Identification and Analysis of Unstructured, Linear B-Cell Epitopes in SARS-CoV-2 Virion Proteins for Vaccine Development
Abstract
1. Introduction
2. Materials and Methods
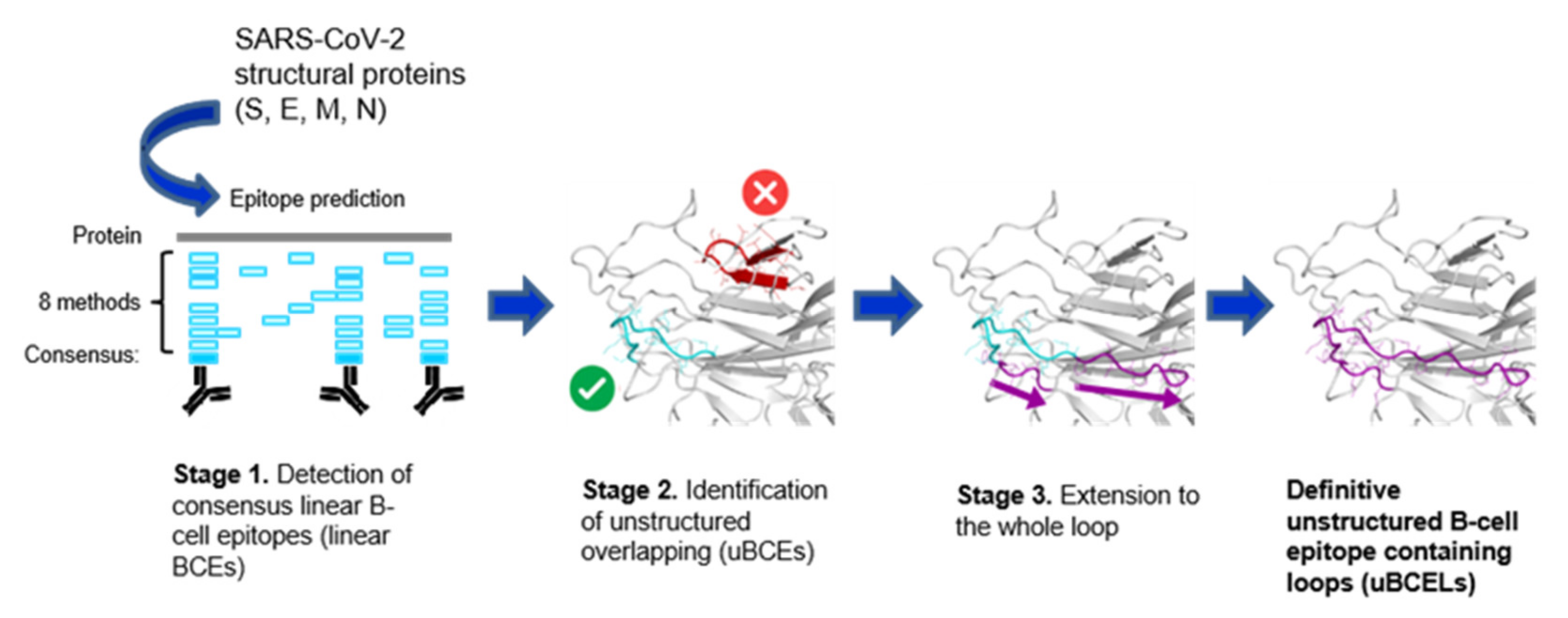
2.1. Identification of uBCELs
2.2. Structural and Accessory Analysis of Protein Antigens
2.3. Phylogenetic Analyses
2.4. Epitope Collection in SARS-CoV-2
2.5. Statistical Analyses
3. Results
3.1. SARS-CoV-2 Epitope Catalogue
3.2. Unstructured Epitope Selection to Design Antigenic Peptides and Chimera Proteins
3.3. S Protein uBCEL Analysis
3.4. E Protein Epitope Analysis
3.5. M Protein Epitope Analysis
3.6. N Protein Epitope Analysis
3.7. Assessment of the Agreement between uBCELs in SARS-CoV-2 and Linear B-Cell Epitopes Previously Reported for SARS-CoV
3.8. Epitope Conservation in Bat Coronaviruses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BCE | B-cell epitope |
| BCEH | B-cell epitope in an alpha-helix section |
| E | envelope protein |
| IEDB | immune epitope database |
| M | membrane protein |
| N | nucleocapsid protein |
| S | spike protein |
| SARS-CoV | Severe Acute Respiratory Syndrome Coronavirus |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| SDP | specificity determining position |
| TMH | transmembrane helix |
| uBCE | unstructured B-cell epitope |
| uBCEL | unstructured B-cell epitope-containing loop |
| UPR | unfolded protein response |
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.K.J.; Shan, J. Novel coronavirus: Where we are and what we know. Infection 2019. [Google Scholar] [CrossRef]
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of viruses from a high proportion of patients with colds. Lancet 1966, 1, 76–77. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Peiris, J.S.; Guan, Y.; Yuen, K.Y. Severe acute respiratory syndrome. Nat. Med. 2004, 10, S88–S97. [Google Scholar] [CrossRef]
- Assiri, A.; McGeer, A.; Perl, T.M.; Price, C.S.; Al Rabeeah, A.A.; Cummings, D.A.; Alabdullatif, Z.N.; Assad, M.; Almulhim, A.; Makhdoom, H.; et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013, 369, 407–416. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.Y.; Guo, D.Y. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- He, Y.X.; Li, J.J.; Du, L.Y.; Yan, X.X.; Hu, G.G.; Zhou, Y.S.; Jiang, S.B. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: Revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine 2006, 24, 5498–5508. [Google Scholar] [CrossRef]
- Lien, S.P.; Shih, Y.P.; Chen, H.W.; Tsai, J.P.; Leng, C.H.; Lin, M.H.; Lin, L.H.; Liu, H.Y.; Chou, A.H.; Chang, Y.W.; et al. Identification of synthetic vaccine candidates against SARS CoV infection. Biochem. Biophys. Res. Commun. 2007, 358, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Hua, R.H.; Zhou, Y.J.; Wang, Y.F.; Hua, Y.Z.; Tong, G.Z. Identification of two antigenic epitopes on SARS-CoV spike protein. Biochem. Biophys. Res. Commun. 2004, 319, 929–935. [Google Scholar] [CrossRef]
- Wang, X.H.; Xu, W.; Tong, D.Y.; Ni, J.; Gao, H.F.; Wang, Y.; Chu, Y.W.; Li, P.P.; Yang, X.M.; Xiong, S.D. A chimeric multi-epitope DNA vaccine elicited specific antibody response against severe acute respiratory syndrome-associated coronavirus which attenuated the virulence of SARS-CoV in vitro. Immunol. Lett. 2008, 119, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.F.; Liang, L.H.; She, M.; Liao, X.L.; Gu, J.; Li, Y.H.; Han, Z.C. Production of a monoclonal antibody against SARS-CoV spike protein with single intrasplenic immunization of plasmid DNA. Immunol. Lett. 2005, 100, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 019–1182. [Google Scholar] [CrossRef]
- Guo, J.P.; Petric, M.; Campbell, W.; McGeer, P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology 2004, 324, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Y.; Qi, J.; Chu, F.; Wu, H.; Gao, F.; Li, T.; Yan, J.; Gao, G.F. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010, 202, 1171–1180. [Google Scholar] [CrossRef]
- Chow, S.C.S.; Ho, C.Y.S.; Tam, T.T.Y.; Wu, C.; Cheung, T.; Chan, P.K.S.; Ng, M.H.L.; Hui, P.K.; Ng, H.K.; Au, D.M.Y.; et al. Specific epitopes of the structural and hypothetical proteins elicit variable humoral responses in SARS patients. J. Clin. Pathol. 2006, 59, 468–476. [Google Scholar] [CrossRef]
- Qian, C.; Qin, D.; Tang, Q.; Zeng, Y.; Tang, G.X.; Lu, C. Identification of a B-cell antigenic epitope at the N-terminus of SARS-CoV M protein and characterization of monoclonal antibody against the protein. Virus Genes 2006, 33, 147–156. [Google Scholar] [CrossRef]
- He, Y.X.; Zhou, Y.S.; Siddiqui, P.; Niu, J.K.; Jiang, S.B. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 2005, 43, 3718–3726. [Google Scholar] [CrossRef]
- Kannan, S.; Ali, P.S.S.; Sheeza, A.; Hemalatha, K. COVID-19 (Novel Coronavirus 2019)—Recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2006–2011. [Google Scholar] [PubMed]
- Cheung, Y.K.; Cheng, S.C.; Sin, F.W.; Chan, K.T.; Xie, Y. Induction of T-cell response by a DNA vaccine encoding a novel HLA-A*0201 severe acute respiratory syndrome coronavirus epitope. Vaccine 2007, 25, 6070–6077. [Google Scholar] [CrossRef] [PubMed]
- Bussmann, B.M.; Reiche, S.; Jacob, L.H.; Braun, J.M.; Jassoy, C. Antigenic and cellular localisation analysis of the severe acute respiratory syndrome coronavirus nucleocapsid protein using monoclonal antibodies. Virus Res. 2006, 122, 119–126. [Google Scholar] [CrossRef]
- Shin, G.C.; Chung, Y.S.; Kim, I.S.; Cho, H.W.; Kang, C. Preparation and characterization of a novel monoclonal antibody specific to severe acute respiratory syndrome-coronavirus nucleocapsid protein. Virus Res. 2006, 122, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Che, X.Y.; Hao, W.; Wang, Y.; Di, B.; Yin, K.; Xu, Y.C.; Feng, C.S.; Wan, Z.Y.; Cheng, V.C.; Yuen, K.Y. Nucleocapsid protein as early diagnostic marker for SARS. Emerg. Infect. Dis. 2004, 10, 1947–1949. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Guo, S.; Sun, J.; Tan, L.; Lu, C.; Ma, Z. Advances in In-silico B-cell Epitope Prediction. Curr. Top. Med. Chem. 2019, 19, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Sher, G.; Zhi, D.; Zhang, S. DRREP: Deep ridge regressed epitope predictor. BMC Genom. 2017, 18, 55–65. [Google Scholar] [CrossRef]
- Van Regenmortel, M.H.V. Mapping Epitope Structure and Activity: From One-Dimensional Prediction to Four-Dimensional Description of Antigenic Specificity. Methods 1996, 9, 465–472. [Google Scholar] [CrossRef]
- Carpentier, G.S.; Fleury, M.J.J.; Touzé, A.; Sadeyen, J.R.; Tourne, S.; Sizaret, P.Y.; Coursaget, P. Mutations on the FG Surface Loop of Human Papillomavirus Type 16 Major Capsid Protein Affect Recognition by Both Type-Specific Neutralizing Antibodies and Cross-Reactive Antibodies. J. Med. Virol. 2005, 77, 558–565. [Google Scholar] [CrossRef][Green Version]
- Qu, P.; Zhang, C.; Li, M.; Ma, W.; Xiong, P.; Liu, Q.; Zou, G.; Lavillette, D.; Yin, F.; Jin, X.; et al. A New Class of Broadly Neutralizing Antibodies That Target the Glycan Loop of Zika Virus Envelope Protein. Cell Discov. 2020, 6, 5. [Google Scholar] [CrossRef]
- Xu, L.; Zheng, Q.; Li, S.; He, M.; Wu, Y.; Li, Y.; Zhu, R.; Yu, H.; Hong, Q.; Jiang, J.; et al. Atomic Structures of Coxsackievirus A6 and Its Complex With a Neutralizing Antibody. Nat. Commun. 2017, 8, 505. [Google Scholar] [CrossRef] [PubMed]
- Potocnakova, L.; Bhide, M.; Pulzova, L.B. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J. Immunol. Res. 2016, 2016, 6760830. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jiang, L.F.; Fang, D.Y.; Yan, H.J.; Zhou, J.J.; Zhou, J.M.; Liang, Y.; Gao, Y.; Zhao, W.; Long, B.G. Selection of SARS-Coronavirus-specific B cell epitopes by phage peptide library screening and evaluation of the immunological effect of epitope-based peptides on mice. Virology 2007, 359, 264–274. [Google Scholar] [CrossRef] [PubMed]
- He, Y.X.; Li, J.J.; Heck, S.; Lustigman, S.; Jiang, S.B. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: Implication for vaccine design. J. Virol. 2006, 80, 5757–5767. [Google Scholar] [CrossRef]
- Hu, H.B.; Li, L.; Kao, R.Y.; Kou, B.B.; Wang, Z.G.; Zhang, L.; Zhang, H.Y.; Hao, Z.Y.; Tsui, W.H.; Ni, A.P.; et al. Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J. Comb. Chem. 2005, 7, 648–656. [Google Scholar] [CrossRef]
- Lu, W.; Wu, X.D.; De Shi, M.; Yang, R.F.; He, Y.Y.; Bian, C.; Shi, T.L.; Yang, S.; Zhu, X.L.; Jiang, W.H.; et al. Synthetic peptides derived from SARS coronavirus S protein with diagnostic and therapeutic potential. FEBS Lett. 2005, 579, 2130–2136. [Google Scholar] [CrossRef]
- Rubinchik, E.; Chow, A.W. Recombinant expression and neutralizing activity of an MHC class II binding epitope of toxic shock syndrome toxin-1. Vaccine 2000, 18, 2312–2320. [Google Scholar] [CrossRef]
- Qin, E.; Zhu, Q.; Yu, M.; Fan, B.; Chang, G.; Si, B.; Yang, B.; Peng, W.; Jiang, T.; Liu, B.; et al. A complete sequence and comparative analysis of a SARS-associated virus (Isolate BJ01). Chin. Sci. Bull. 2003, 48, 941–948. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef]
- Kumar, S.; Maurya, V.K.; Prasad, A.K.; Bhatt, M.L.B.; Saxena, S.K. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virusdisease 2020, 31, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Tilocca, B.; Soggiu, A.; Musella, V.; Britti, D.; Sanguinetti, M.; Urbani, A.; Roncada, P. Molecular basis of COVID-19 relationships in different species: A one health perspective. Microbes Infect. 2020, 22, 218–220. [Google Scholar] [CrossRef]
- Zheng, M.; Song, L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell. Mol. Immunol. 2020, 17, 536–538. [Google Scholar] [CrossRef] [PubMed]
- Baruah, V.; Bose, S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020, 92, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Robson, B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 2020, 119, 26. [Google Scholar] [CrossRef]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 2020, 12, 30166–30169. [Google Scholar] [CrossRef]
- Shang, W.; Yang, Y.; Rao, Y.; Rao, X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. Npj Vaccines 2020, 5, 1–3. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elfiky, A.A. GRP78: A cell’s response to stress. Life Sci. 2019, 226, 156–163. [Google Scholar] [CrossRef]
- Marra, M.A.; Jones, S.J.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome sequence of the SARS-associated coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chang, C.K.; Chang, Y.W.; Sue, S.C.; Bai, H.I.; Riang, L.; Hsiao, C.D.; Huang, T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007, 368, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Buus, S.; Rockberg, J.; Forsstrom, B.; Nilsson, P.; Uhlen, M.; Schafer-Nielsen, C. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol. Cell. Proteom. 2012, 11, 1790–1800. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, O.V.; Mironov, A.A.; Gelfand, M.S.; Rakhmaninova, A.B. Automated selection of positions determining functional specificity of proteins by comparative analysis of orthologous groups in protein families. Protein Sci. 2004, 13, 443–456. [Google Scholar] [CrossRef]
- Goo, L.; DeMaso, C.R.; Pelc, R.S.; Ledgerwood, J.E.; Graham, B.S.; Kuhn, R.J.; Pierson, T.C. The Zika virus envelope protein glycan loop regulates virion antigenicity. Virology 2018, 515, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Zhu, C.; Ai, L.; He, T.; Wang, Y.; Ye, F.; Yang, L.; Ding, C.; Zhu, X.; Lv, R.; et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microbes Infect. 2018, 7, 154. [Google Scholar] [CrossRef]
- Ng, O.W.; Keng, C.T.; Leung, C.S.W.; Peiris, J.S.M.; Poon, L.L.M.; Tan, Y.J. Substitution at Aspartic Acid 1128 in the SARS Coronavirus Spike Glycoprotein Mediates Escape from a S2 Domain-Targeting Neutralizing Monoclonal Antibody. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef]
- Li, Z.; Song, S.; He, M.; Wang, D.; Shi, J.; Liu, X.; Li, Y.; Chi, X.; Wei, S.; Yang, Y.; et al. Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity. Nat. Commun. 2018, 9, 018–07199. [Google Scholar] [CrossRef]
- Walls, A.C.; Tortorici, M.A.; Snijder, J.; Xiong, X.; Bosch, B.J.; Rey, F.A.; Veesler, D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA 2017, 114, 11157–11162. [Google Scholar] [CrossRef]
- Du, L.; Tai, W.; Yang, Y.; Zhao, G.; Zhu, Q.; Sun, S.; Liu, C.; Tao, X.; Tseng, C.K.; Perlman, S.; et al. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat. Commun. 2016, 7, 13473. [Google Scholar] [CrossRef]
- Enjuanes, L.; Zuniga, S.; Castano-Rodriguez, C.; Gutierrez-Alvarez, J.; Canton, J.; Sola, I. Molecular Basis of Coronavirus Virulence and Vaccine Development. Adv. Virus Res. 2016, 96, 245–286. [Google Scholar] [PubMed]
- Wrapp, D.; Wang, N.S.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Kumar, R.; Deshpande, S.; Samal, S.; Shrivastava, T.; Boliar, S.; Bansal, M.; Chaudhary, N.K.; Srikrishnan, A.K.; Murugavel, K.G.; et al. Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop. J. Virol. 2016, 90, 3446–3457. [Google Scholar] [CrossRef] [PubMed]
- Plant, E.P.; Manukyan, H.; Sanchez, J.L.; Laassri, M.; Ye, Z. Immune Pressure on Polymorphous Influenza B Populations Results in Diverse Hemagglutinin Escape Mutants and Lineage Switching. Vaccines 2020, 8, 125. [Google Scholar] [CrossRef]
- Walls, A.C.; Tortorici, M.A.; Frenz, B.; Snijder, J.; Li, W.; Rey, F.A.; DiMaio, F.; Bosch, B.J.; Veesler, D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016, 23, 899–905. [Google Scholar] [CrossRef]
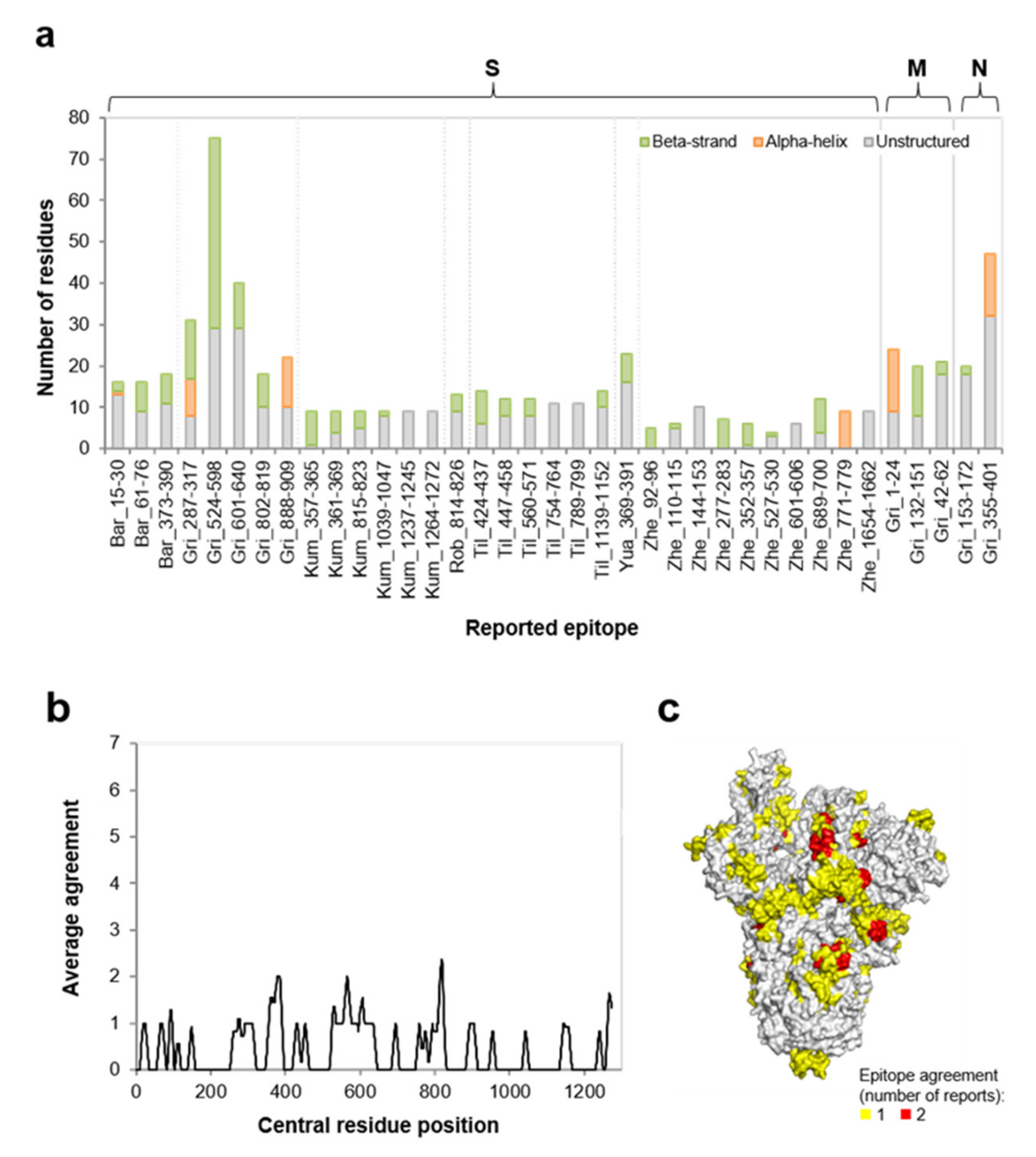
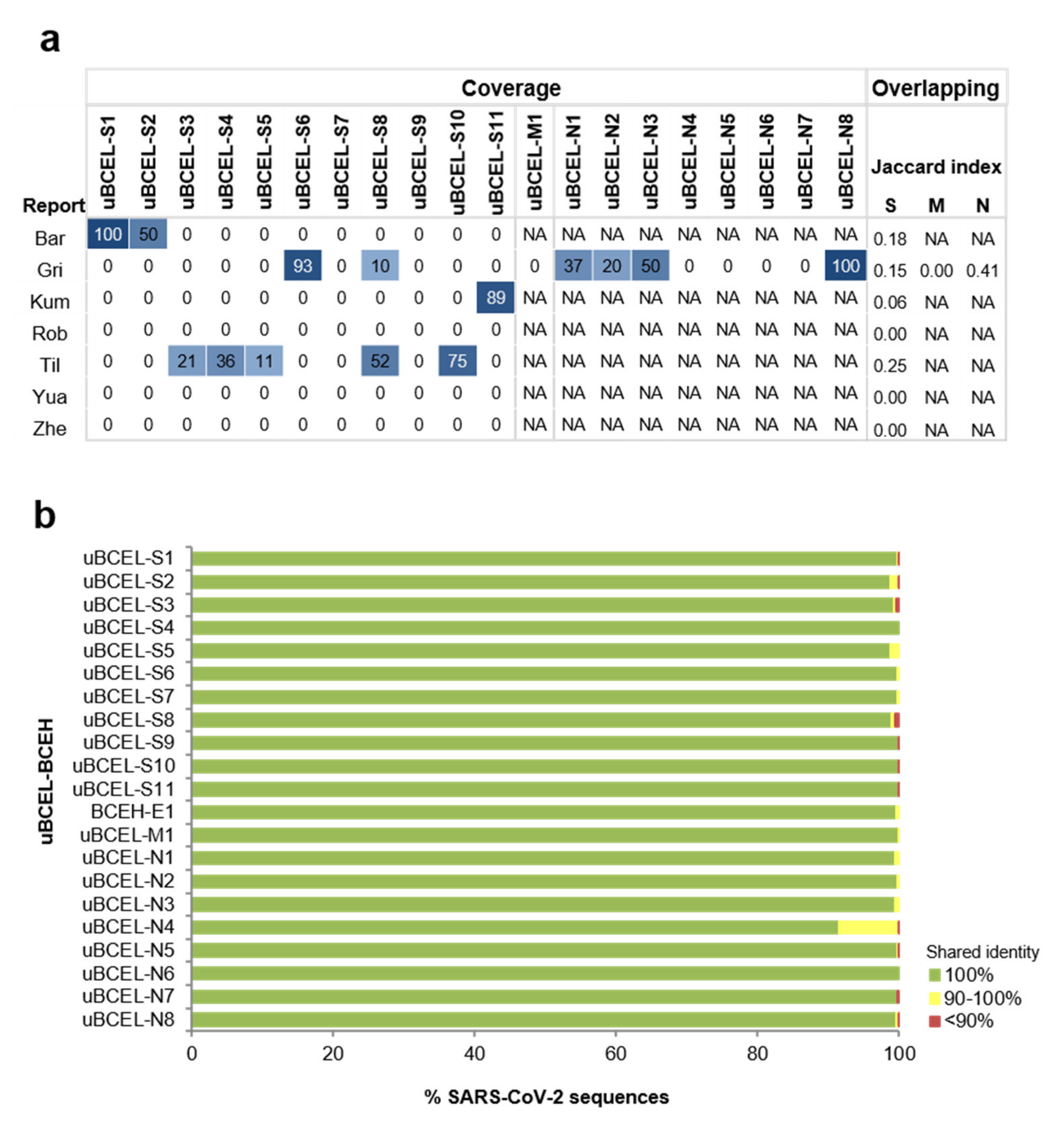
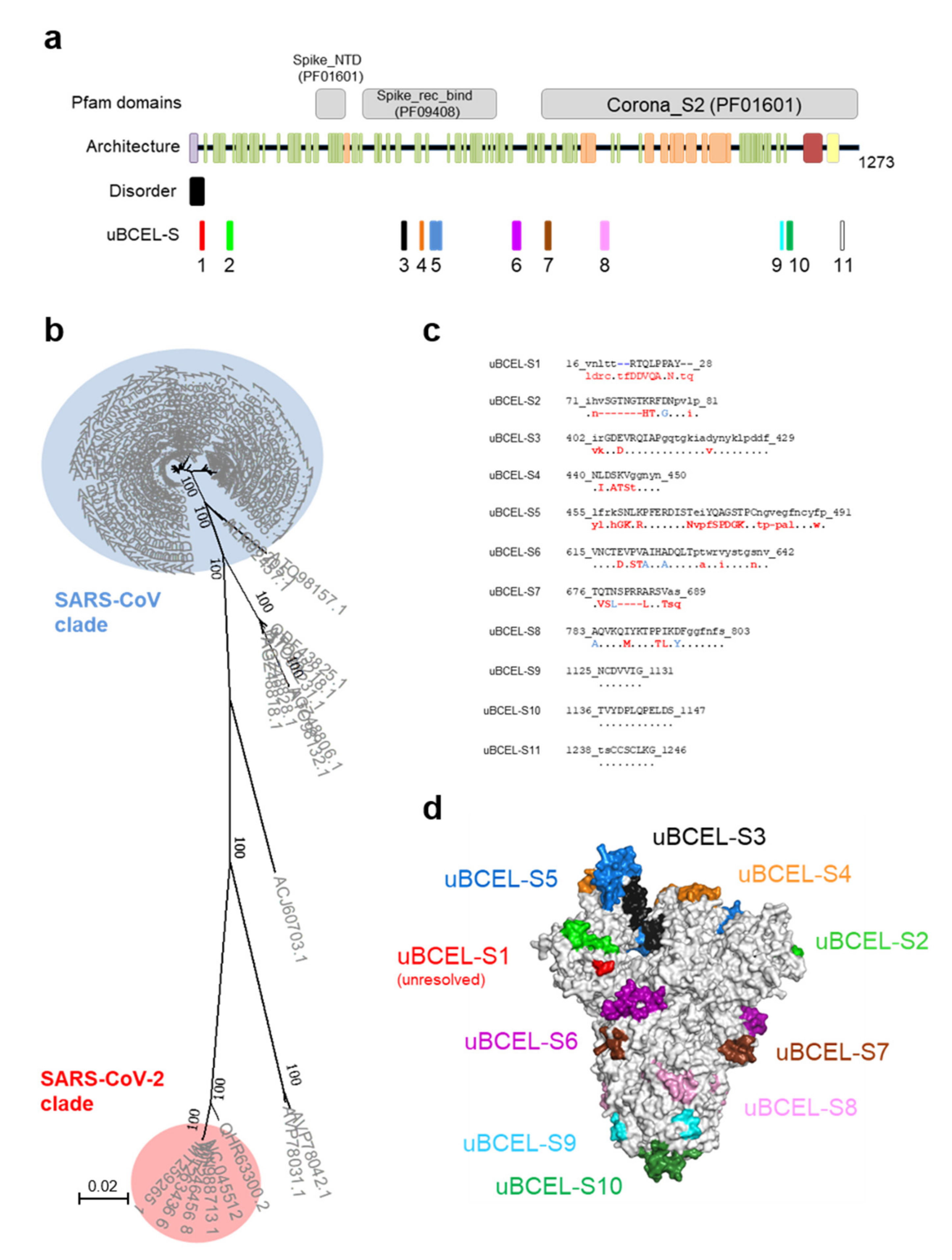
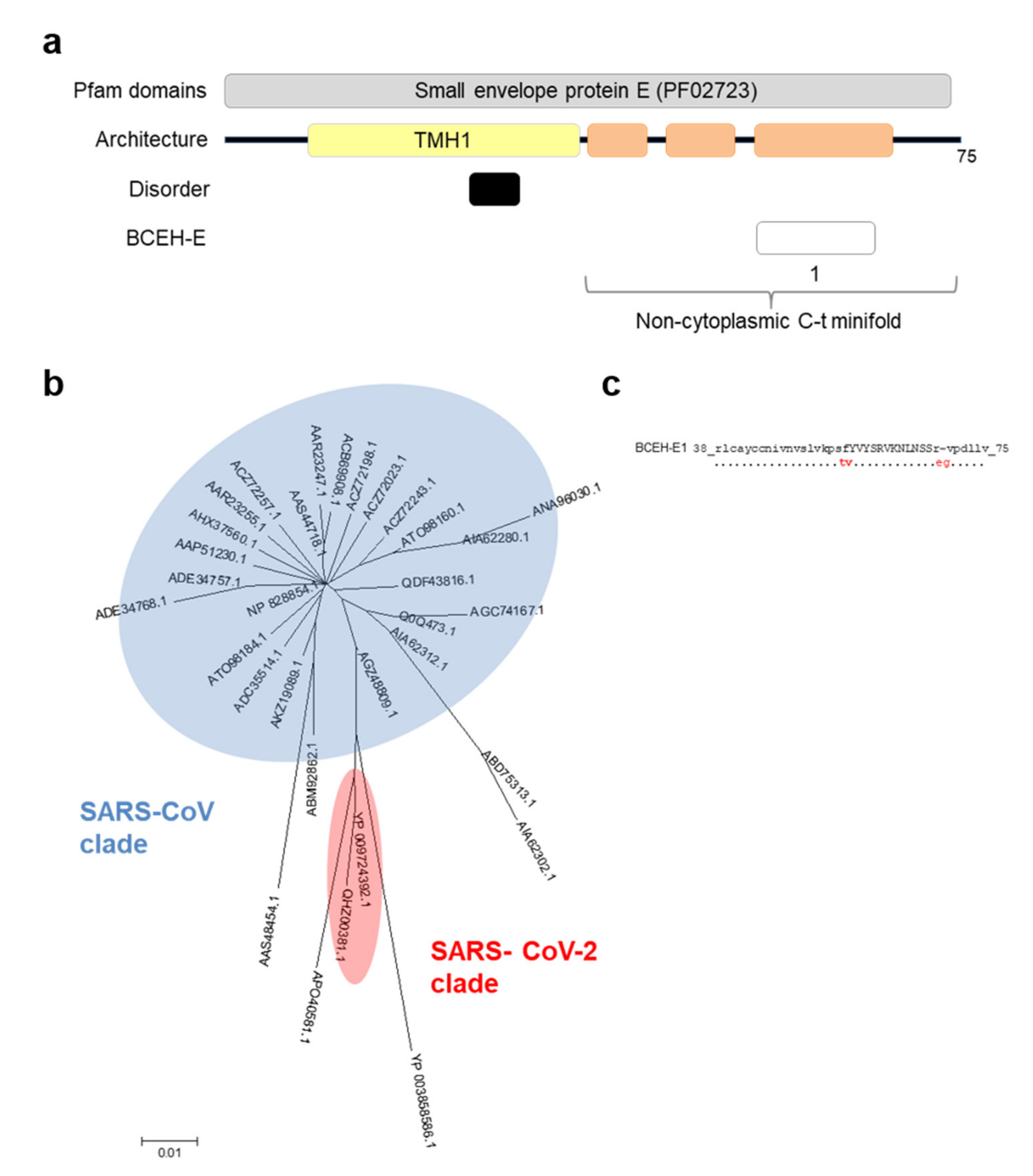
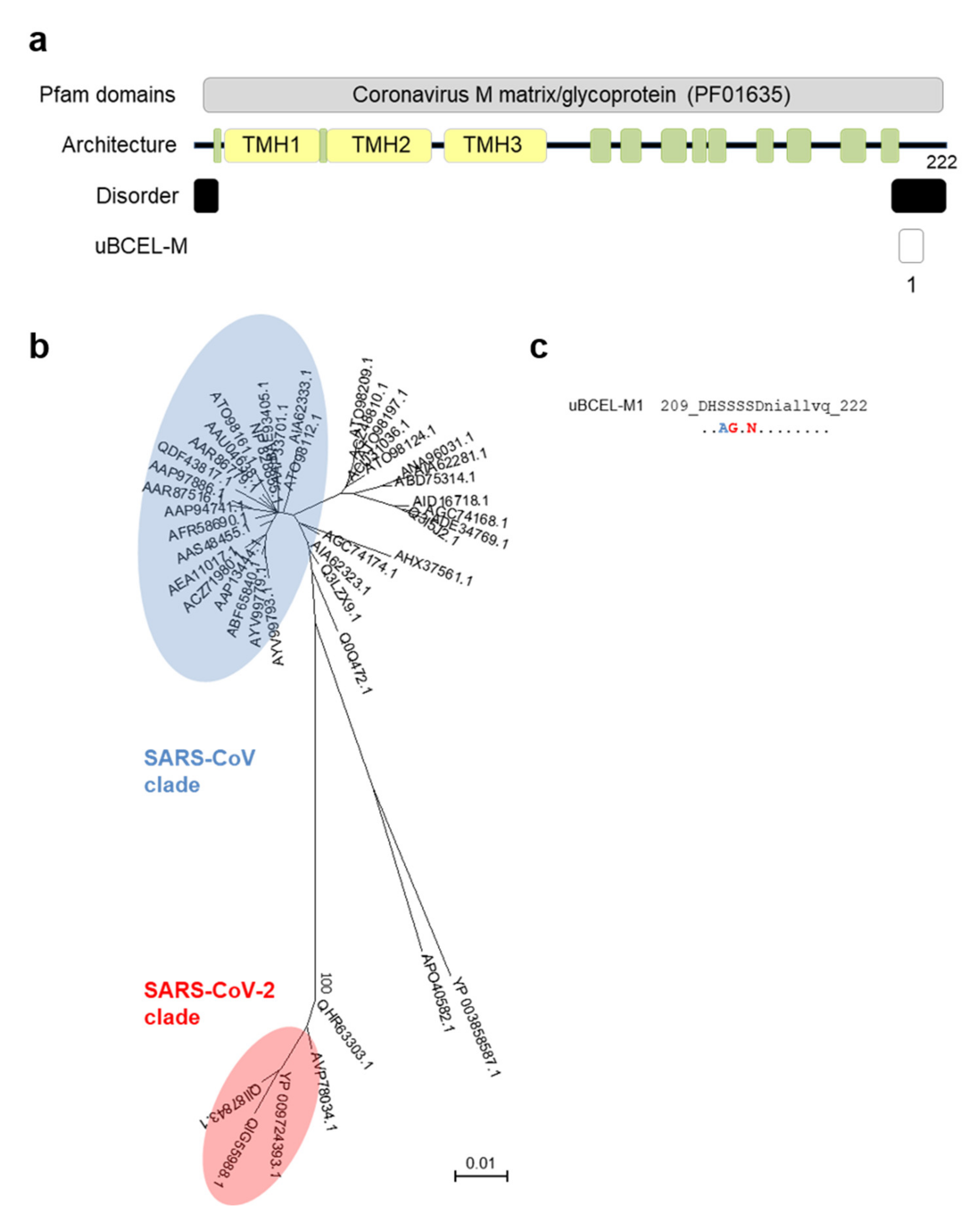
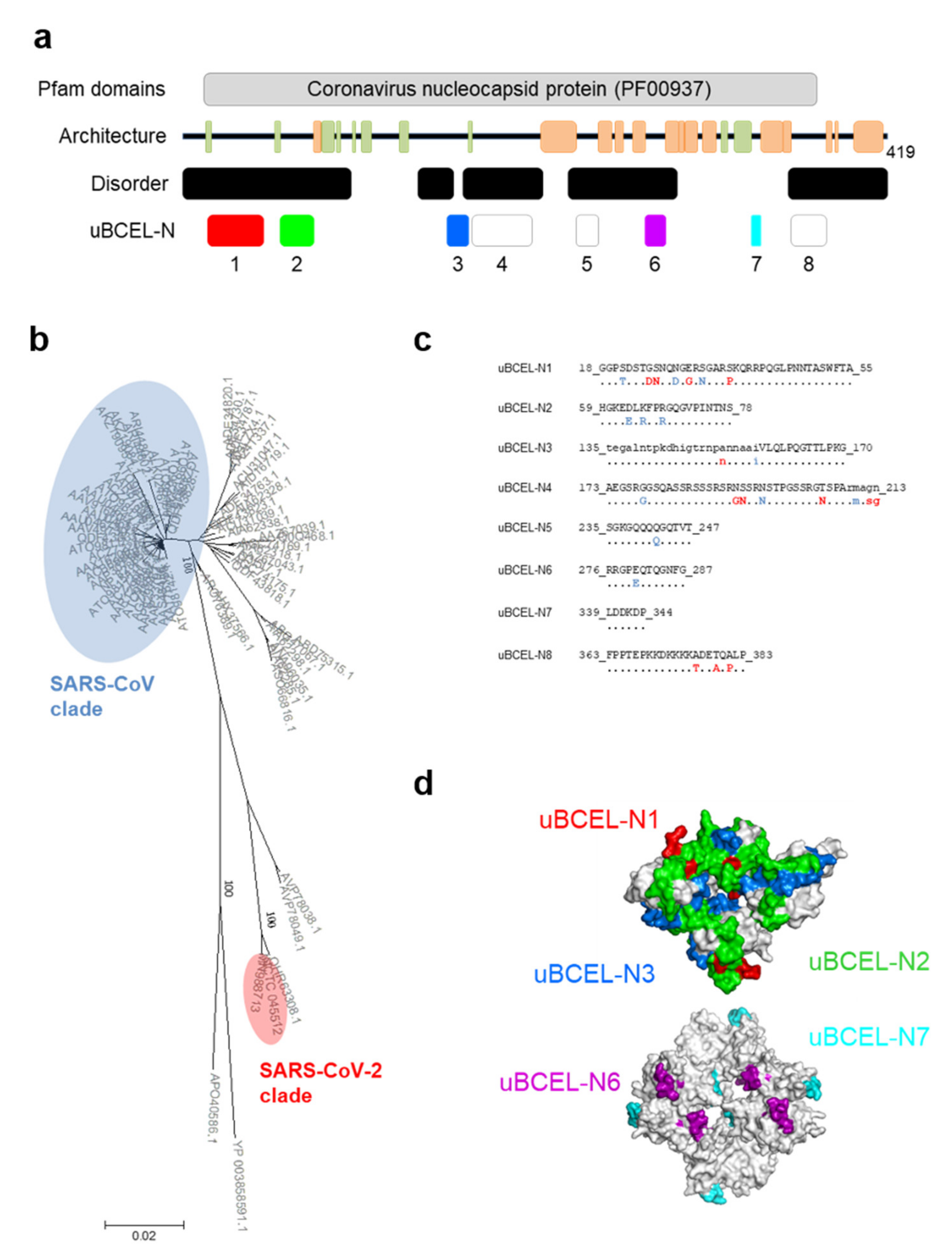
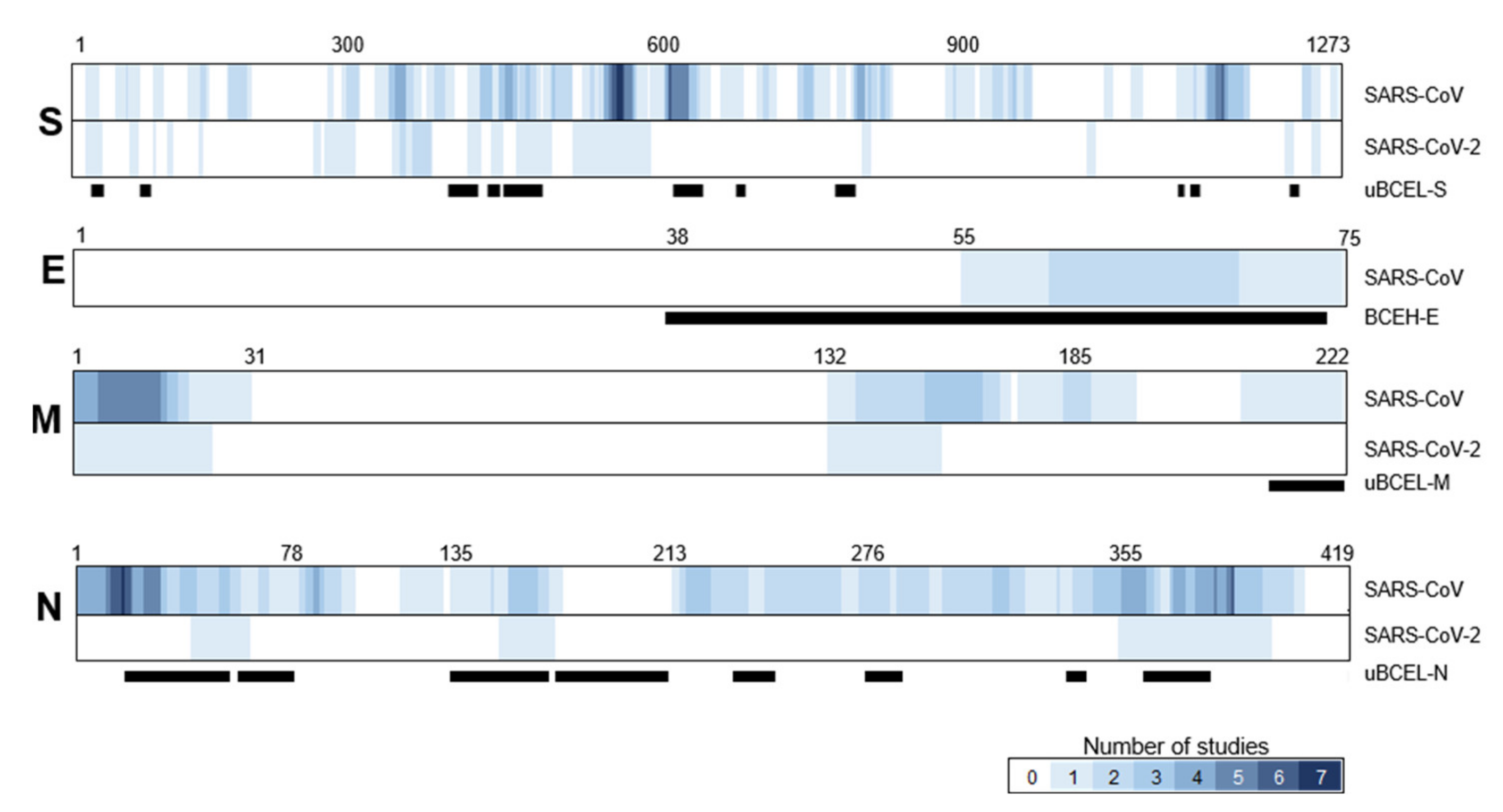
| Protein | uBECL or BCEH a | uBCE b Location | uBCEL or BCEH Location | Flanking SS c | uBCEL Sequence d |
|---|---|---|---|---|---|
| S | uBCEL-S1 | 21–28 | 16–28 | SP-B1 | vnlttRTQLPPAY |
| uBCEL-S2 | 71–81 | 68–85 | B3-B4 | ihvSGTNGTKRFDNpvlp | |
| uBCEL-S3 | 404–412 | 402–429 | B25-B26 | irGDEVRQIAPgqtgkiadynyklpddf | |
| uBCEL-S4 | 440–445 | 440–450 | B26-B27 | NLDSKVggnyn | |
| uBCEL-S5 | 459–470 473–480 | 455–491 | B27-B28 | lfrkSNLKPFERDISTeiYQAGSTPCngvegfncyfp | |
| uBCEL-S6 | 615–630 | 615–642 | B38-B39 | VNCTEVPVAIHADQLTptwrvystgsnv | |
| uBCEL-S7 | 676–687 | 676–689 | B43-B44 | TQTNSPRRARSVas | |
| uBCEL-S8 | 783–797 | 783–803 | H3-B48 | AQVKQIYKTPPIKDFggfnfs | |
| uBCEL-S9 | 1125–1131 | 1125–1131 | B60-B61 | NCDVVIG | |
| uBCEL-S10 | 1137–1147 | 1136–1147 | B61-H12 | TVYDPLQPELDS | |
| uBCEL-S11 | 1240–1246 | 1238–1246 | H15p-H16p | tsCCSCLKG | |
| E | BCEH-E1 | 57–68 | 38–75 | H3p | rlcayccnivnvslvkpsfYVYSRVKNLNSSRvpdllv |
| M | uBCEL-M1 | 209–215 | 209–222 | B10-Ct | DHSSSSDniallvq |
| N | uBCEL-N1 | 16–48 | 18–55 | B1p-B2 | GGPSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTA |
| uBCEL-N2 | 59–78 | 59–78 | B2-H1 | HGKEDLKFPRGQGVPINTNS | |
| uBCEL-N3 | 158–170 | 135–170 | B8-B9 | tegalntpkdhigtrnpannaaiVLQLPQGTTLPKG | |
| uBCEL-N4 | 173–208 | 173–213 | B9-H2p | AEGSRGGSQASSRSSSRSRNSSRNSTPGSSRGTSPArmagn | |
| uBCEL-N5 | 235–247 | 235–247 | H2p-H3p | SGKGQQQQGQTVT | |
| uBCEL-N6 | 276–287 | 276–287 | H4-H5 | RRGPEQTQGNFG | |
| uBCEL-N7 | 339–344 | 339–344 | B11-H9 | LDDKDP | |
| uBCEL-N8 | 363–383 | 363–383 | H10-H11p | FPPTEPKKDKKKKADETQALP |
| uBCEL | Change(s) | n | Date of First Isolation | Geolocation |
|---|---|---|---|---|
| uBCEL-S2 | I68- | 11 | 15/03/2020 | USA: WA |
| N74K | 2 | 20/01/2020 | Brasil; China | |
| D80Y | 2 | 31/03/2020 | USA: WA | |
| uBCEL-S5 | G476S | 7 | 10/03/2020 | USA: WA |
| V483A | 11 | 05/03/2020 | USA: WA | |
| uBCEL-S7 | Q677H | 2 | 19/03/2020 | USA: UT |
| uBCEL-S8 | T791I | 6 | 26/02/2020 | Taiwan |
| BCEH-E1 | P71L | 2 | 19/03/2020 | USA: WA |
| uBCEL-N2 | P67S | 2 | 17/03/2020 | USA: NY; USA: WA |
| uBCEL-N3 | A152S | 2 | 13/03/2020 | USA: UT |
| uBCEL-N4 | S180I | 2 | 31/03/2020 | USA: WA |
| S183Y | 4 | 17/03/2020 | USA | |
| R185C | 5 | 15/03/2020 | USA | |
| R185L | 2 | 19/03/2020 | USA | |
| S188L | 3 | 18/03/2020 | USA | |
| S188P | 2 | 13/03/2020 | Taiwan | |
| S190I | 3 | 17/03/2020 | USA: NY | |
| S196L | 6 | 29/02/2020 | USA | |
| S197L | 17 | 26/02/2020 | Greece; Spain; USA | |
| S202N | 7 | 30/01/2020 | China; USA | |
| R203K,G204R | 62 | 27/02/2020 | Czech Republic; Greece; India; Israel; Peru; Spain; Sri Lanka; Taiwan; USA | |
| T205I | 10 | 29/01/2020 | China; USA | |
| A208G | 4 | 16/03/2020 | USA: WA; USA: NY | |
| uBCEL-N7 | P344S | 2 | ?/01/2020 | Japan |
| uBCEL-N8 | E367- | 2 | 16/03/2020 | SA: UT; USA: WA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corral-Lugo, A.; López-Siles, M.; López, D.; McConnell, M.J.; Martin-Galiano, A.J. Identification and Analysis of Unstructured, Linear B-Cell Epitopes in SARS-CoV-2 Virion Proteins for Vaccine Development. Vaccines 2020, 8, 397. https://doi.org/10.3390/vaccines8030397
Corral-Lugo A, López-Siles M, López D, McConnell MJ, Martin-Galiano AJ. Identification and Analysis of Unstructured, Linear B-Cell Epitopes in SARS-CoV-2 Virion Proteins for Vaccine Development. Vaccines. 2020; 8(3):397. https://doi.org/10.3390/vaccines8030397
Chicago/Turabian StyleCorral-Lugo, Andrés, Mireia López-Siles, Daniel López, Michael J. McConnell, and Antonio J. Martin-Galiano. 2020. "Identification and Analysis of Unstructured, Linear B-Cell Epitopes in SARS-CoV-2 Virion Proteins for Vaccine Development" Vaccines 8, no. 3: 397. https://doi.org/10.3390/vaccines8030397
APA StyleCorral-Lugo, A., López-Siles, M., López, D., McConnell, M. J., & Martin-Galiano, A. J. (2020). Identification and Analysis of Unstructured, Linear B-Cell Epitopes in SARS-CoV-2 Virion Proteins for Vaccine Development. Vaccines, 8(3), 397. https://doi.org/10.3390/vaccines8030397







