Histone Methyltransferase DOT1L Is Involved in Larval Molting and Second Stage Nymphal Feeding in Ornithodoros moubata
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Primers Design, Gene Amplification, Cloning, and Sequencing of Omdot1l
2.3. Quantitative PCR
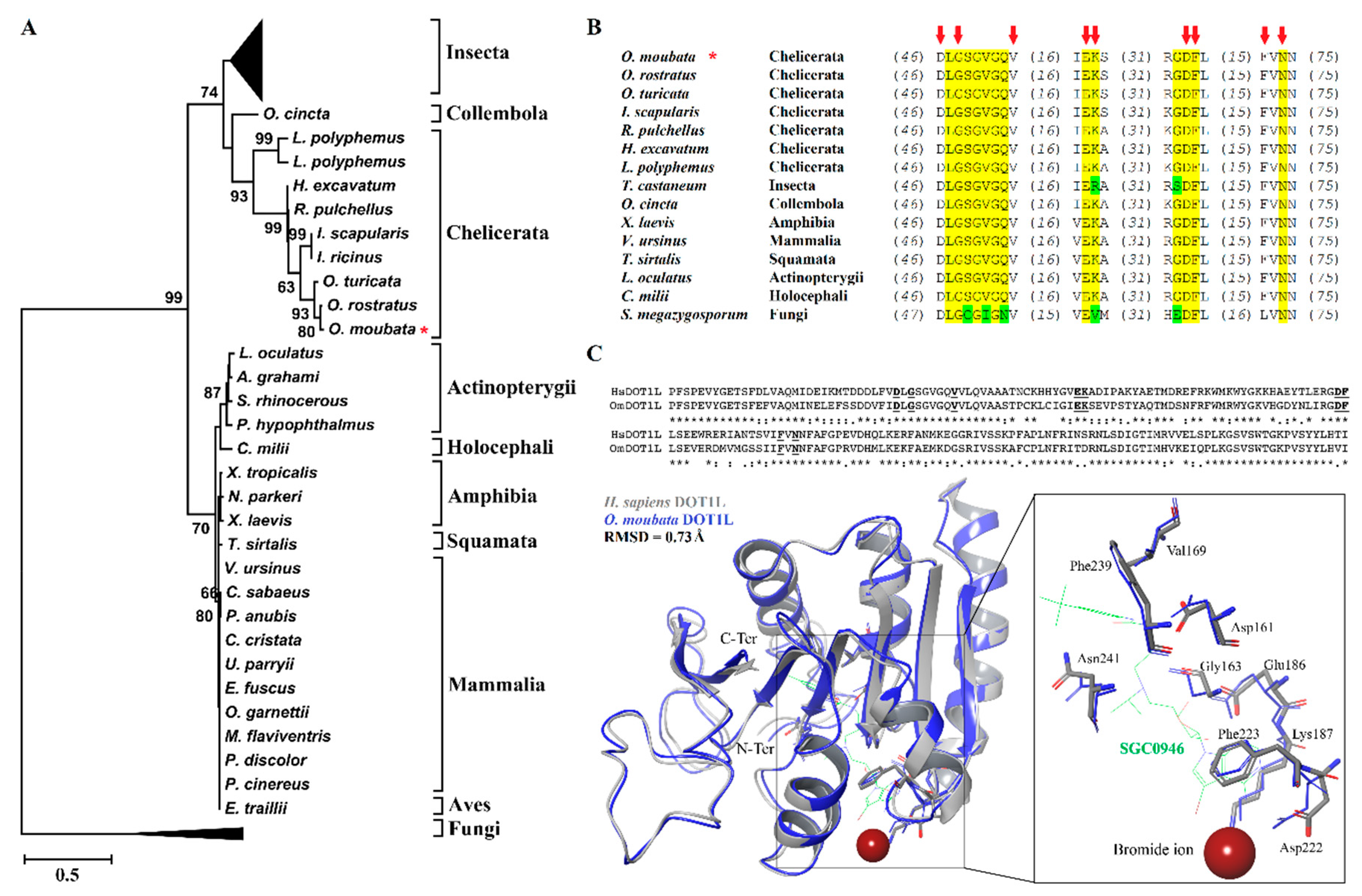
2.4. Phylogenetic Tree
2.5. Tertiary Structure Prediction
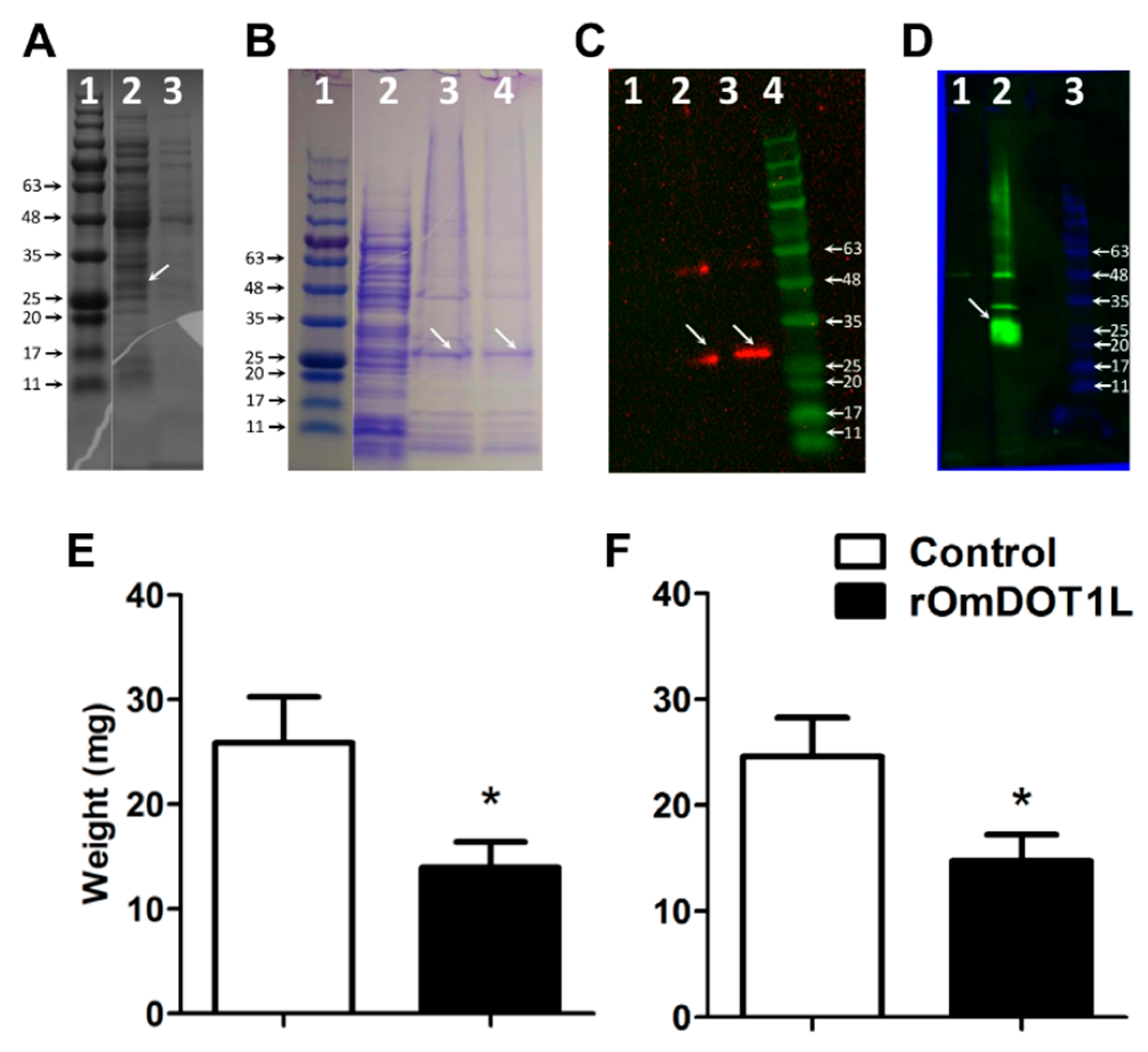
2.6. Recombinant OmDOT1L Expression
2.7. Detection of rOmDOT1L by Western Blot
2.8. Rabbit Immunization and Tick Infestation
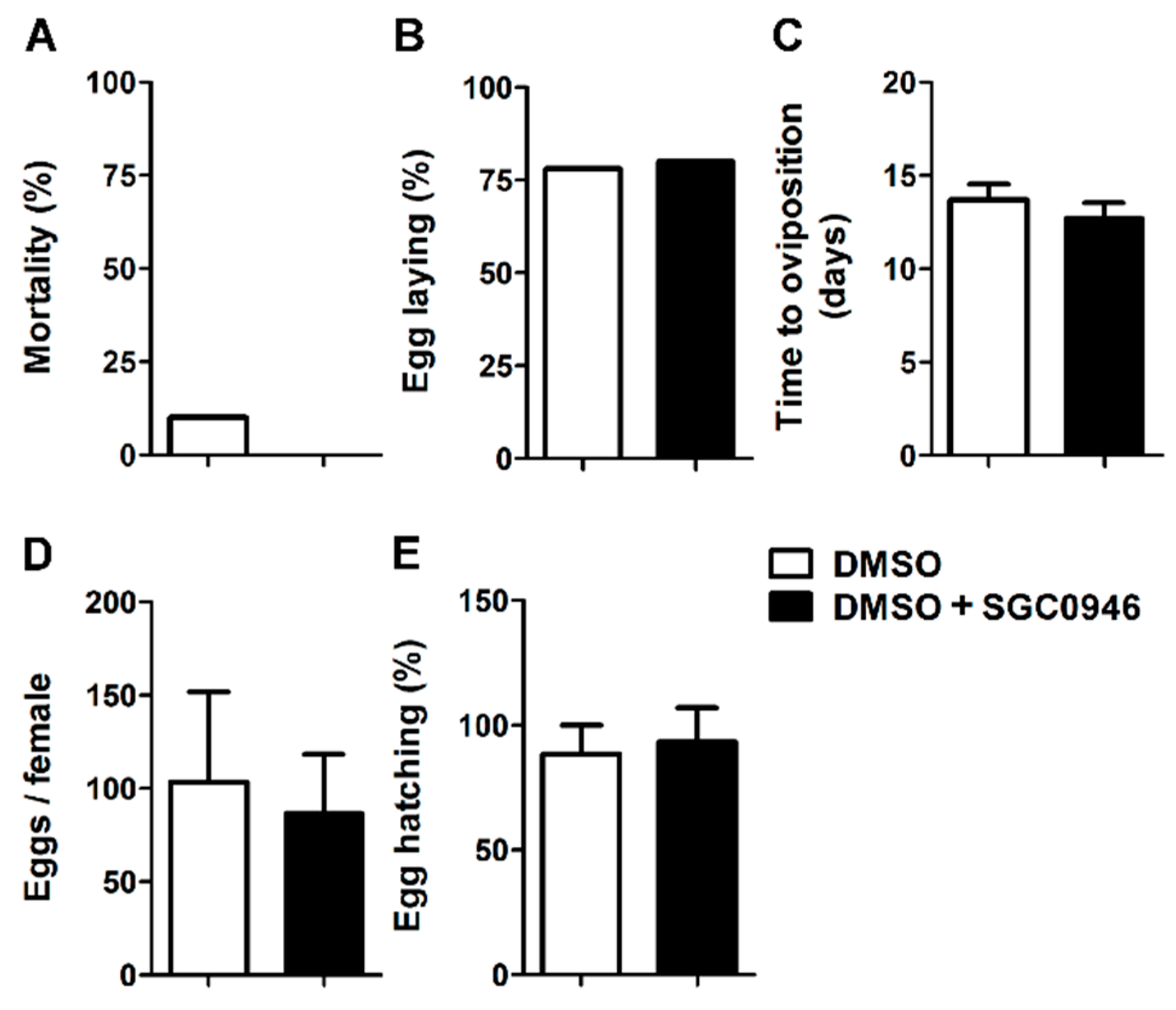
2.9. Membrane Feeding of Adult Females O. moubata
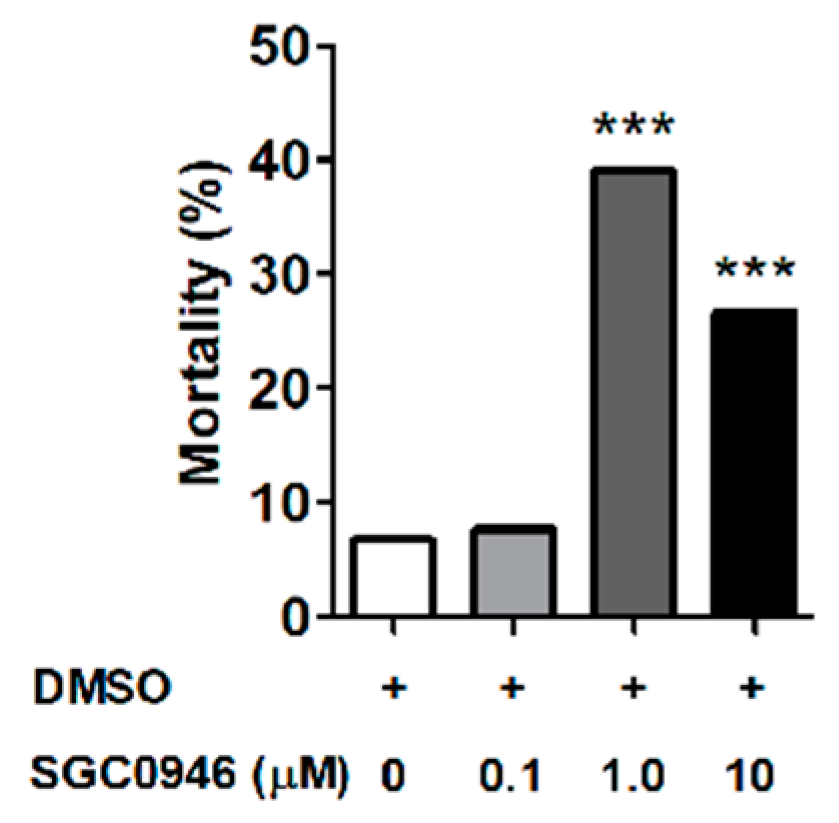
2.10. Larval Immersion Test Using the DOT1L Inhibitor SGC0946
3. Results and Discussion
3.1. Phylogenetic and Molecular Characterization of OmDOT1L Catalytic Domain
3.2. Omdot1l Gene is Upregulated in Unfed First Stage Nymphs
3.3. The DOT1L Inhibitor SGC0946 Reduces Larval Molting Success in O. moubata
3.4. Immunity against Recombinant OmDOT1L in Rabbits Impairs Second-Stage Nymphal Feeding
3.5. The DOT1L Inhibitor SGC0946 Does not Affect the Reproductive Performance of Adult Females O. moubata
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manzano-Román, R.; Díaz-Martín, V.; de la Fuente, J.; Pérez-Sánchez, R. Soft Ticks as Pathogen Vectors: Distribution, Surveillance and Control. TechOpen 2012, 7, 125–162. [Google Scholar]
- Brites-Neto, J.; Duarte, K.M.R.; Martins, T.F. Tick-borne infections in human and animal population worldwide. Vet. World 2015, 8, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Singer, M.S.; Kahana, A.; Wolf, A.J.; Meisinger, L.L.; Peterson, S.E.; Goggin, C.; Mahowald, M.; Gottschling, D.E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 1998, 150, 613–632. [Google Scholar] [PubMed]
- Jenuwein, T.; Laible, G.; Dorn, R.; Reuter, G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. 1998, 54, 80–93. [Google Scholar] [CrossRef]
- Feng, Q.; Wang, H.; Ng, H.H.; Erdjument-Bromage, H.; Tempst, P.; Struhl, K.; Zhang, Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002, 12, 1052–1058. [Google Scholar] [CrossRef]
- Min, J.; Feng, Q.; Li, Z.; Zhang, Y.; Xu, R.; Hill, C.; Carolina, N. Structure of the Catalytic Domain of Human DOT1L, a Non-SET Domain Nucleosomal Histone Methyltransferase. Cell 2003, 112, 711–723. [Google Scholar] [CrossRef]
- Cheng, X.; Collins, R.E.; Zhang, X. Structural and Sequence Motifs of Protein (Histone) Methylation Enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 267–294. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, F.; Gafken, P.R.; Gottschling, D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 2002, 109, 745–756. [Google Scholar] [CrossRef]
- Shanower, G.A. Characterization of the grappa Gene, the Drosophila Histone H3 Lysine 79 Methyltransferase. Genetics 2004, 169, 173–184. [Google Scholar] [CrossRef]
- Jones, B.; Su, H.; Bhat, A.; Lei, H.; Bajko, J.; Hevi, S.; Baltus, G.A.; Kadam, S.; Zhai, H.; Valdez, R.; et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Shilatifard, A. Posttranslational Modifications of Histones by Methylation. In Advances in Protein Chemistry; Academic Press: Cambridge, MA, USA, 2004; pp. 201–222. [Google Scholar]
- Yu, W.; Chory, E.J.; Wernimont, A.K.; Tempel, W.; Scopton, A.; Federation, A.; Marineau, J.J.; Qi, J.; Barsyte-Lovejoy, D.; Yi, J.; et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat. Commun. 2012, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kari, V.; Raul, S.K.; Henck, J.M.; Kitz, J.; Kramer, F.; Kosinsky, R.L.; Übelmesser, N.; Mansour, W.Y.; Eggert, J.; Spitzner, M.; et al. The histone methyltransferase DOT1L is required for proper DNA damage response, DNA repair, and modulates chemotherapy responsiveness. Clin. Epigenetics 2019, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Fu, L.; Guo, X.; Chen, Y.; Shi, Y.-B. Histone methyltransferase Dot1L plays a role in postembryonic development in Xenopus tropicalis. FASEB J. 2015, 29, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Fu, L.; Shi, Y.-B. Histone methyltransferase Dot1L is a coactivator for thyroid hormone receptor during Xenopus development. FASEB J. 2017, 31, 4821–4831. [Google Scholar] [CrossRef]
- Matsuura, K.; Fujimoto, K.; Das, B.; Fu, L.; Lu, C.D.; Shi, Y.-B. Histone H3K79 methyltransferase Dot1L is directly activated by thyroid hormone receptor during Xenopus metamorphosis. Cell Biosci. 2012, 2, 25. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Alberdi, P.; Ayllón, N.; Valdés, J.J.; Pierce, R.; Villar, M.; de la Fuente, J. Anaplasma phagocytophilum increases the levels of histone modifying enzymes to inhibit cell apoptosis and facilitate pathogen infection in the tick vector Ixodes scapularis. Epigenetics 2016, 11, 303–319. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Koci, J.; Simo, L.; Park, Y. Validation of internal reference genes for real-time quantitative polymerase chain reaction studies in the tick, Ixodes scapularis (Acari: Ixodidae). J. Med. Entomol. 2013, 50, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, 526–531. [Google Scholar] [CrossRef]
- Schrödinger Release 2020-1: Maestro; Schrödinger, LLC: New York, NY, USA, 2020.
- Krull, C.; Böhme, B.; Clausen, P.-H.; Nijhof, A.M. Optimization of an artificial tick feeding assay for Dermacentor reticulatus. Parasites Vectors 2017, 10, 60. [Google Scholar] [CrossRef]
- Knorr, S.; Anguita, J.; Cortazar, J.T.; Hajdusek, O.; Kopáček, P.; Trentelman, J.J.; Kershaw, O.; Hovius, J.W.; Nijhof, A.M. Preliminary Evaluation of Tick Protein Extracts and Recombinant Ferritin 2 as Anti-tick Vaccines Targeting Ixodes ricinus in Cattle. Front. Physiol. 2018, 9, 1696. [Google Scholar] [CrossRef]
- Klafke, G.M.; Sabatini, G.A.; de Albuquerque, T.A.; Martins, J.R.; Kemp, D.H.; Miller, R.J.; Schumaker, T.T.S. Larval immersion tests with ivermectin in populations of the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae) from State of Sao Paulo, Brazil. Vet. Parasitol. 2006, 142, 386–390. [Google Scholar] [CrossRef]
- Santos, T.R.B.; Klafke, G.M.; Pappen, F.G.; Nizoli, L.Q.; Biegelmeyer, P.; Farias, N.A.R. Comparison of three larval bioassays to evaluate susceptibility of Rhipicephalus (Boophilus) microplus to amitraz. Brazilian J. Vet. Parasitol. 2013, 22, 495–501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Webster, A.; Souza, U.A.; Martins, J.R.; Klafke, G.; Reck, J.; Schrank, A. Comparative study between Larval Packet Test and Larval Immersion Test to assess the effect of Metarhizium anisopliae on Rhipicephalus microplus tick larvae. Exp. Appl. Acarol. 2018, 74, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, D.; Šíma, R.; Konvičková, J.; Perner, J.; Kopáček, P.; Sojka, D. Multiple Legumain Isoenzymes in Ticks. Int. J. Parasitol. 2018, 48, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Artigas-Jerónimo, S.; Villar, M.; Cabezas-Cruz, A.; Valdés, J.J.; Estrada-Peña, A.; Alberdi, P.; de la Fuente, J. Functional Evolution of Subolesin/Akirin. Front Physiol. 2018, 9, 1612. [Google Scholar] [CrossRef] [PubMed]
- Jasinskas, A.; Barbour, A.G. The Fc Fragment Mediates the Uptake of Immunoglobulin C From the Midgut to Hemolymph in the Ixodid Tick Amblyomma Americanum (Acari: Ixodidae). J. Med. Entomol. 2005, 42, 359–366. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Román, R.; Pérez-Sánchez, R. Functional Annotation and Analysis of the Ornithodoros moubata Midgut Genes Differentially Expressed After Blood Feeding. Ticks Tick Borne Dis. 2017, 8, 693–708. [Google Scholar] [CrossRef]
- Buczek, A.; Bartosik, K.; Kuczyński, P. Evaluation of the Effect of Various Concentrations of Selected Pyrethroids on the Development of Dermacentor Reticulatus Eggs and Larvae. Ann. Agric. Environ. Med. 2013, 20, 447–451. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gobl, J.; Kumar Sinha, D.; Sima, R.; Perner, J.; Kopáček, P.; Valdés, J.J.; Rego, R.O.M.; Cabezas-Cruz, A. Histone Methyltransferase DOT1L Is Involved in Larval Molting and Second Stage Nymphal Feeding in Ornithodoros moubata. Vaccines 2020, 8, 157. https://doi.org/10.3390/vaccines8020157
Gobl J, Kumar Sinha D, Sima R, Perner J, Kopáček P, Valdés JJ, Rego ROM, Cabezas-Cruz A. Histone Methyltransferase DOT1L Is Involved in Larval Molting and Second Stage Nymphal Feeding in Ornithodoros moubata. Vaccines. 2020; 8(2):157. https://doi.org/10.3390/vaccines8020157
Chicago/Turabian StyleGobl, Julia, Deepak Kumar Sinha, Radek Sima, Jan Perner, Petr Kopáček, James J Valdés, Ryan O. M. Rego, and Alejandro Cabezas-Cruz. 2020. "Histone Methyltransferase DOT1L Is Involved in Larval Molting and Second Stage Nymphal Feeding in Ornithodoros moubata" Vaccines 8, no. 2: 157. https://doi.org/10.3390/vaccines8020157
APA StyleGobl, J., Kumar Sinha, D., Sima, R., Perner, J., Kopáček, P., Valdés, J. J., Rego, R. O. M., & Cabezas-Cruz, A. (2020). Histone Methyltransferase DOT1L Is Involved in Larval Molting and Second Stage Nymphal Feeding in Ornithodoros moubata. Vaccines, 8(2), 157. https://doi.org/10.3390/vaccines8020157






