Search of Potential Vaccine Candidates against Trueperella pyogenes Infections through Proteomic and Bioinformatic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. “Shaving” of Bacterial Live Cells and Peptide Extraction
2.3. Liquid Chromatography-Mass Spectrometry (LC-MS/MS) Analysis
2.4. Database Searching and Protein Identification
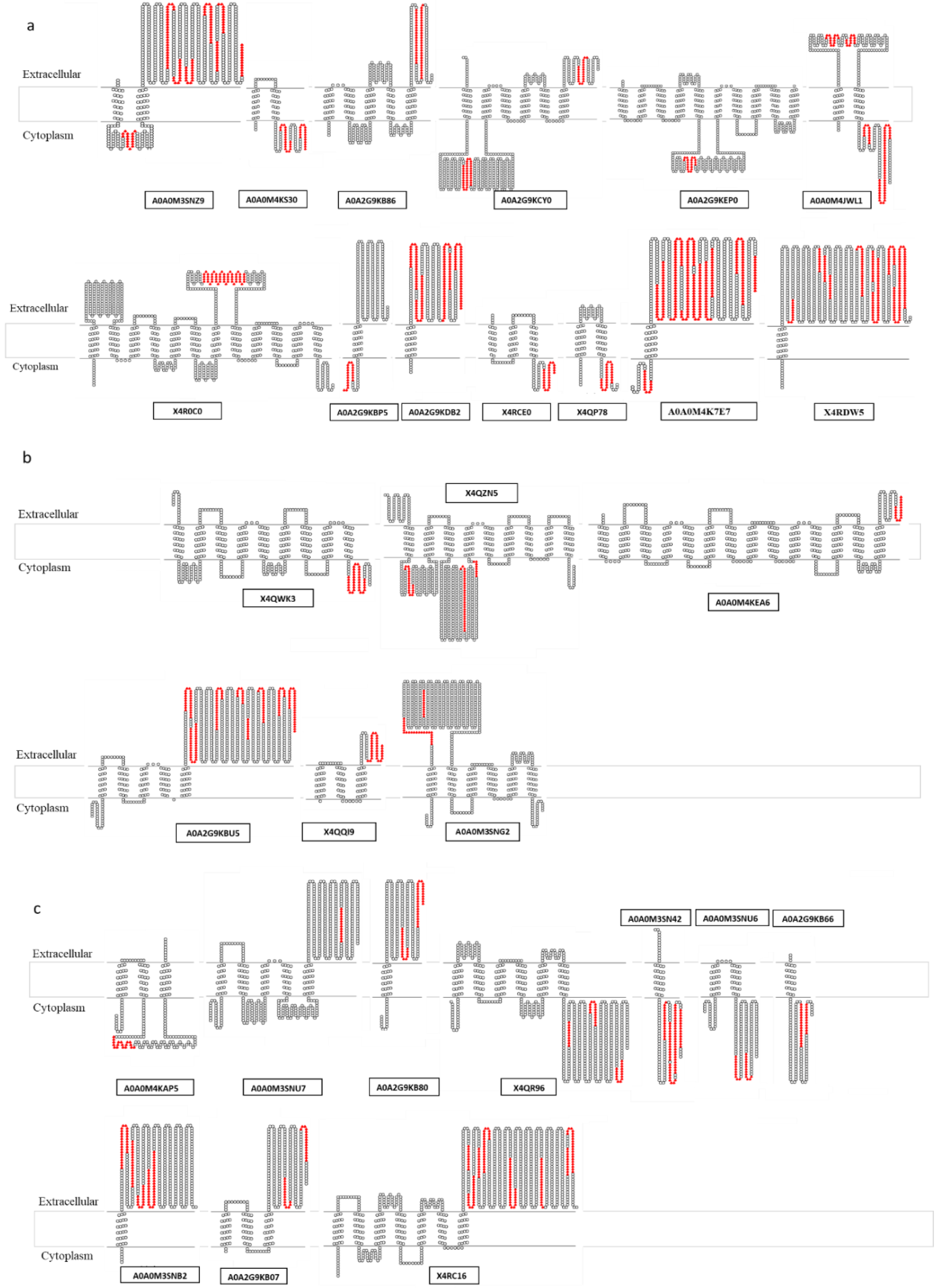
2.5. Bioinformatic Analysis of Protein Sequences
2.6. Data Analysis and Statistics
3. Results
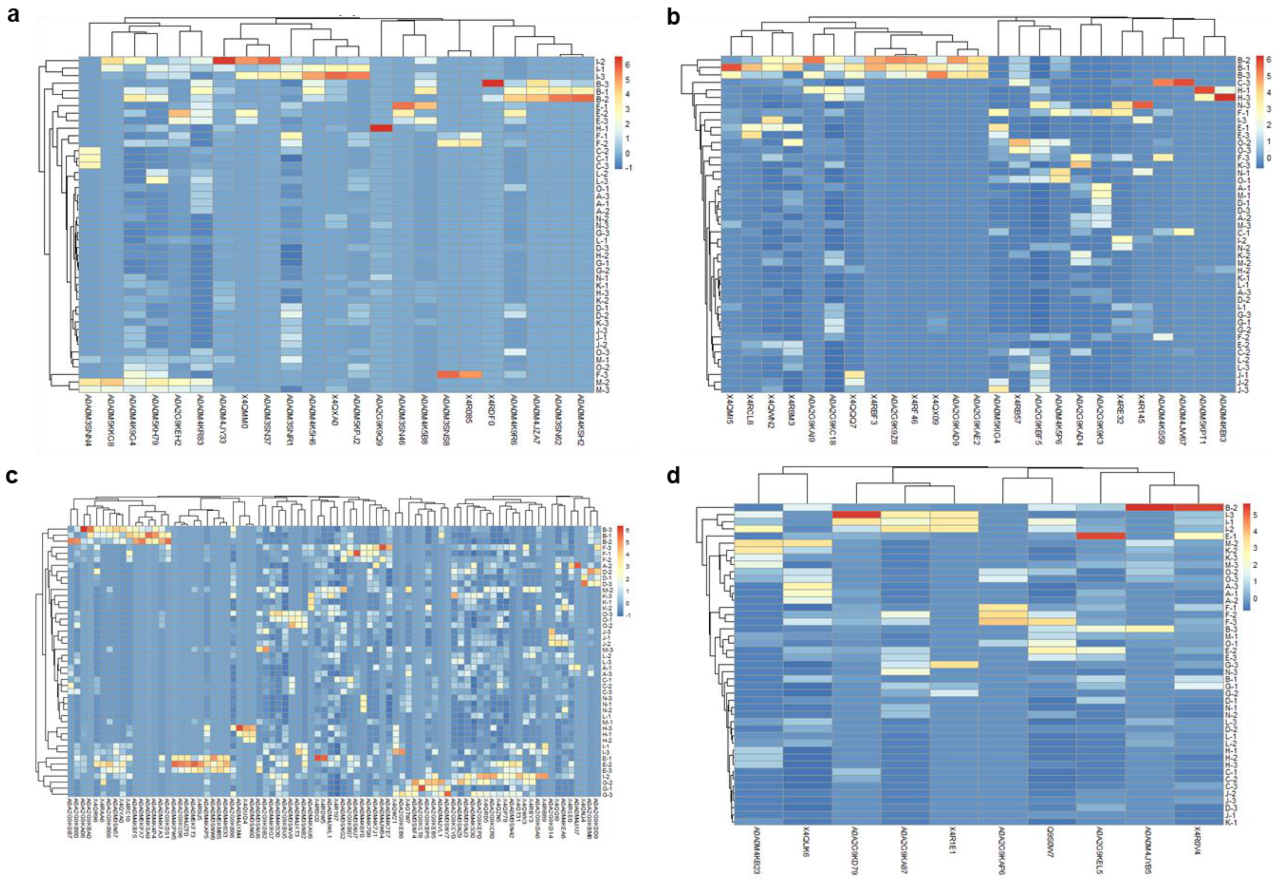
3.1. Describing the “Pan-Surfome” of T. pyogenes
3.2. Analysis of Differences in Surface Protein Abundances of T. pyogenes Clinical Isolates
3.3. Ranking of Proteins from the “Pan-Surfome” of T. pyogenes Based on Their Potential as Putative Vaccine Candidates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Wareth, G.; El-Diasty, M.; Melzer, F.; Murugaiyan, J.; Abdulmawjood, A.; Sprague, L.D.; Neubauer, H. Trueperella pyogenes and brucella abortus coinfection in a dog and a cat on a dairy farm in Egypt with recurrent cases of mastitis and abortion. Vet. Med. Int. 2018, 2018, 2056436. [Google Scholar] [CrossRef] [PubMed]
- Rzewuska, M.; Kwiecień, E.; Chrobak-Chmiel, D.; Kizerwetter-Świda, M.; Stefańska, I.; Gieryńska, M. Pathogenicity and Virulence of Trueperella pyogenes: A Review. Int. J. Mol. Sci. 2019, 20, 2737. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.G.; Risseti, R.M.; Bolaños, C.A.D.; Caffaro, K.A.; de Morais, A.C.B.; Lara, G.H.B.; Zamprogna, T.O.; Paes, A.C.; Listoni, F.J.P.; Franco, M.M.J. Trueperella pyogenes multispecies infections in domestic animals: A retrospective study of 144 cases (2002 to 2012). Vet. Q. 2015, 35, 82–87. [Google Scholar] [CrossRef]
- Galán-Relaño, Á.; Gómez-Gascón, L.; Luque, I.; Barrero-Domínguez, B.; Casamayor, A.; Cardoso-Toset, F.; Vela, A.I.; Fernández-Garayzábal, J.F.; Tarradas, C. Antimicrobial susceptibility and genetic characterization of Trueperella pyogenes isolates from pigs reared under intensive and extensive farming practices. Vet. Microbiol. 2019, 232, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Cameron, C.M.; Botha, W.F.; Smit, B.H.J. Antibody response to and immunity induced by Corynebacterium pyogenes vaccine. Onderstepoort J. Vet. Res. 1976, 43, 97–103. [Google Scholar] [PubMed]
- Derbyshire, J.B.; Matthews, P.R.J. Immunological Studies with Corynebacterium pyogenes in Mice. Res. Vet. Sci. 1963, 4, 537–542. [Google Scholar] [CrossRef]
- Hunter, P.; Van der Lugt, J.J.; Gouws, J.J. Failure of an Actinomyces pyogenes vaccine to protect sheep against an intravenous challenge. Onderstepoort J. Vet. Res. 1990, 57, 239–241. [Google Scholar]
- Desvaux, M.; Candela, T.; Serror, P. Surfaceome and Proteosurfaceome in Parietal Monoderm Bacteria: Focus on Protein Cell-Surface Display. Front. Microbiol. 2018, 9, 100. [Google Scholar] [CrossRef]
- Baums, C.G.; Valentin-Weigand, P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim. Health Res. Rev. 2009, 10, 65–83. [Google Scholar] [CrossRef]
- Cordwell, S.J. Technologies for bacterial surface proteomics. Curr. Opin. Microbiol. 2006, 9, 320–329. [Google Scholar] [CrossRef]
- Navarre, W.W.; Schneewind, O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef] [PubMed]
- Olaya-Abril, A.; Gómez-Gascón, L.; Jiménez-Munguía, I.; Obando, I.; Rodríguez-Ortega, M.J. Another turn of the screw in shaving Gram-positive bacteria: Optimization of proteomics surface protein identification in Streptococcus pneumoniae. J. Proteom. 2012, 75, 3733–3746. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gascón, L.; Luque, I.; Tarradas, C.; Olaya-Abril, A.; Astorga, R.J.; Huerta, B.; Rodríguez-Ortega, M.J. Comparative immunosecretome analysis of prevalent Streptococcus suis serotypes. Comp. Immunol. Microbiol. Infect. Dis. 2018, 57, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Merrill, C.; Ensermu, D.B.; Abdi, R.D.; Gillespie, B.E.; Vaughn, J.; Headrick, S.I.; Hash, K.; Walker, T.B.; Stone, E.; Kerro Dego, O. Immunological responses and evaluation of the protection in dairy cows vaccinated with staphylococcal surface proteins. Vet. Immunol. Immunopathol. 2019, 214, 109890. [Google Scholar] [CrossRef]
- Żakowska, D.; Graniak, G.; Rutyna, P.; Naylor, K.; Głowacka, P.; Niemcewicz, M. Protective antigen domain 4 of Bacillus anthracis as a candidate for use as vaccine for anthrax. Ann. Agric. Environ. Med. 2019, 26, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Song, X.; Jing, J.; Zhao, K.; Shen, Y.; Zhang, X.; Yue, B. Chitosan-DNA nanoparticles enhanced the immunogenicity of multivalent DNA vaccination on mice against Trueperella pyogenes infection. J. Nanobiotechnol. 2018, 16, 8. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, W.; Bao, J.; Wu, Y.; Yan, M.; Xiao, Y.; Yang, L.; Zhang, Y.; Wang, J. A chimeric protein composed of the binding domains of Clostridium perfringens phospholipase C and Trueperella pyogenes pyolysin induces partial immunoprotection in a mouse model. Res. Vet. Sci. 2016, 107, 106–115. [Google Scholar] [CrossRef]
- Yang, L.; Liang, H.; Wang, B.; Ma, B.; Wang, J.; Zhang, W. Evaluation of the potency of two pyolysin-derived recombinant proteins as vaccine candidates of trueperella pyogenes in a mouse model: Pyolysin oligomerization and structural change affect the efficacy of pyolysin-based vaccines. Vaccines 2020, 8, 79. [Google Scholar] [CrossRef]
- Gómez-Gascón, L.; Luque, I.; Olaya-Abril, A.; Jiménez-Munguía, I.; Orbegozo-Medina, R.A.; Peralbo, E.; Tarradas, C.; Rodríguez-Ortega, M.J. Exploring the pan-surfome of Streptococcus suis: Looking for common protein antigens. J. Proteom. 2012, 75, 5654–5666. [Google Scholar] [CrossRef]
- Rodríguez-Ortega, M.J. “Shaving” live bacterial cells with proteases for proteomic analysis of surface proteins. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1722, pp. 21–29. [Google Scholar]
- Santos, T.; Viala, D.; Chambon, C.; Esbelin, J.; Hébraud, M. Listeria monocytogenes Biofilm Adaptation to Different Temperatures Seen through Shotgun Proteomics. Front. Nutr. 2019, 6, 89. [Google Scholar] [CrossRef]
- Espino, E.; Koskenniemi, K.; Mato-Rodriguez, L.; Nyman, T.A.; Reunanen, J.; Koponen, J.; Öhman, T.; Siljamäki, P.; Alatossava, T.; Varmanen, P.; et al. Uncovering surface-exposed antigens of Lactobacillus rhamnosus by cell shaving proteomics and two-dimensional immunoblotting. J. Proteome Res. 2015, 14, 1010–1024. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Schwarz, S. Antimicrobial Resistance in Corynebacterium spp., Arcanobacterium spp., and Trueperella pyogenes. Microbiol. Spectr. 2017, 5, 395–408. [Google Scholar] [CrossRef]
- van de Rijn, I.; Kessler, R.E. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 1980, 27, 444–448. [Google Scholar] [CrossRef] [PubMed]
- García-Angulo, V.A.; Kalita, A.; Kalita, M.; Lozano, L.; Torres, A.G. Comparative genomics and immunoinformatics approach for the identification of vaccine candidates for enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2014, 82, 2016–2026. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Solis, N.; Cordwell, S.J. Current methodologies for proteomics of bacterial surface-exposed and cell envelope proteins. Proteomics 2011, 11, 3169–3189. [Google Scholar] [CrossRef]
- Grandi, G. Genomics and proteomics in reverse vaccines. Methods Biochem. Anal. 2006, 49, 379–393. [Google Scholar]
- Rodríguez-Ortega, M.J.; Luque, I.; Tarradas, C.; Bárcena, J.A. Overcoming function annotation errors in the Gram-positive pathogen Streptococcus suis by a proteomics-driven approach. BMC Genom. 2008, 9, 588. [Google Scholar] [CrossRef]
- Rodríguez-Ortega, M.J.; Norais, N.; Bensi, G.; Liberatori, S.; Capo, S.; Mora, M.; Scarselli, M.; Doro, F.; Ferrari, G.; Garaguso, I.; et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 2006, 24, 191–197. [Google Scholar] [CrossRef]
- Bøhle, L.A.; Riaz, T.; Egge-Jacobsen, W.; Skaugen, M.; Busk, Ø.L.; Eijsink, V.G.H.; Mathiesen, G. Identification of surface proteins in Enterococcus faecalis V583. BMC Genom. 2011, 12, 135. [Google Scholar] [CrossRef] [PubMed]
- Olaya-Abril, A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Rodríguez-Ortega, M.J. Surfomics: Shaving live organisms for a fast proteomic identification of surface proteins. J Proteom. 2014, 97, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, W.J.; Davies, A.H.; Chambers, C.J.; Roberts, A.K.; Shone, C.C.; Acharya, K.R. Molecular features of the sortase enzyme family. FEBS J. 2015, 282, 2097–2114. [Google Scholar] [CrossRef] [PubMed]
- Spirig, T.; Weiner, E.M.; Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011, 82, 1044–1059. [Google Scholar] [CrossRef] [PubMed]
- Susmitha, A.; Nampoothiri, K.M.; Bajaj, H. Insights into the biochemical and functional characterization of sortase E transpeptidase of Corynebacterium glutamicum. Biochem. J. 2019, 476, 3835–3847. [Google Scholar] [CrossRef]
- Das, S.; Pawale, V.S.; Dadireddy, V.; Singh, A.K.; Ramakumar, S.; Roy, R.P. Structure and specificity of a new class of Ca2+-independent housekeeping sortase from Streptomyces avermitilis provide insights into its non-canonical substrate preference. J. Biol. Chem. 2017, 292, 7244–7257. [Google Scholar] [CrossRef]
- Goudarzi, M.; Kobayashi, N.; Hashemi, A.; Fazeli, M.; Navidinia, M. Genetic variability of methicillin resistant staphylococcus aureus strains isolated from burns patients. Osong Public Heal. Res. Perspect. 2019, 10, 170–176. [Google Scholar] [CrossRef]
- Prados de la Torre, E.; Rodríguez-Franco, A.; Rodríguez-Ortega, M.J. Proteomic and bioinformatic analysis of streptococcus suis human isolates: Combined prediction of potential vaccine candidates. Vaccines 2020, 8, 188. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Obando, I.; Rodríguez-Ortega, M.J. Identification of potential new protein vaccine candidates through pan-surfomic analysis of pneumococcal clinical isolates from adults. PLoS ONE 2013, 8, e70365. [Google Scholar] [CrossRef]
- Jost, B.H.; Trinh, H.T.; Songer, J.G.; Billington, S.J. Immunization with genetic toxoids of the Arcanobacterium pyogenes cholesterol-dependent cytolysin, pyolysin, protects mice against infection. Infect. Immun. 2003, 71, 2966–2969. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, P.; Wang, B.; Ma, B.; Wang, J. A combined Clostridium perfringens/Trueperella pyogenes inactivated vaccine induces complete immunoprotection in a mouse model. Biologicals 2017, 47, 1–10. [Google Scholar] [CrossRef]
- Gómez-Gascón, L.; Cardoso-Toset, F.; Tarradas, C.; Gómez-Laguna, J.; Maldonado, A.; Nielsen, J.; Olaya-Abril, A.; Rodríguez-Ortega, M.J.; Luque, I. Characterization of the immune response and evaluation of the protective capacity of rSsnA against Streptococcus suis infection in pigs. Comp. Immunol. Microbiol. Infect. Dis. 2016, 47, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Zhao, K.; Zhang, Z.; Tang, C.; Zhang, X.; Yue, B. DNA vaccination based on pyolysin co-immunized with IL-1β enhances host antibacterial immunity against Trueperella pyogenes infection. Vaccine 2016, 34, 3469–3477. [Google Scholar] [CrossRef] [PubMed]
| Proteomics Reference | Origin a | Rearing System b | PFGE Pattern c | Cluster d |
|---|---|---|---|---|
| A | Liver | Extensive | 43 | B |
| B | Lung | Intensive | 28 | A |
| C | Lymph nodes | Intensive | 43 | B |
| D | Brain | Intensive | 53 | C |
| E | Lung | Intensive | 37 | B |
| F | Joint | Intensive | 37 | B |
| G | Lymph nodes | Extensive | 12 | A |
| H | Lymph nodes | Extensive | 1 | A |
| I | Liver | Extensive | 12 | A |
| J | Spleen | Extensive | 44 | B |
| K | Lymph nodes | Extensive | 43 | B |
| L | Heart | Intensive | 43 | B |
| M | Joint | Intensive | 13 | A |
| N | Heart | Intensive | 44 | B |
| O | Abscess | Extensive | 64 | C |
| Protein Category a | Identified Proteins among All the Strains | Range of Identified Proteins Per Strain |
|---|---|---|
| Surface proteins | 140 | 35–73 |
| Lipoprotein | 23 (16.4%) | 1–13 |
| Cell Wall | 25 (17.9%) | 13–25 |
| Secretory | 10 (7.1%) | 5–10 |
| Membrane | 82 (58.6%) | 22–50 |
| Ranking | Proteins |
|---|---|
| A (16) a | Cell wall: X4QWN2, X4R8M3 Lipoproteins: A0A0M3SNR1, A0A0M4K9G4, A0A0M4JY33 Secreted proteins: X4R0V4, A0A2G9KEL5, Q9S0W7, X4QUK6 Membrane proteins (1 TMD): A0A0M4K7E7, A0A2G9KDB2, X4RDW5 Membrane proteins (>1 TMD): A0A0M3SNZ9, A0A0M4KS30, A0A2G9KB86, A0A2G9KCY0, A0A0M4JWL1 |
| B (9) | Cell wall: A0A2G9KC18, A0A2G9KBF5 Lipoproteins: A0A0M5KH79, A0A0M4KR83 Secreted proteins: A0A2G9KD79, A0A2G9KA87 Membrane proteins (>1 TMD): X4QZN5, A0A2G9KBU5, A0A0M3SNG2 |
| C (15) | Cell wall: X4RE32, X4RB57, A0A0M5KIG4, X4RCL8, X4QMI5, A0A2G9KAD4, A0A0M4K5P6 Lipoproteins: A0A0M5KPJ2, A0A0M4K9R6, X4QXA0, A0A2G9KEH2, A0A0M4K5B8 Secreted proteins: A0A0M4KB23, A0A0M4JYB5 Membrane proteins (1 TMD): A0A2G9KB80, A0A0M3SN42, A0A2G9KB66, A0A0M3SNB2 Membrane proteins (>1 TMD): X4QR96, A0A0M3SNU6, A0A2G9KB07, X4RC16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galán-Relaño, Á.; Gómez-Gascón, L.; Rodríguez-Franco, A.; Luque, I.; Huerta, B.; Tarradas, C.; Rodríguez-Ortega, M.J. Search of Potential Vaccine Candidates against Trueperella pyogenes Infections through Proteomic and Bioinformatic Analysis. Vaccines 2020, 8, 314. https://doi.org/10.3390/vaccines8020314
Galán-Relaño Á, Gómez-Gascón L, Rodríguez-Franco A, Luque I, Huerta B, Tarradas C, Rodríguez-Ortega MJ. Search of Potential Vaccine Candidates against Trueperella pyogenes Infections through Proteomic and Bioinformatic Analysis. Vaccines. 2020; 8(2):314. https://doi.org/10.3390/vaccines8020314
Chicago/Turabian StyleGalán-Relaño, Ángela, Lidia Gómez-Gascón, Antonio Rodríguez-Franco, Inmaculada Luque, Belén Huerta, Carmen Tarradas, and Manuel J. Rodríguez-Ortega. 2020. "Search of Potential Vaccine Candidates against Trueperella pyogenes Infections through Proteomic and Bioinformatic Analysis" Vaccines 8, no. 2: 314. https://doi.org/10.3390/vaccines8020314
APA StyleGalán-Relaño, Á., Gómez-Gascón, L., Rodríguez-Franco, A., Luque, I., Huerta, B., Tarradas, C., & Rodríguez-Ortega, M. J. (2020). Search of Potential Vaccine Candidates against Trueperella pyogenes Infections through Proteomic and Bioinformatic Analysis. Vaccines, 8(2), 314. https://doi.org/10.3390/vaccines8020314








