Attenuated Salmonella enterica Serovar Typhimurium, Strain NC983, Is Immunogenic, and Protective against Virulent Typhimurium Challenges in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Bacterial Growth and Preparation of Cell Suspensions
2.3. Animals
2.4. Determination of Dose Required to Kill 50% of Mice (LD50)
2.5. Fitness of NC983 In Vivo
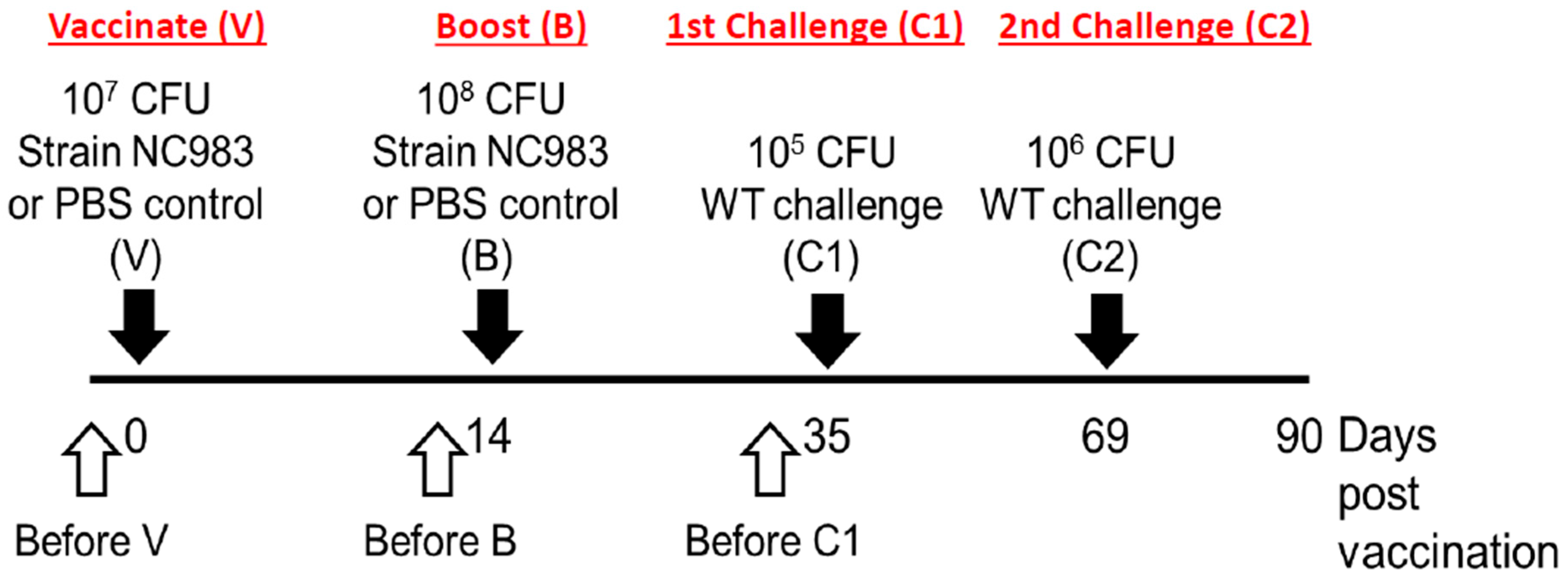
2.6. Vaccination and Challenge Protocols
2.7. Measurement of the Anti-Salmonella IgG Response by Elisa
2.8. Measurement of the Anti-Salmonella IgG Response by Immunoblot
2.9. Statistical Analysis
2.10. Ethics Statement
3. Results
3.1. Strain NC983 Exhibits a Fitness Defect in the Colonization of the Liver and Spleen
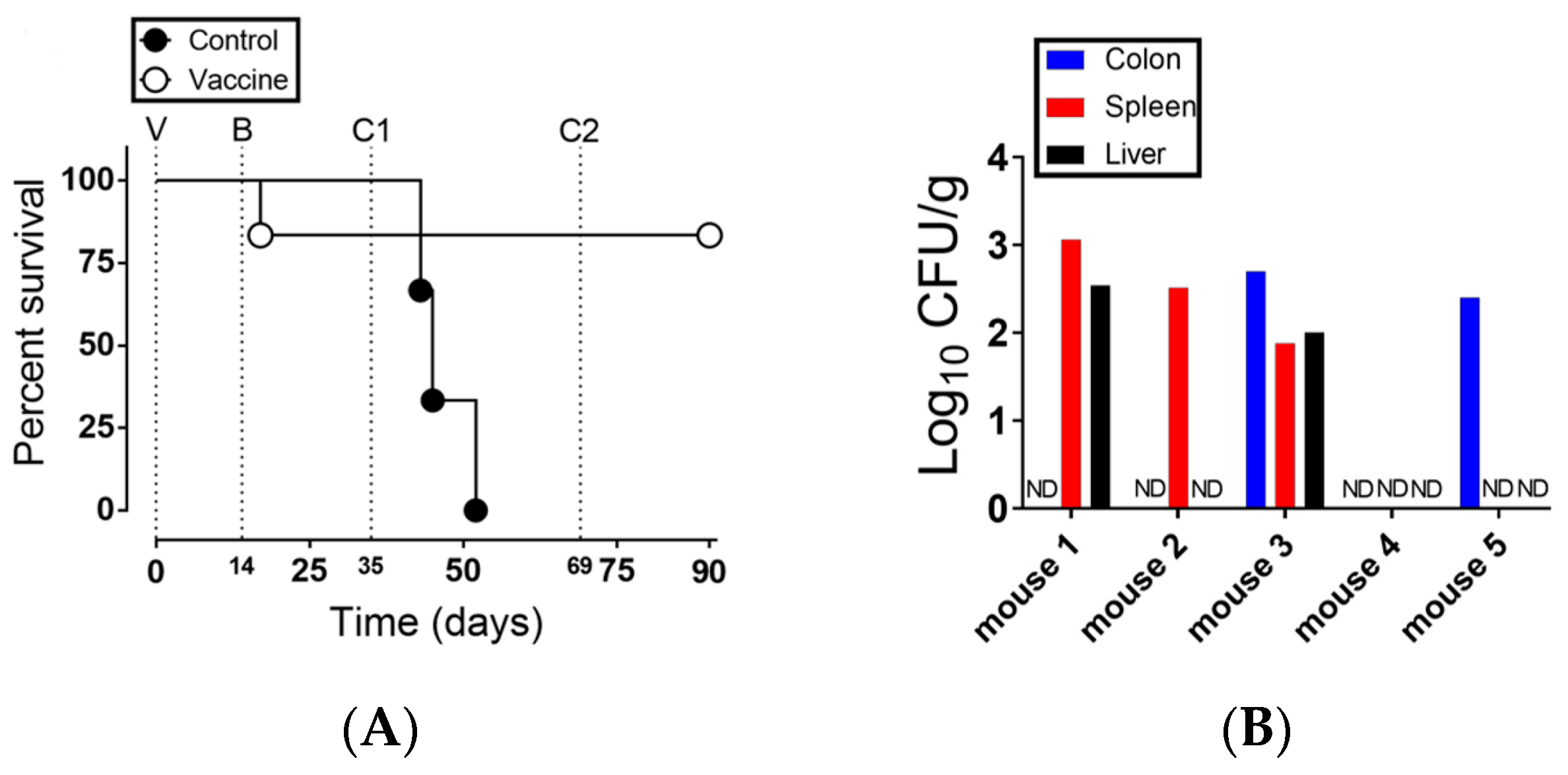
3.2. Strain NC983 Is a Live Attenuated Salmonella Strain that Protects against Virulent S. typhimurium and Is Immunogenic in Mice
4. Discussion
5. Patents
Author Contributions
Funding
Conflicts of Interest
Availability of Data and Materials
References
- CDC. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2010. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 41–47. [Google Scholar]
- Parry, C.M.; Thomas, S.; Aspinall, E.J.; Cooke, R.P.; Rogerson, S.J.; Harries, A.D.; Beeching, N.J. A retrospective study of secondary bacteraemia in hospitalised adults with community acquired non-typhoidal Salmonella gastroenteritis. BMC Infect. Dis. 2013, 13, 107. [Google Scholar] [CrossRef]
- Crump, J.A.; Sjolund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, Clinical Presentation, Laboratory Diagnosis, Antimicrobial Resistance, and Antimicrobial Management of Invasive Salmonella Infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef]
- Medalla, F.; Hoekstra, R.M.; Whichard, J.M.; Barzilay, E.J.; Chiller, T.M.; Joyce, K.; Rickert, R.; Krueger, A.; Stuart, A.; Griffin, P.M. Increase in resistance to ceftriaxone and nonsusceptibility to ciprofloxacin and decrease in multidrug resistance among Salmonella strains, United States, 1996–2009. Foodborne Pathog. Dis. 2013, 10, 302–309. [Google Scholar] [CrossRef]
- Glenn, L.M.; Lindsey, R.L.; Folster, J.P.; Pecic, G.; Boerlin, P.; Gilmour, M.W.; Harbottle, H.; Zhao, S.; McDermott, P.F.; Fedorka-Cray, P.J.; et al. Antimicrobial resistance genes in multidrug-resistant Salmonella enterica isolated from animals, retail meats, and humans in the United States and Canada. Microb. Drug Resist. 2013, 19, 175–184. [Google Scholar] [CrossRef]
- Germanier, R. Immunity in experimental salmonellosis. 3. Comparative immunization with viable and heat-inactivated cells of Salmonella typhimurium. Infect. Immun. 1972, 5, 792–797. [Google Scholar] [CrossRef]
- Hoiseth, S.K.; Stocker, B.A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 1981, 291, 238–239. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.P.; Reina-Guerra, M.; Hoiseth, S.K.; Stocker, B.A.; Habasha, F.; Johnson, E.; Merritt, F. Aromatic-dependent Salmonella typhimurium as modified live vaccines for calves. Am. J. Vet. Res. 1984, 45, 59–66. [Google Scholar]
- Smith, B.P.; Reina-Guerra, M.; Stocker, B.A.; Hoiseth, S.K.; Johnson, E. Aromatic-dependent Salmonella dublin as a parenteral modified live vaccine for calves. Am. J. Vet. Res. 1984, 45, 2231–2235. [Google Scholar]
- Mukkur, T.K.; McDowell, G.H.; Stocker, B.A.; Lascelles, A.K. Protection against experimental salmonellosis in mice and sheep by immunisation with aromatic-dependent Salmonella typhimurium. J. Med. Microbiol. 1987, 24, 11–19. [Google Scholar] [CrossRef]
- Nnalue, N.A.; Stocker, B.A. Test of the virulence and live-vaccine efficacy of auxotrophic and galE derivatives of Salmonella choleraesuis. Infect. Immun. 1987, 55, 955–962. [Google Scholar] [CrossRef]
- Edwards, M.F.; Stocker, B.A. Construction of delta aroA his delta pur strains of Salmonella typhi. J. Bacteriol. 1988, 170, 3991–3995. [Google Scholar] [CrossRef]
- Levine, M.M.; Herrington, D.; Murphy, J.R.; Morris, J.G.; Losonsky, G.; Tall, B.; Lindberg, A.A.; Svenson, S.; Baqar, S.; Edwards, M.F.; et al. Safety, infectivity, immunogenicity, and in vivo stability of two attenuated auxotrophic mutant strains of Salmonella typhi, 541Ty and 543Ty, as live oral vaccines in humans. J. Clin. Investig. 1987, 79, 888–902. [Google Scholar] [CrossRef]
- Formal, S.B.; Baron, L.S.; Kopecko, D.J.; Washington, O.; Powell, C.; Life, C.A. Construction of a potential bivalent vaccine strain: Introduction of Shigella sonnei form I antigen genes into the galE Salmonella typhi Ty21a typhoid vaccine strain. Infect. Immun. 1981, 34, 746–750. [Google Scholar] [CrossRef]
- Clements, J.D.; El-Morshidy, S. Construction of a potential live oral bivalent vaccine for typhoid fever and cholera-Escherichia coli-related diarrheas. Infect. Immun. 1984, 46, 564–569. [Google Scholar] [CrossRef]
- Baron, L.S.; Kopecko, D.J.; Formal, S.B.; Seid, R.; Guerry, P.; Powell, C. Introduction of Shigella flexneri 2a type and group antigen genes into oral typhoid vaccine strain Salmonella typhi Ty21a. Infect. Immun. 1987, 55, 2797–2801. [Google Scholar] [CrossRef]
- Hone, D.; Attridge, S.; van den Bosch, L.; Hackett, J. A chromosomal integration system for stabilization of heterologous genes in Salmonella based vaccine strains. Microb. Pathog. 1988, 5, 407–418. [Google Scholar] [CrossRef]
- Ji, Z.; Shang, J.; Li, Y.; Wang, S.; Shi, H. Live attenuated Salmonella enterica serovar Choleraesuis vaccine vector displaying regulated delayed attenuation and regulated delayed antigen synthesis to confer protection against Streptococcus suis in mice. Vaccine 2015, 33, 4858–4867. [Google Scholar] [CrossRef]
- Laniewski, P.; Kuczkowski, M.; Chrzastek, K.; Wozniak, A.; Wyszynska, A.; Wieliczko, A.; Jagusztyn-Krynicka, E.K. Evaluation of the immunogenicity of Campylobacter jejuni CjaA protein delivered by Salmonella enterica sv. Typhimurium strain with regulated delayed attenuation in chickens. World J. Microbiol. Biotechnol. 2014, 30, 281–292. [Google Scholar] [CrossRef]
- Jiang, Y.; Mo, H.; Willingham, C.; Wang, S.; Park, J.Y.; Kong, W.; Roland, K.L.; Curtiss, R., 3rd. Protection against Necrotic Enteritis in Broiler Chickens by Regulated Delayed Lysis Salmonella Vaccines. Avian Dis. 2015, 59, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Rodriguez, M.D.; Yang, J.; Kader, R.; Alamuri, P.; Curtiss, R., 3rd; Clark-Curtiss, J.E. Live attenuated Salmonella vaccines displaying regulated delayed lysis and delayed antigen synthesis to confer protection against Mycobacterium tuberculosis. Infect. Immun. 2012, 80, 815–831. [Google Scholar] [CrossRef]
- Wang, S.; Li, Y.; Scarpellini, G.; Kong, W.; Shi, H.; Baek, C.H.; Gunn, B.; Wanda, S.Y.; Roland, K.L.; Zhang, X.; et al. Salmonella vaccine vectors displaying delayed antigen synthesis in vivo to enhance immunogenicity. Infect. Immun. 2010, 78, 3969–3980. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Scarpellini, G.; Gunn, B.; Xin, W.; Wanda, S.Y.; Roland, K.L.; Curtiss, R., 3rd. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc. Natl. Acad. Sci. USA 2009, 106, 593–598. [Google Scholar] [CrossRef]
- Kong, Q.; Liu, Q.; Jansen, A.M.; Curtiss, R., 3rd. Regulated delayed expression of rfc enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Vaccine 2010, 28, 6094–6103. [Google Scholar] [CrossRef]
- Shi, H.; Wang, S.; Roland, K.L.; Gunn, B.M.; Curtiss, R., 3rd. Immunogenicity of a live recombinant Salmonella enterica serovar typhimurium vaccine expressing pspA in neonates and infant mice born from naive and immunized mothers. Clin. Vaccine Immunol. 2010, 17, 363–371. [Google Scholar] [CrossRef]
- Curtiss, R., 3rd; Wanda, S.Y.; Gunn, B.M.; Zhang, X.; Tinge, S.A.; Ananthnarayan, V.; Mo, H.; Wang, S.; Kong, W. Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo. Infect. Immun. 2009, 77, 1071–1082. [Google Scholar] [CrossRef]
- Moustafa, D.A.; Scarff, J.M.; Garcia, P.P.; Cassidy, S.K.; DiGiandomenico, A.; Waag, D.M.; Inzana, T.J.; Goldberg, J.B. Recombinant Salmonella Expressing Burkholderia mallei LPS O Antigen Provides Protection in a Murine Model of Melioidosis and Glanders. PLoS ONE 2015, 10, e0132032. [Google Scholar] [CrossRef]
- Micoli, F.; Rondini, S.; Alfini, R.; Lanzilao, L.; Necchi, F.; Negrea, A.; Rossi, O.; Brandt, C.; Clare, S.; Mastroeni, P.; et al. Comparative immunogenicity and efficacy of equivalent outer membrane vesicle and glycoconjugate vaccines against nontyphoidal Salmonella. Proc. Natl. Acad. Sci. USA 2018, 115, 10428–10433. [Google Scholar] [CrossRef]
- Fass, E.; Groisman, E.A. Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 2009, 12, 199–204. [Google Scholar] [CrossRef]
- Grille, S.; Moreno, M.; Bascuas, T.; Marques, J.M.; Munoz, N.; Lens, D.; Chabalgoity, J.A. Salmonella enterica serovar Typhimurium immunotherapy for B-cell lymphoma induces broad anti-tumour immunity with therapeutic effect. Immunology 2014, 143, 428–437. [Google Scholar] [CrossRef]
- Grille, S.; Moreno, M.; Brugnini, A.; Lens, D.; Chabalgoity, J.A. A therapeutic vaccine using Salmonella-modified tumor cells combined with interleukin-2 induces enhanced antitumor immunity in B-cell lymphoma. Leuk. Res. 2013, 37, 341–348. [Google Scholar] [CrossRef]
- Arrach, N.; Cheng, P.; Zhao, M.; Santiviago, C.A.; Hoffman, R.M.; McClelland, M. High-throughput screening for salmonella avirulent mutants that retain targeting of solid tumors. Cancer Res. 2010, 70, 2165–2170. [Google Scholar] [CrossRef]
- Arrach, N.; Zhao, M.; Porwollik, S.; Hoffman, R.M.; McClelland, M. Salmonella promoters preferentially activated inside tumors. Cancer Res. 2008, 68, 4827–4832. [Google Scholar] [CrossRef] [PubMed]
- Pawelek, J.M.; Sodi, S.; Chakraborty, A.K.; Platt, J.T.; Miller, S.; Holden, D.W.; Hensel, M.; Low, K.B. Salmonella pathogenicity island-2 and anticancer activity in mice. Cancer Gene Ther. 2002, 9, 813–818. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xiong, G.; Husseiny, M.I.; Song, L.; Erdreich-Epstein, A.; Shackleford, G.M.; Seeger, R.C.; Jackel, D.; Hensel, M.; Metelitsa, L.S. Novel cancer vaccine based on genes of Salmonella pathogenicity island 2. Int. J. Cancer 2010, 126, 2622–2634. [Google Scholar] [CrossRef] [PubMed]
- Manuel, E.R.; Blache, C.A.; Paquette, R.; Kaltcheva, T.I.; Ishizaki, H.; Ellenhorn, J.D.; Hensel, M.; Metelitsa, L.; Diamond, D.J. Enhancement of cancer vaccine therapy by systemic delivery of a tumor-targeting Salmonella-based STAT3 shRNA suppresses the growth of established melanoma tumors. Cancer Res. 2011, 71, 4183–4191. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Hegazy, W.A.; Guo, L.; Gao, X.; Courtney, A.N.; Kurbanov, S.; Liu, D.; Tian, G.; Manuel, E.R.; Diamond, D.J.; et al. Effective cancer vaccine platform based on attenuated salmonella and a type III secretion system. Cancer Res. 2014, 74, 6260–6270. [Google Scholar] [CrossRef]
- Frahm, M.; Felgner, S.; Kocijancic, D.; Rohde, M.; Hensel, M.; Curtiss, R., 3rd; Erhardt, M.; Weiss, S. Efficiency of conditionally attenuated Salmonella enterica serovar Typhimurium in bacterium-mediated tumor therapy. MBio 2015, 6. [Google Scholar] [CrossRef]
- Available online: http://www.cdc.gov/salmonella/ (accessed on 21 September 2020).
- NASS/USDA. Chicken and Eggs 2011 Summary; USDA: Washington, DC, USA, 2012. [Google Scholar]
- ERS/USDA Data Foodborne Illness Cost Calculator. Available online: http://webarchives.cdlib.org/sw1rf5mh0k/http://www.ers.usda.gov/Data/FoodborneIllness/ (accessed on 21 September 2020).
- Galen, J.E.; Curtiss, R., 3rd. The delicate balance in genetically engineering live vaccines. Vaccine 2014, 32, 4376–4385. [Google Scholar] [CrossRef]
- Bochner, B.R.; Huang, H.C.; Schieven, G.L.; Ames, B.N. Positive selection for loss of tetracycline resistance. J. Bacteriol. 1980, 143, 926–933. [Google Scholar] [CrossRef]
- Fink, R.C.; Evans, M.R.; Porwollik, S.; Vazquez-Torres, A.; Jones-Carson, J.; Troxell, B.; Libby, S.J.; McClelland, M.; Hassan, H.M. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 2007, 189, 2262–2273. [Google Scholar] [CrossRef]
- Troxell, B.; Petri, N.; Daron, C.; Pereira, R.; Mendoza, M.; Hassan, H.M.; Koci, M.D. Poultry body temperature contributes to invasion control through reduced expression of Salmonella pathogenicity island 1 genes in Salmonella enterica serovars Typhimurium and Enteritidis. Appl. Environ. Microbiol. 2015, 81, 8192–8201. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Finney, D.J. Probit Analysis: A Statistical Treatment of the Sigmoid Response Curve; Cambridge University Press: Cambridge, UK, 1952. [Google Scholar]
- Freter, R.; O’Brien, P.C.; Macsai, M.S. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: In vivo studies. Infect. Immun. 1981, 34, 234–240. [Google Scholar] [CrossRef]
- Ullman-Cullere, M.H.; Foltz, C.J. Body condition scoring: A rapid and accurate method for assessing health status in mice. Lab. Anim. Sci. 1999, 49, 319–323. [Google Scholar]
- Troxell, B.; Ye, M.; Yang, Y.; Carrasco, S.E.; Lou, Y.; Yang, X.F. Manganese and zinc regulate virulence determinants in Borrelia burgdorferi. Infect. Immun. 2013, 81, 2743–2752. [Google Scholar] [CrossRef]
- Troxell, B.; Xu, H.; Yang, X.F. Borrelia burgdorferi, a pathogen that lacks iron, encodes manganese-dependent superoxide dismutase essential for resistance to streptonigrin. J. Biol. Chem. 2012, 287, 19284–19293. [Google Scholar] [CrossRef]
- Graham, S.M. Nontyphoidal salmonellosis in Africa. Curr. Opin. Infect. Dis. 2010, 23, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.A. Invasive nontyphoidal Salmonella disease: Epidemiology, pathogenesis and diagnosis. Curr. Opin. Infect. Dis. 2011, 24, 484–489. [Google Scholar] [CrossRef]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg. Infect. Dis. 2015, 21. [Google Scholar] [CrossRef]
- Feasey, N.A.; Dougan, G.; Kingsley, R.A.; Heyderman, R.S.; Gordon, M.A. Invasive non-typhoidal salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 2012, 379, 2489–2499. [Google Scholar] [CrossRef]
- Porwollik, S.; Santiviago, C.A.; Cheng, P.; Long, F.; Desai, P.; Fredlund, J.; Srikumar, S.; Silva, C.A.; Chu, W.; Chen, X.; et al. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS ONE 2014, 9, e99820. [Google Scholar] [CrossRef]
- Jarvik, T.; Smillie, C.; Groisman, E.A.; Ochman, H. Short-term signatures of evolutionary change in the Salmonella enterica serovar typhimurium 14028 genome. J. Bacteriol. 2010, 192, 560–567. [Google Scholar] [CrossRef]
- Troxell, B.; Fink, R.C.; Dickey, A.N.; Scholl, E.H.; Hassan, H.M. Complete Genome Sequence of NC983, a Live Attenuated Strain of Salmonella enterica Serovar Typhimurium. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Fields, P.I.; Swanson, R.V.; Haidaris, C.G.; Heffron, F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 1986, 83, 5189–5193. [Google Scholar] [CrossRef] [PubMed]
- Baumler, A.J.; Kusters, J.G.; Stojiljkovic, I.; Heffron, F. Salmonella typhimurium loci involved in survival within macrophages. Infect. Immun. 1994, 62, 1623–1630. [Google Scholar] [CrossRef]
- Lindgren, S.W.; Stojiljkovic, I.; Heffron, F. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 1996, 93, 4197–4201. [Google Scholar] [CrossRef]
- Buchmeier, N.A.; Heffron, F. Induction of Salmonella stress proteins upon infection of macrophages. Science 1990, 248, 730–732. [Google Scholar] [CrossRef] [PubMed]
- Souwer, Y.; Griekspoor, A.; Jorritsma, T.; de Wit, J.; Janssen, H.; Neefjes, J.; van Ham, S.M. B cell receptor-mediated internalization of salmonella: A novel pathway for autonomous B cell activation and antibody production. J. Immunol. 2009, 182, 7473–7481. [Google Scholar] [CrossRef] [PubMed]
- Nauciel, C. Role of CD4+ T cells and T-independent mechanisms in acquired resistance to Salmonella typhimurium infection. J. Immunol. 1990, 145, 1265–1269. [Google Scholar]
- Crotzer, V.L.; Matute, J.D.; Arias, A.A.; Zhao, H.; Quilliam, L.A.; Dinauer, M.C.; Blum, J.S. Cutting edge: NADPH oxidase modulates MHC class II antigen presentation by B cells. J. Immunol. 2012, 189, 3800–3804. [Google Scholar] [CrossRef]
- Nanton, M.R.; Way, S.S.; Shlomchik, M.J.; McSorley, S.J. Cutting edge: B cells are essential for protective immunity against Salmonella independent of antibody secretion. J. Immunol. 2012, 189, 5503–5507. [Google Scholar] [CrossRef]
- Toso, J.F.; Gill, V.J.; Hwu, P.; Marincola, F.M.; Restifo, N.P.; Schwartzentruber, D.J.; Sherry, R.M.; Topalian, S.L.; Yang, J.C.; Stock, F.; et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol. 2002, 20, 142–152. [Google Scholar] [CrossRef]
- Tacket, C.O.; Hone, D.M.; Curtiss, R., 3rd; Kelly, S.M.; Losonsky, G.; Guers, L.; Harris, A.M.; Edelman, R.; Levine, M.M. Comparison of the safety and immunogenicity of delta aroC delta aroD and delta cya delta crp Salmonella typhi strains in adult volunteers. Infect. Immun. 1992, 60, 536–541. [Google Scholar] [CrossRef]
- Tacket, C.O.; Sztein, M.B.; Wasserman, S.S.; Losonsky, G.; Kotloff, K.L.; Wyant, T.L.; Nataro, J.P.; Edelman, R.; Perry, J.; Bedford, P.; et al. Phase 2 clinical trial of attenuated Salmonella enterica serovar typhi oral live vector vaccine CVD 908-htrA in U.S. volunteers. Infect. Immun. 2000, 68, 1196–1201. [Google Scholar] [CrossRef]
- Galen, J.E.; Buskirk, A.D.; Tennant, S.M.; Pasetti, M.F. Live Attenuated Human Salmonella Vaccine Candidates: Tracking the Pathogen in Natural Infection and Stimulation of Host Immunity. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Fritz, S.E.; Henson, M.S.; Greengard, E.; Winter, A.L.; Stuebner, K.M.; Yoon, U.; Wilk, V.L.; Borgatti, A.; Augustin, L.B.; Modiano, J.F.; et al. A phase I clinical study to evaluate safety of orally administered, genetically engineered Salmonella enterica serovar Typhimurium for canine osteosarcoma. Vet. Med. Sci. 2016, 2, 179–190. [Google Scholar] [CrossRef]
- McWhorter, A.R.; Chousalkar, K.K. A Long-Term Efficacy Trial of a Live, Attenuated Salmonella Typhimurium Vaccine in Layer Hens. Front. Microbiol. 2018, 9, 1380. [Google Scholar] [CrossRef]
- Ku, Y.W.; McDonough, S.P.; Palaniappan, R.U.; Chang, C.F.; Chang, Y.F. Novel attenuated Salmonella enterica serovar Choleraesuis strains as live vaccine candidates generated by signature-tagged mutagenesis. Infect. Immun. 2005, 73, 8194–8203. [Google Scholar] [CrossRef]




| Strain | Genotype a | Source |
|---|---|---|
| Salmonella enterica serovar Typhimurium 14028s | Wild-Type | ATCC b |
| NC983 | Fusaric Acid Resistant | [44] |
| NC1040 | ATCC 14028s fnr’:ha (Kan R) | [45] |
| NC1189 | ATCC 14028s (Rif R) | This study |
| NC1190 | NC983 (Rif R) | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troxell, B.; Mendoza, M.; Ali, R.; Koci, M.; Hassan, H. Attenuated Salmonella enterica Serovar Typhimurium, Strain NC983, Is Immunogenic, and Protective against Virulent Typhimurium Challenges in Mice. Vaccines 2020, 8, 646. https://doi.org/10.3390/vaccines8040646
Troxell B, Mendoza M, Ali R, Koci M, Hassan H. Attenuated Salmonella enterica Serovar Typhimurium, Strain NC983, Is Immunogenic, and Protective against Virulent Typhimurium Challenges in Mice. Vaccines. 2020; 8(4):646. https://doi.org/10.3390/vaccines8040646
Chicago/Turabian StyleTroxell, Bryan, Mary Mendoza, Rizwana Ali, Matthew Koci, and Hosni Hassan. 2020. "Attenuated Salmonella enterica Serovar Typhimurium, Strain NC983, Is Immunogenic, and Protective against Virulent Typhimurium Challenges in Mice" Vaccines 8, no. 4: 646. https://doi.org/10.3390/vaccines8040646
APA StyleTroxell, B., Mendoza, M., Ali, R., Koci, M., & Hassan, H. (2020). Attenuated Salmonella enterica Serovar Typhimurium, Strain NC983, Is Immunogenic, and Protective against Virulent Typhimurium Challenges in Mice. Vaccines, 8(4), 646. https://doi.org/10.3390/vaccines8040646







