-Verschuren, S.B.E.; Gonzales, J.L.; Bossers, A.; Poen, M.J.; Dutta, J.; Khan, Z.; Kriti, D.; van Bakel, H.; Bouwstra, R.; et al. Circulation of low pathogenic avian influenza (LPAI) viruses in wild birds and poultry in the Netherlands, 2006–2016. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Fouchier, R.A.M.; Munster, V.J. Epidemiology of low pathogenic avian influenza viruses in wild birds. OIE Revue Scientifique Technique 2009, 28, 49–58. [Google Scholar] [CrossRef]
- Pavade, G.; Weber-vintzel, L.; Hamilton, K.; Dehove, A. OFFLU Review of Avian Influenza Surveillance and Epidemiological Projects in some European, African, and Asian Countries. 2011. Available online: https://rr-africa.oie.int/en/news/offlu-review-of-avian-influenza-surveillance-and-epidemiological-projects/ (accessed on 11 May 2020).
- Guan, Y.; Smith, G.J.D. The emergence and diversification of panzootic H5N1 influenza viruses. Virus Res. 2013, 178, 35–43. [Google Scholar] [CrossRef]
- Teru, V.C.; Manu, S.A.; Ahmed, G.I.; Junaidu, K.; Newman, S.; Nyager, J.; Iwar, V.N.; Mshelbwala, G.M.; Joannis, T.; Maina, J.A.; et al. Situation-based survey of avian influenza viruses in possible “ bridge ” species of wild and domestic birds in Nigeria. Influenza Res. Treatment 2012, 2012, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; Hicks, J.T.; Young, S.G.; Taweel, A.N.; El Kayed, A.S.; Moatasim, Y.; Kutkat, O.; Bagato, O.; Mckenzie, P.P.; Cai, Z.; et al. Active surveillance and genetic evolution of avian influenza viruses in Egypt. Emerg. Microbes Infect. 2019, 8, 1370–1382. [Google Scholar] [CrossRef]
- Rohaim, M.A.; El-Naggar, R.F.; Hamoud, M.M.; Nasr, S.A.; Ismael, E.; Laban, S.E.; Ahmed, H.A.; Munir, M. Re-emergence of a novel H5N1 avian influenza virus variant subclade 2.2.1.1 in Egypt during 2014. Transbound. Emerg. Dis. 2017, 64, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Gurley, E.S.; Gerloff, N.; Rahman, M.Z.; Simpson, N.; Rahman, M.; Haider, M.; Chowdhury, S.; Balish, A.; Zaman, R.U.; et al. Avian influenza surveillance in domestic waterfowl and environment of live bird markets in Bangladesh, 2007–2012. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Hill, E.M.; House, T.; Dhingra, M.S.; Kalpravidh, W.; Morzaria, S.; Osmani, M.G.; Brum, E.; Yamage, M.; Kalam, M.A.; Prosser, D.J.; et al. The impact of surveillance and control on highly pathogenic avian influenza outbreaks in poultry in Dhaka division, Bangladesh. PLoS Comput. Biol. 2018, 14, 1–27. [Google Scholar] [CrossRef]
- Peacock, T.P.; James, J.; Sealy, J.E.; Iqbal, M. A global perspective on H9N2 avian influenza virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef]
- Su, S.; Bi, Y.; Wong, G.; Gray, G.C.; Gao, G.F.; Li, S. Epidemiology, Evolution, and Recent Outbreaks of Avian Influenza Virus in China. J. Virol. 2015, 89, 8671–8676. [Google Scholar] [CrossRef] [PubMed]
- Skalickova, S.; Heger, Z.; Krejcova, L.; Pekarik, V.; Bastl, K.; Janda, J.; Kostolansky, F.; Vareckova, E.; Zitka, O.; Adam, V.; et al. Perspective of use of antiviral peptides against influenza virus. Viruses 2015, 7, 5428–5442. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.M.; Hafez, H.M. Insight into alternative approaches for control of avian influenza in poultry, with emphasis on highly pathogenic H5N1. Viruses 2012, 4, 3179–3208. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.; Narayan, O.; Rouse, B.T. Prevention of malignant avian iniluenza by 1-adamantanamine hydrochloride. Arch. Ges. Virusforsch. 1970, 32, 171–184. [Google Scholar] [CrossRef]
- Beard, C.W.; Brugh, M.; Webster, R.G. Emergence of amantadine-resistant H5N2 avian influenza virus during a simulated layer flock treatment program. Avian Dis. 1987, 31, 533–537. [Google Scholar] [CrossRef]
- Rollinson, E.A. Prospects for antiviral chemotherapy in veterinary medicine: 2. Avian, piscine, canine, porcine, bovine and equine virus diseases. Antivir. Chem. Chemo. 1992, 3, 311–326. [Google Scholar] [CrossRef]
- Meijer, A.; van der Goot, J.A.; Koch, G.; van Boven, M.; Kimman, T.G. Oseltamivir reduces transmission, morbidity, and mortality of highly pathogenic avian influenza in chickens. Int. Cong. 2004, 1263, 495–498. [Google Scholar] [CrossRef]
- Abdelwhab, E.M.; Hafez, H.M. Control of avian influenza in poultry with antivirals and molecular manipulation control of avian influenza in poultry with antivirals and molecular manipulation. In Epidemiology II Theory, Research and Practice Publisher; iConcept Press: Hong Kong, China, 2015; ISBN 978-1-922227-76-8. [Google Scholar]
- Barbour, E.K.; El-hakim, R.G.; Kaadi, M.S.; Shaib, H.A.; Gerges, D.D.; Nehme, P.A. Evaluation of the histopathology of the respiratory system in essential oil-treated broilers following a challenge with Mycoplasma gallisepticum and/or H9N2 influenza virus. Intern. J. Appl. Res. Vet. Med. 2006, 4, 293–300. [Google Scholar]
- Barbour, E.K.; El-hakim, R.G.; Kaadi, M.S.; Shaib, H.A.; Gerges, D.D.; Nehme, P.A. Evaluation of essential oils in the treatment of broilers co-infected with multiple respiratory etiologic agents. Intern. J. Appl. Res. Vet. Med. 2009, 9, 317–323. [Google Scholar]
- Lee, H.J.; Lee, Y.N.; Youn, H.N.; Lee, D.H.; Kwak, J.H.; Seong, B.L.; Lee, J.B.; Park, S.Y.; Choi, I.S.; Song, C.S. Immunology, health, and disease, anti-influenza virus activity of green tea by-products in vitro and efficacy against influenza virus infection in chickens. Poult. Sci. 2012, 91, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.F.; Liang, J.P.; Na, Z.Y.; Yang, H.J.; Lu, Y.; Hua, L.Y.; Guo, W.Z.; Cui, Y.; Wang, L. In vivo inhibition of NAS preparation on H9N2 Subtype AIV. Virol. Sin. 2010, 25, 145–150. [Google Scholar] [CrossRef]
- Ehrhardt, C.; Dudek, S.E.; Holzberg, M.; Urban, S.; Hrincius, E.R.; Haasbach, E.; Seyer, R.; Lapuse, J.; Planz, O.; Ludwig, S. A Plant Extract of Ribes nigrum folium possesses anti-influenza virus activity in vitro and in vivo by preventing virus entry to host cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, Y.; Zhong, S.; Zhao, S.; Zeng, X.; Mo, Z.; Qin, S.; Guan, W.; Li, C.; Zhong, N. In vitro inhibition of influenza virus infection by a crude extract from Isatis indigotica root resulting in the prevention of viral attachment. Mol. Med. Rep. 2012, 5, 793–799. [Google Scholar] [CrossRef]
- OIE. OIE Situation Report for Highly Pathogenic Avian Influenza, Latest Update 31 August 2018; OIE World Organisation for Animal Health: Paris, France, 2018; pp. 1–9. [Google Scholar]
- Swayne, D.E.; Spackman, E. Current status and future needs in diagnostics and vaccines for high pathogenicity avian influenza. Dev. Biol. 2013, 135, 79–94. [Google Scholar] [CrossRef]
- Capua, I.; Alexander, D.J. Avian influenza vaccines and vaccination in birds. Vaccine 2008, 26, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Senne, D.A.; Suarez, D.L. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J. Virol. 2004, 78, 8372–8381. [Google Scholar] [CrossRef] [PubMed]
- Ellis, T.M.; Leung, C.Y.H.C.; Chow, M.K.W.; Bissett, L.A.; Wong, W.; Guan, Y.; Peiris, J.S.M. Vaccination of chickens against H5N1 avian influenza in the face of an outbreak interrupts virus transmission. Avian Pathol. 2004, 33, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Philippa, J.D.W.; Munster, V.J.; Bolhuis, H.; Van Bestebroer, T.M.; Schaftenaar, W.; Beyer, W.E.P.; Fouchier, R.A.M.; Kuiken, T.; Osterhaus, A.D.M.E. Highly pathogenic avian influenza (H7N7): Vaccination of zoo birds and transmission to non-poultry species. Vaccine 2005, 23, 5743–5750. [Google Scholar] [CrossRef]
- Lone, N.A.; Spackman, E.; Kapczynski, D. Immunologic evaluation of 10 different adjuvants for use in vaccines for chickens against highly pathogenic avian influenza virus. Vaccine 2017, 35, 3401–3408. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Ghosh, P.P.; Azim, K.F.; Mukta, S.; Abir, R.A.; Nahar, J.; Hasan Khan, M.M. Reverse vaccinology approach to design a novel multi-epitope subunit vaccine against avian influenza A (H7N9) virus. Microb. Pathog. 2019, 130, 19–37. [Google Scholar] [CrossRef]
- Tian, G.; Zhang, S.; Li, Y.; Bu, Z.; Liu, P.; Zhou, J.; Li, C.; Shi, J.; Yu, K.; Chen, H. Protective efficacy in chickens, geese and ducks of an H5N1-inactivated vaccine developed by reverse genetics. Virology 2005, 341, 153–162. [Google Scholar] [CrossRef]
- Swayne, D.E.; Beck, J.R.; Kinney, N. Failure of a recombinant fowl poxvirus vaccine containing an avian influenza hemagglutinin gene to provide consistent protection against influenza in chickens preimmunized with a fowl pox vaccine. Avian Dis. 2000, 44, 132. [Google Scholar] [CrossRef]
- Roldão, A.; Mellado, M.C.M.; Castilho, L.R.; Carrondo, M.J.T.; Alves, P.M. Virus-like particles in vaccine development. Expert Rev. Vaccines 2010, 9, 1149–1176. [Google Scholar] [CrossRef]
- Kumar, A.; Meldgaard, T.S.; Bertholet, S. Novel platforms for the development of a universal influenza vaccine. Front. Immunol. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Noad, R.; Roy, P. Virus-like particles as immunogens. Trends Microbiol. 2003, 11, 438–444. [Google Scholar] [CrossRef]
- Pushko, P.; Pumpens, P.; Grens, E. Development of virus-like particle technology from small highly symmetric to large complex virus-like particle structures. Intervirology 2013, 56, 141–165. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Song, J.M.; Quan, F.S.; Compans, R.W. Influenza vaccines based on virus-like particles. Virus Res. 2009, 143, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction between virus-like particles (VLPs) and pattern recognition receptors (PRRs) from dendritic cells (DCs): Toward better engineering of VLPs. Front. Immunol. 2020, 11, 1–22. [Google Scholar] [CrossRef]
- Krammer, F. The quest for a universal flu vaccine: Headless HA 2.0. Cell Host Microbe 2015, 18, 395–397. [Google Scholar] [CrossRef]
- Ninyio, N.N.; Ho, K.L.; Ong, H.K.; Yong, C.Y.; Chee, H.Y.; Hamid, M.; Tan, W.S. Immunological analysis of the hepatitis b virus ‘a’ determinant displayed on chimeric virus-like particles of Macrobrachium rosenbergii nodavirus capsid protein produced in Sf9 cells. Vaccines 2020, 8, 275. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Gamvrellis, A.; Leong, D.; Hanley, J.C.; Xiang, S.D.; Mottram, P.; Plebanski, M. Vaccines that facilitate antigen entry into dendritic cells. Immunol. Cell Biol. 2004, 82, 506–516. [Google Scholar] [CrossRef]
- Hütter, J.; Rödig, J.V.; Höper, D.; Seeberger, P.H.; Reichl, U.; Rapp, E.; Lepenies, B. Toward animal cell culture–based influenza vaccine design: Viral hemagglutinin N-glycosylation markedly impacts immunogenicity. J. Immunol. 2013, 190, 220–230. [Google Scholar] [CrossRef]
- Thompson, C.M.; Petiot, E.; Mullick, A.; Aucoin, M.G.; Henry, O.; Kamen, A.A. Critical assessment of influenza VLP production in Sf9 and HEK293 expression systems. BMC Biotechnol. 2015, 15, 31. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, J.; Rapp, E.; Reichl, U. N-glycan analysis by CGE-LIF: Profiling influenza A virus hemagglutinin N-glycosylation during vaccine production. Electrophoresis 2008, 29, 4203–4214. [Google Scholar] [CrossRef] [PubMed]
- Ravin, N.V.; Kotlyarov, R.Y.; Mardanova, E.S.; Kuprianov, V.V.; Migunov, A.I.; Stepanova, L.A.; Tsybalova, L.M.; Kiselev, O.I.; Skryabin, K.G. Plant-produced recombinant influenza vaccine based on virus-like HBc particles carrying an extracellular domain of M2 protein. Biochemistry 2012, 77, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.J.; Landry, N.; Trépanier, S.; Mercier, G.; Dargis, M.; Couture, M.; D’Aoust, M.A.; Vézina, L.P. Human antibody response to N-glycans present on plant-made influenza virus-like particle (VLP) vaccines. Vaccine 2014, 32, 6098–6106. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; McCullers, J.A.; Alymova, I.; Parsons, L.M.; Cipollo, J.F. Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J. Proteome Res. 2015, 14, 3957–3969. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liang, L.; Wang, S.; Nakao, T.; Li, Y.; Liu, L.; Guan, Y.; Fukuyama, S.; Bu, Z.; Kawaoka, Y.; et al. Glycosylation of the hemagglutinin protein of H5N1 influenza virus increases its virulence in mice by exacerbating the host immune response. J. Virol. 2017, 91, 1–14. [Google Scholar] [CrossRef]
- Hyakumura, M.; Walsh, R.; Thaysen-Andersen, M.; Kingston, N.J.; La, M.; Lu, L.; Lovrecz, G.; Packer, N.H.; Locarnini, S.; Netter, H.J. Modification of asparagine-linked glycan density for the design of hepatitis B virus virus-like particles with enhanced immunogenicity. J. Virol. 2015, 89, 11312–11322. [Google Scholar] [CrossRef]
- Wu, C.Y.; Yeh, Y.C.; Yang, Y.C.; Chou, C.; Liu, M.T.; Wu, H.S.; Chan, J.T.; Hsiao, P.W. Mammalian expression of virus-like particles for advanced mimicry of authentic influenza virus. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Buffin, S.; Peubez, I.; Barrière, F.; Nicolaï, M.C.; Tapia, T.; Dhir, V.; Forma, E.; Sève, N.; Legastelois, I. Influenza A and B virus-like particles produced in mammalian cells are highly immunogenic and induce functional antibodies. Vaccine 2019, 37, 6857–6867. [Google Scholar] [CrossRef]
- D’Aoust, M.A.; Couture, M.M.J.; Charland, N.; Trépanier, S.; Landry, N.; Ors, F.; Vézina, L.P. The production of hemagglutinin-based virus-like particles in plants: A rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 2010, 8, 607–619. [Google Scholar] [CrossRef]
- Takeyama, N.; Kiyono, H.; Yuki, Y. Plant-based vaccines for animals and humans: Recent advances in technology and clinical trials. Ther. Adv. Vaccines 2015, 3, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Mena, J.A.; Kamen, A.A. Insect cell technology is a versatile and robust vaccine manufacturing platform. Expert Rev. Vaccines 2011, 10, 1063–1081. [Google Scholar] [CrossRef] [PubMed]
- Yee, C.M.; Zak, A.J.; Hill, B.D.; Wen, F. The Coming Age of Insect Cells for Manufacturing and Development of Protein Therapeutics. Ind. Eng. Chem. Res. 2018, 57, 10061–10070. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Park, J.K.; Lee, Y.N.; Song, J.M.; Kang, S.M.; Lee, J.B.; Park, S.Y.; Choi, I.S.; Song, C.S. H9N2 avian influenza virus-like particle vaccine provides protective immunity and a strategy for the differentiation of infected from vaccinated animals. Vaccine 2011, 29, 4003–4007. [Google Scholar] [CrossRef]
- McCraw, D.M.; Gallagher, J.R.; Torian, U.; Myers, M.L.; Conlon, M.T.; Gulati, N.M.; Harris, A.K. Structural analysis of influenza vaccine virus-like particles reveals a multicomponent organization. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.B.; Van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Van Meersbergen, R.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306. [Google Scholar] [CrossRef]
- Bose, S.; Zokarkar, A.; Welch, B.D.; Leser, G.P.; Jardetzky, T.S.; Lamb, R.A. Fusion activation by a headless parainfluenza virus 5 hemagglutinin- neuraminidase stalk suggests a modular mechanism for triggering. Proc. Natl. Acad. Sci. USA 2012, 109, 2625–2634. [Google Scholar] [CrossRef]
- Fiers, W.; De Filette, M.; El Bakkouri, K.; Schepens, B.; Roose, K.; Schotsaert, M.; Birkett, A.; Saelens, X. M2e-based universal influenza A vaccine. Vaccine 2009, 27, 6280–6283. [Google Scholar] [CrossRef] [PubMed]
- Mezhenskaya, D.; Isakova-Sivak, I.; Rudenko, L. M2e-based universal influenza vaccines: A historical overview and new approaches to development. J. Biomed. Sci. 2019, 26, 1–15. [Google Scholar] [CrossRef]
- Beljanski, V.; Chiang, C.; Kirchenbaum, G.A.; Olagnier, D.; Bloom, C.E.; Wong, T.; Haddad, E.K.; Trautmann, L.; Ross, T.M.; Hiscott, J. Enhanced influenza virus-like particle vaccination with a structurally optimized RIG-I agonist as adjuvant. J. Virol. 2015, 89, 10612–10624. [Google Scholar] [CrossRef]
- Khalaj-Hedayati, A.; Chua, C.L.L.; Smooker, P.; Lee, K.W. Nanoparticles in influenza subunit vaccine development: Immunogenicity enhancement. Influenza Other Respi. Viruses 2020, 14, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Chu, K.B.; Lee, D.H.; Lee, S.H.; Park, B.R.; Kim, M.C.; Kang, S.M.; Quan, F.S. Influenza M2 virus-like particle vaccination enhances protection in combination with avian influenza HA VLPs. PLoS ONE 2018, 14, 1–14. [Google Scholar] [CrossRef]
- Crisci, E.; Bárcena, J.; Montoya, M. Virus-like particles: The new frontier of vaccines for animal viral infections. Vet. Immunol. Immunopathol. 2012, 148, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Morón, V.G.; Rueda, P.; Sedlik, C.; Leclerc, C. In vivo, dendritic cells can cross-present virus-like particles using an endosome-to-cytosol pathway. J. Immunol. 2003, 171, 2242–2250. [Google Scholar] [CrossRef]
- Guillén, G.; Aguilar, J.C.; Dueñas, S.; Hermida, L.; Guzmán, M.G.; Penton, E.; Iglesias, E.; Junco, J.; Torrens, I.; Iglesias, E.; et al. Virus-like particles as vaccine antigens and adjuvants: Application to chronic disease, cancer immunotherapy and infectious disease preventive strategies. Procedia Vaccinol. 2010, 2, 128–133. [Google Scholar] [CrossRef]
- Fifis, T.; Gamvrellis, A.; Crimeen-Irwin, B.; Pietersz, G.A.; Li, J.; Mottram, P.L.; McKenzie, I.F.C.; Plebanski, M. Size-dependent immunogenicity: Therapeutic and protective properties of nano-vaccines against tumors. J. Immunol. 2004, 173, 3148–3154. [Google Scholar] [CrossRef]
- Nayak, D.P.; Hui, E.K.W.; Barman, S. Assembly and budding of influenza virus. Vir. Res. 2004, 106, 147–165. [Google Scholar] [CrossRef]
- Pielak, R.M.; Chou, J.J. Influenza M2 proton channels. Biochim. Biophys. Acta 2011, 1808, 522–529. [Google Scholar] [CrossRef]
- Lamb, R.A.; Lai, C.; Choppin, P.W. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: Colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. USA 1981, 78, 4170–4174. [Google Scholar] [CrossRef] [PubMed]
- Mozdzanowska, K.; Maiese, K.; Furchner, M.; Gerhard, W. Treatment of influenza virus-infected SCID mice with nonneutralizing antibodies specific for the transmembrane proteins matrix 2 and neuraminidase reduces the pulmonary virus titer but fails to clear the infection. Virology 1999, 146, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Liang, X.; Horton, M.S.; Perry, H.C.; Citron, M.P.; Heidecker, G.J.; Fu, T.M.; Joyce, J.; Przysiecki, C.T.; Keller, P.M.; et al. Preclinical study of influenza virus a M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 2004, 22, 2993–3003. [Google Scholar] [CrossRef] [PubMed]
- De Filette, M.; Ramne, A.; Birkett, A.; Lycke, N.; Löwenadler, B.; Min Jou, W.; Saelens, X.; Fiers, W. The universal influenza vaccine M2e-HBc administered intranasally in combination with the adjuvant CTA1-DD provides complete protection. Vaccine 2006, 24, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Elaish, M.; Kang, K.I.; Xia, M.; Ali, A.; Shany, S.A.S.; Wang, L.; Jiang, X.; Lee, C.W. Immunogenicity and protective efficacy of the norovirus P particle-M2e chimeric vaccine in chickens. Vaccine 2015, 33, 4901–4909. [Google Scholar] [CrossRef] [PubMed]
- Matić, S.; Rinaldi, R.; Masenga, V.; Noris, E. Efficient production of chimeric Human papillomavirus 16 L1 protein bearing the M2e influenza epitope in Nicotiana benthamiana plants. BMC Biotechnol. 2011, 11, 1–12. [Google Scholar] [CrossRef]
- Denis, J.; Acosta-Ramirez, E.; Zhao, Y.; Hamelin, M.E.; Koukavica, I.; Baz, M.; Abed, Y.; Savard, C.; Pare, C.; Lopez Macias, C.; et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine 2008, 26, 3395–3403. [Google Scholar] [CrossRef]
- Petukhova, N.; Gasanova, T.; Stepanova, L.; Rusova, O.; Potapchuk, M.; Korotkov, A.; Jiang, X.; Atabekov, J. Immunogenicity and protective efficacy of candidate universal influenza A nanovaccines produced in plants by tobacco mosaic virus-based vectors. Curr. Pharm. Des. 2013, 19, 5587–5600. [Google Scholar] [CrossRef]
- Tissot, A.C.; Renhofa, R.; Schmitz, N.; Cielens, I.; Meijerink, E.; Ose, V.; Jennings, G.T.; Saudan, P.; Pumpens, P.; Bachmann, M.F. Versatile Virus-Like Particle Carrier for Epitope Based Vaccines. PLoS ONE 2010, 5, e9809. [Google Scholar] [CrossRef]
- Pascual, E.; Mata, C.P.; Gómez-Blanco, J.; Moreno, N.; Bárcena, J.; Blanco, E.; Rodríguez-Frandsen, A.; Nieto, A.; Carrascosa, J.L.; Castón, J.R. Structural basis for the development of avian virus capsids that display influenza virus proteins and induce protective immunity. J Virol. 2015, 89, 2563–2574. [Google Scholar] [CrossRef] [PubMed]
- Moreno, N.; Mena, I.; Angulo, I.; Gómez, Y.; Crisci, E.; Montoya, M.; Castón, J.R.; Blanco, E.; Bárcena, J. Rabbit hemorrhagic disease virus capsid, a versatile platform for foreign B-cell epitope display inducing protective humoral immune responses. Sci Rep. 2016, 6, 31844. [Google Scholar] [CrossRef] [PubMed]
- Graves, P.N.; Schulman, J.L.; Young, J.F.; Palese, P. Preparation of influenza virus subviral particles lacking the HA1 subunit of hemagglutinin: Unmasking of cross-reactive HA2 determinants. Virology 1983, 126, 106–116. [Google Scholar] [CrossRef]
- Erbelding, E.J.; Post, D.; Stemmy, E.; Roberts, P.C.; Deckhut, A.; Ferguson, A.S.; Paules, C.I.; Graham, B.S.; Fauci, E.E.; Anthony, S.; et al. A universal influenza vaccine: The strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Vareckova, E.; Mucha, V.; Ciampor, F.; Betakova, T.; Russ, G. Monoclonal antibodies demonstrate accessible HA2 epitopes in minor subpopulation of native influenza virus haemagglutinin molecules. Arch. Virol. 1993, 130, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Neirynck, S.; Deroo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Huber, V.C.; Lynch, J.M.; Bucher, D.J.; Le, J.; Metzger, D.W. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 2001, 166, 7381–7388. [Google Scholar] [CrossRef]
- Jegerlehner, A.; Schmitz, N.; Storni, T.; Bachmann, M.F. Influenza A vaccine based on the extracellular domain of M2: Weak protection mediated via antibody-dependent nk cell activity. J. Immunol. 2004, 172, 5598–5605. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Moki, T.; Takizawa, T.; Shiratsuchi, A.; Nakanishi, Y. Evidence for phagocytosis of Influenza Virus-Infected, Apoptotic Cells by Neutrophils and Macrophages in Mice. J. Immunol. 2007, 178, 2448–2457. [Google Scholar] [CrossRef]
- Guilliams, M.; Bruhns, P.; Saeys, Y.; Hammad, H.; Lambrecht, B.N. The function of Fcγ receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 2014, 14, 94–108. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, K.; Ko, E.; Kim, M.; Lee, Y.; Jung, Y.; Lee, Y.; Kwon, Y.; Song, J.; Kanga, S. Complement C3 plays a key role in inducing humoral and cellular immune responses to influenza virus strain-specific hemagglutinin-based or cross- protective M2 extracellular domain-based vaccination. J. Virol. 2018, 92, 1–14. [Google Scholar] [CrossRef]
- Rajão, D.S.; Pérez, D.R. Universal vaccines and vaccine platforms to protect against influenza viruses in humans and agriculture. Front. Microb. 2018, 9, 1–21. [Google Scholar] [CrossRef]
- Jegerlehner, A.; Wiesel, M.; Dietmeier, K.; Zabel, F.; Gatto, D.; Saudan, P.; Bachmann, M.F. Carrier induced epitopic suppression of antibody responses induced by virus-like particles is a dynamic phenomenon caused by carrier-specific antibodies. Vaccine 2010, 28, 5503–5512. [Google Scholar] [CrossRef]
- Pride, M.W.; Shi, H.; Anchin, J.M.; Scott, L.D.; LoVerde, P.T.; Thakur, A.; Thanavala, Y. Molecular mimicry of hepatitis B surface antigen by an anti-idiotype-derived synthetic peptide. Proc. Natl. Acad. Sci. USA 1992, 89, 11900–11904. [Google Scholar] [CrossRef] [PubMed]
- Schädlich, L.; Senger, T.; Kirschning, C.J.; Müller, M.; Gissmann, L. Refining HPV 16 L1 purification from E. coli: Reducing endotoxin contaminations and their impact on immunogenicity. Vaccine 2009, 27, 1511–1522. [Google Scholar] [CrossRef]
- Fuenmayor, J.; Gòdia, F.; Cervera, L. Production of virus-like particles for vaccines. New Biotechnol. 2017, 39, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Vicente, T.; Roldão, A.; Peixoto, C.; Carrondo, M.J.T.; Alves, P.M. Large-scale production and purification of VLP-based vaccines. J. Invertebr. Pathol. 2011, 107, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Chroboczek, J.; Szurgot, I.; Szolajska, E. Virus-like particles as vaccine. Acta Biochim. Pol. 2014, 61, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Manolova, V.; Flace, A.; Bauer, M.; Schwarz, K.; Saudan, P.; Bachmann, M.F. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008, 38, 1404–1413. [Google Scholar] [CrossRef]
- Wang, B.Z.; Quan, F.S.; Kang, S.M.; Bozja, J.; Skountzou, I.; Compans, R.W. Incorporation of membrane-anchored flagellin into influenza virus-like particles enhances the breadth of immune responses. J. Virol. 2008, 82, 11813–11823. [Google Scholar] [CrossRef]
- Sesterhenn, F.; Yang, C.; Bonet, J.; Cramer, J.T.; Wen, X.; Wang, Y.; Chiang, C.I.; Abriata, L.A.; Kucharska, I.; Castoro, G.; et al. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 2020, 368, 1–10. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ninyio, N.N.; Ho, K.L.; Omar, A.R.; Tan, W.S.; Iqbal, M.; Mariatulqabtiah, A.R. Virus-like Particle Vaccines: A Prospective Panacea Against an Avian Influenza Panzootic. Vaccines 2020, 8, 694. https://doi.org/10.3390/vaccines8040694
Ninyio NN, Ho KL, Omar AR, Tan WS, Iqbal M, Mariatulqabtiah AR. Virus-like Particle Vaccines: A Prospective Panacea Against an Avian Influenza Panzootic. Vaccines. 2020; 8(4):694. https://doi.org/10.3390/vaccines8040694
Chicago/Turabian StyleNinyio, Nathaniel Nyakaat, Kok Lian Ho, Abdul Rahman Omar, Wen Siang Tan, Munir Iqbal, and Abdul Razak Mariatulqabtiah. 2020. "Virus-like Particle Vaccines: A Prospective Panacea Against an Avian Influenza Panzootic" Vaccines 8, no. 4: 694. https://doi.org/10.3390/vaccines8040694
APA StyleNinyio, N. N., Ho, K. L., Omar, A. R., Tan, W. S., Iqbal, M., & Mariatulqabtiah, A. R. (2020). Virus-like Particle Vaccines: A Prospective Panacea Against an Avian Influenza Panzootic. Vaccines, 8(4), 694. https://doi.org/10.3390/vaccines8040694






