Influenza H1 Mosaic Hemagglutinin Vaccine Induces Broad Immunity and Protection in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Viruses
2.3. Mosaic Gene Design
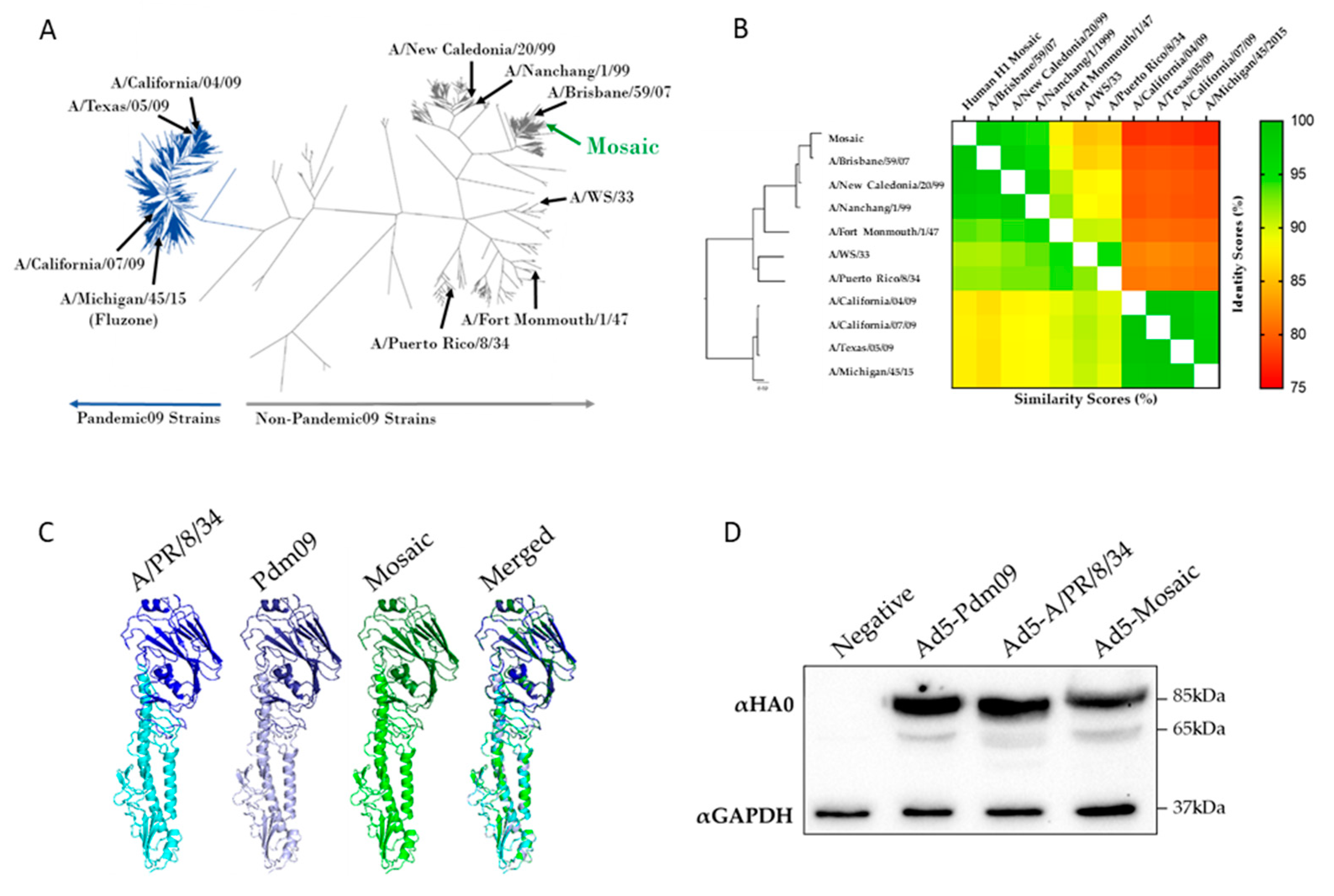
2.4. Phylogenetic and Sequence Analysis of Mosaic Gene
2.5. Structural Analysis of Mosaic Gene
2.6. Recombinant Adenovirus Type 5 Plasmid Construction
2.7. Recombinant Adenovirus Rescue, Purification, and Quantification
2.8. Western Blotting
2.9. Tissues for Humoral and Cellular Assays
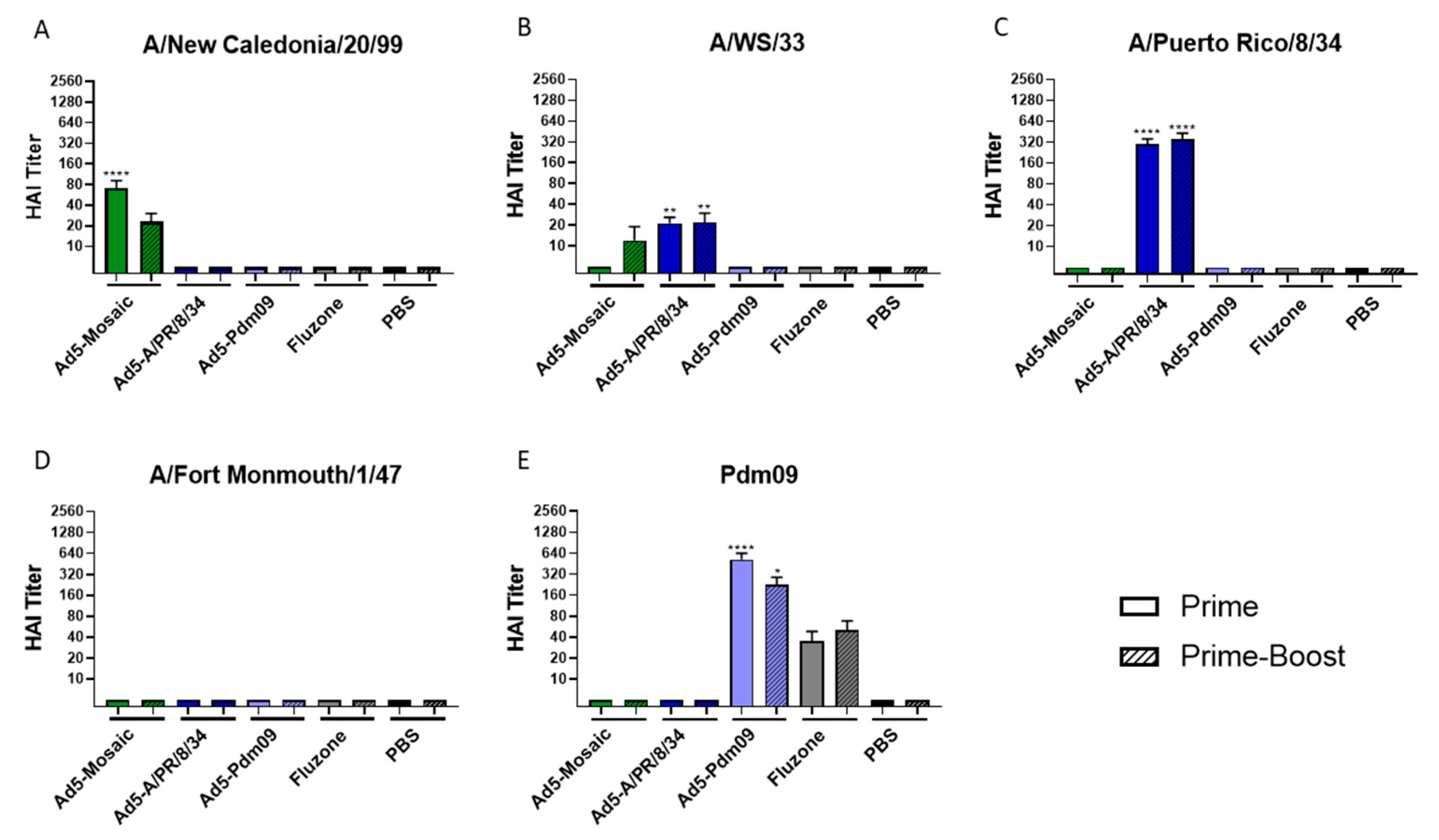
2.10. Hemagglutination Inhibition Titers
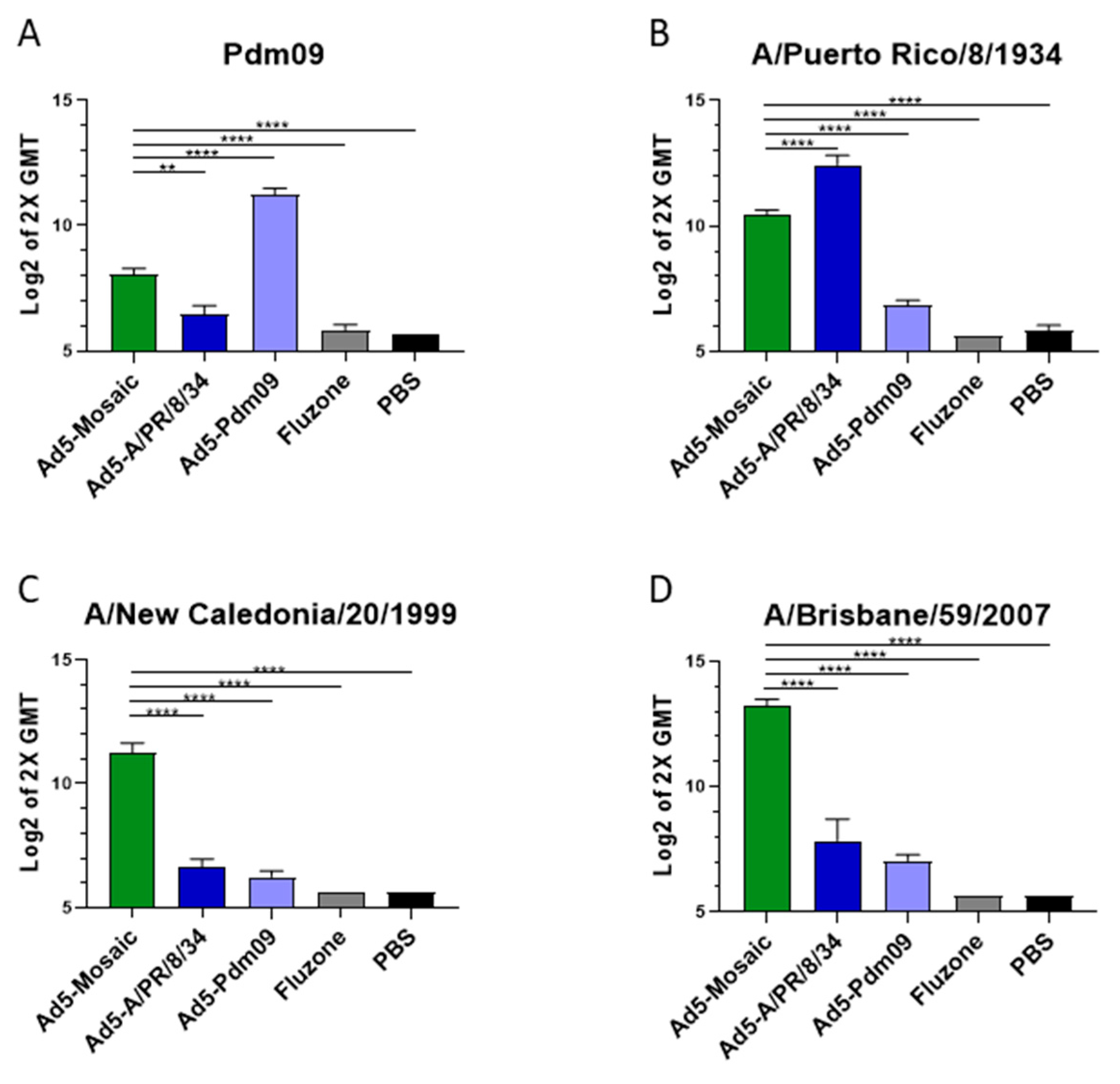
2.11. ELISA
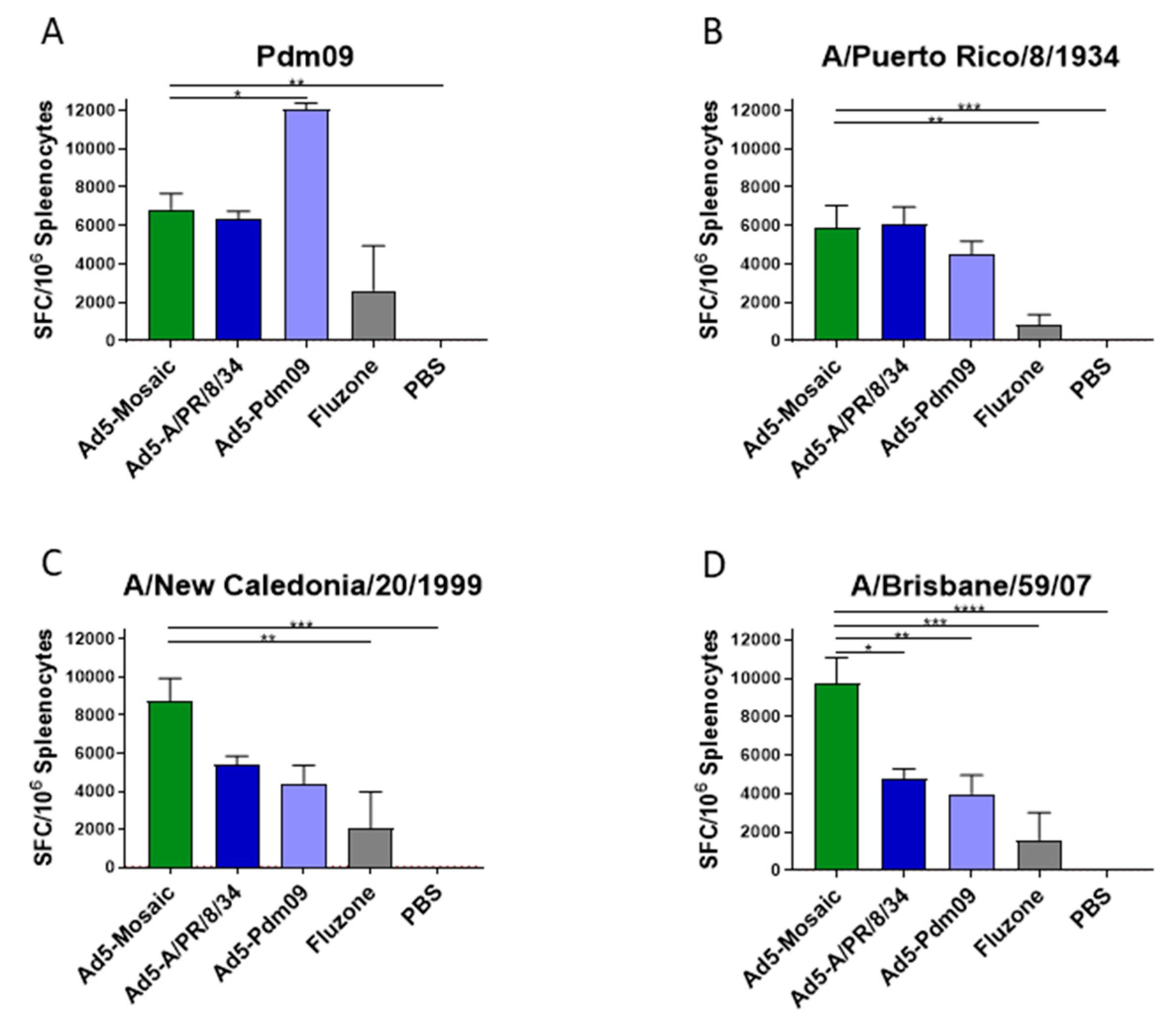
2.12. ELISpot Assay
2.13. Mouse Challenge Studies
2.14. Statistical Analysis
3. Results
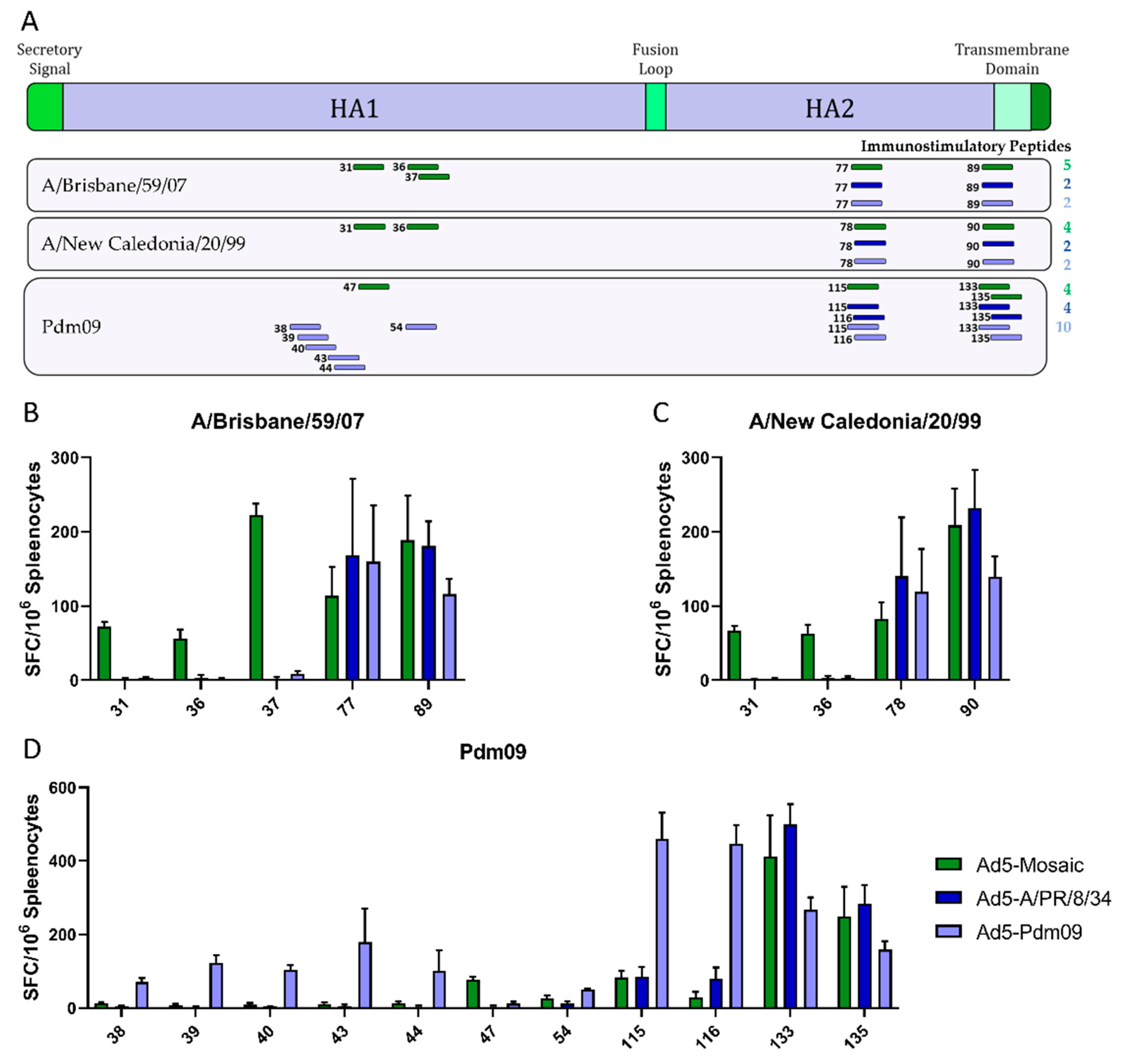
3.1. Design and Characteristics of the Human H1 HA Mosaic Immunogen
3.2. Vaccine-Induced Antibody Responses
3.3. Vaccine Induced T Cell Responses
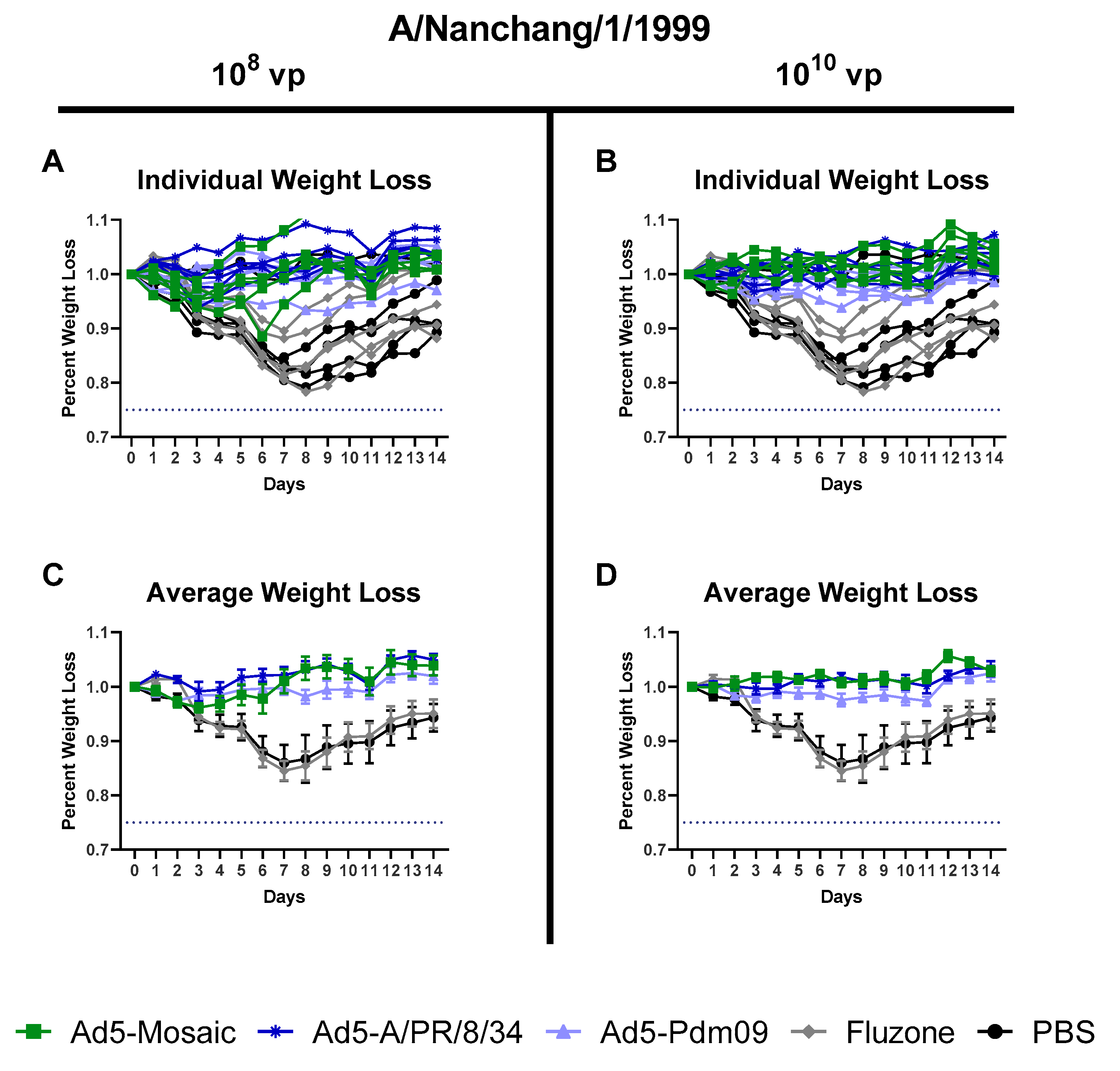
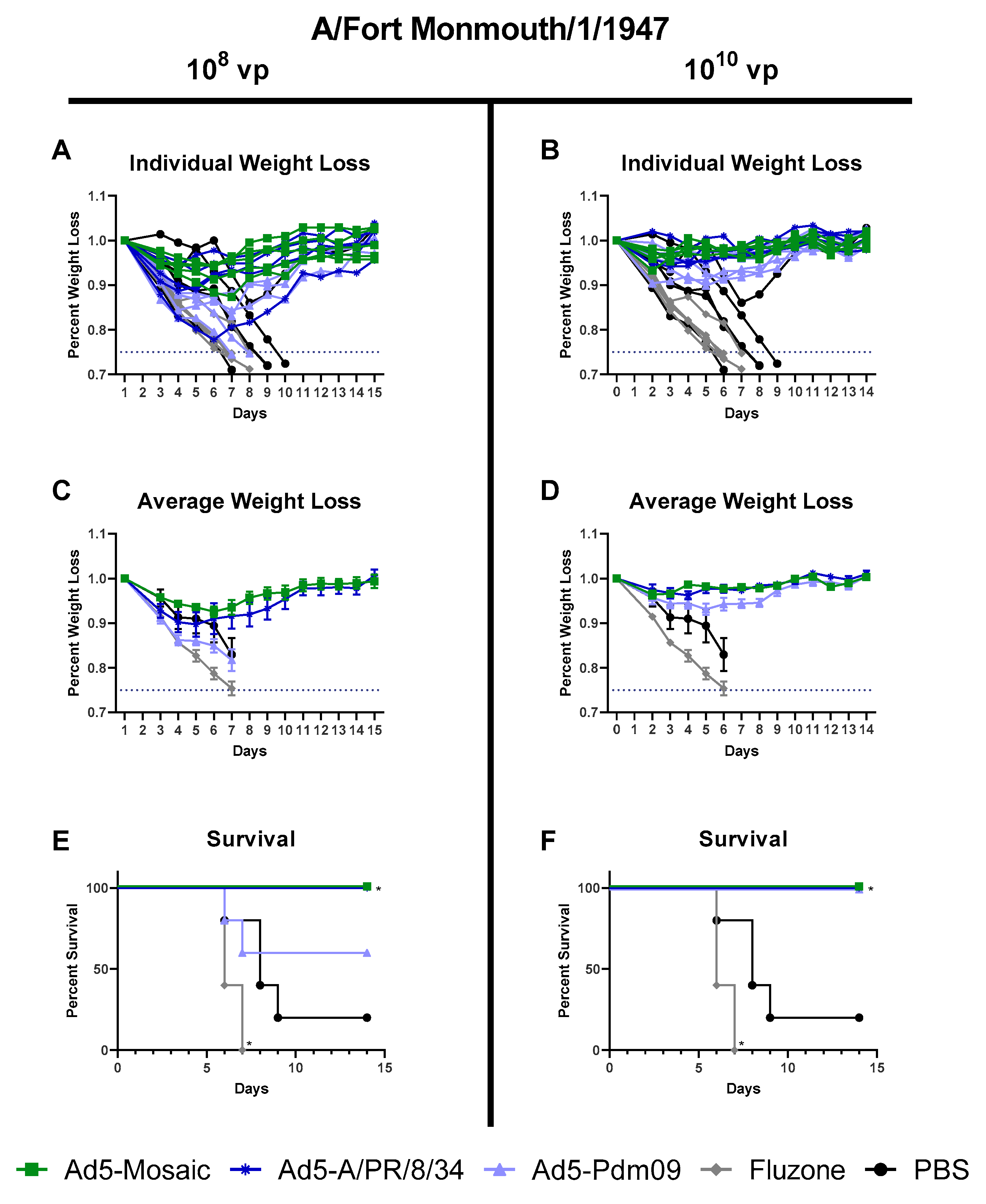
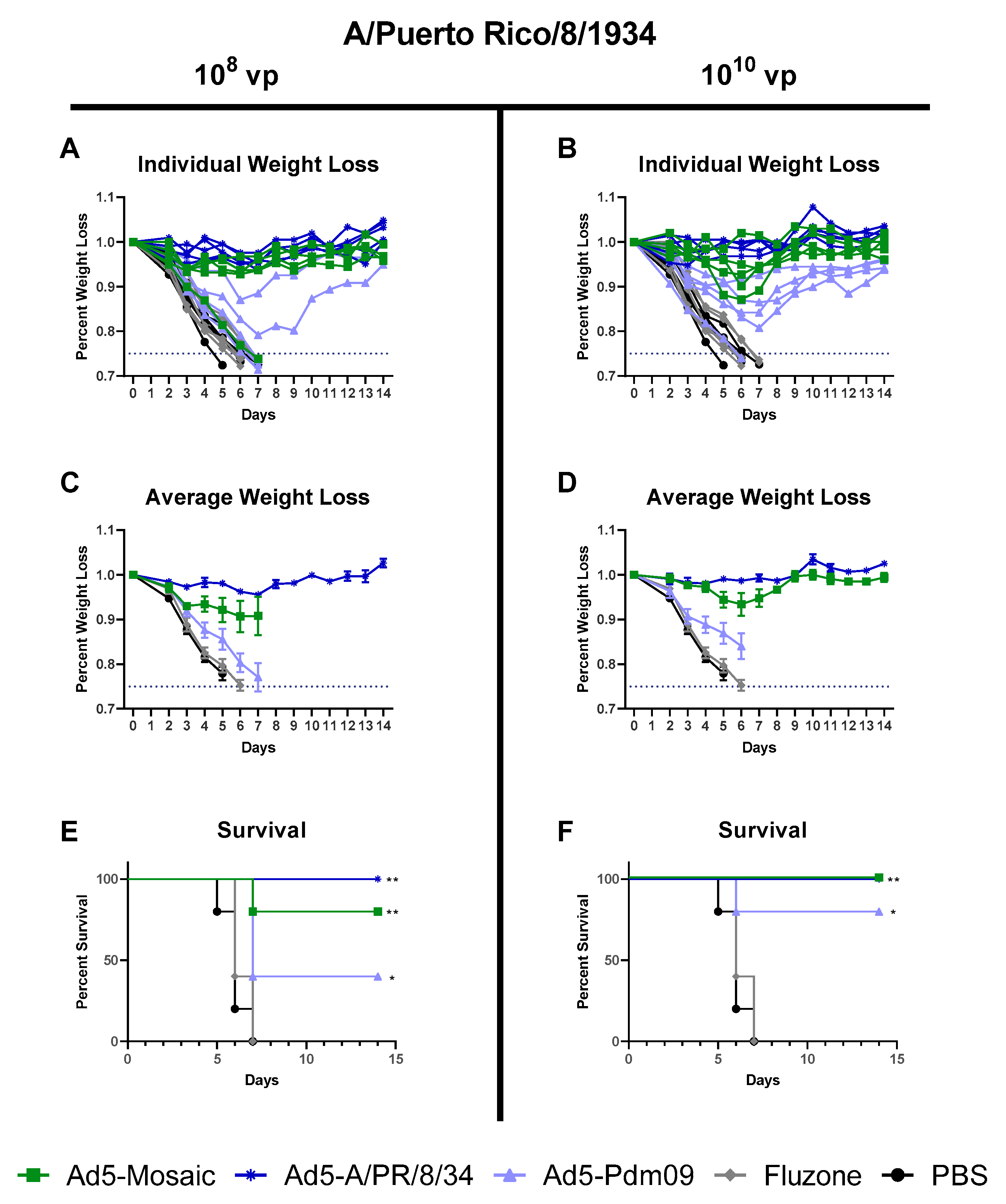
3.4. Protection Against Divergent Influenza Virus Challenges
3.4.1. Influenza Virus A/Nanchang/1/99 challenge
3.4.2. Influenza Virus A/Fort Monmouth/1/47 challenge
3.4.3. Influenza Virus A/Puerto Rico/8/34 challenge
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bresee, J.; Fitzner, J.; Campbell, H.; Cohen, C.; Cozza, V.; Jara, J.; Krishnan, A.; Lee, V.; The WHO Working Group on the Burden of Influenza Disease. Progress and Remaining Gaps in Estimating the Global Disease Burden of Influenza. Emerg. Infect. Dis. 2018, 24, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- WHO. Seasonal Influenza and Influenza A(H1N1). Available online: https://www.who.int/ith/diseases/si_iAh1n1/en/ (accessed on 9 October 2019).
- Rafeek, R.A.M.; Divarathna, M.V.M.; Noordeen, F. History and current trends in influenza virus infections with special reference to Sri Lanka. Virusdisease 2017, 28, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Jain, S.; Finelli, L.; Shaw, M.W.; Lindstrom, S.; Garten, R.J.; Gubareva, L.V.; Xu, X.; Bridges, C.B.; Uyeki, T.M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. 2009, 360, 2605–2615. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef]
- Young, B.; Sadarangani, S.; Jiang, L.; Wilder-Smith, A.; Chen, M.I. Duration of Influenza Vaccine Effectiveness: A Systematic Review, Meta-analysis, and Meta-regression of Test-Negative Design Case-Control Studies. J. Infect. Dis. 2018, 217, 731–741. [Google Scholar] [CrossRef]
- Ferdinands, J.M.; Fry, A.M.; Reynolds, S.; Petrie, J.; Flannery, B.; Jackson, M.L.; Belongia, E.A. Intraseason waning of influenza vaccine protection: Evidence from the US Influenza Vaccine Effectiveness Network, 2011-12 through 2014-15. Clin. Infect. Dis. 2017, 64, 544–550. [Google Scholar] [CrossRef]
- CDC. CDC Seasonal Flu Vaccine Effectiveness Studies. Available online: https://www.cdc.gov/flu/vaccines-work/effectiveness-studies.htm (accessed on 4 September 2019).
- Vemula, S.V.; Sayedahmed, E.E.; Sambhara, S.; Mittal, S.K. Vaccine approaches conferring cross-protection against influenza viruses. Exp. Rev.Vvaccines 2017, 16, 1141–1154. [Google Scholar] [CrossRef]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virology 2013, 87, 6542–6550. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P.; Steel, J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr. Top. Microbiol. Immunol. 2015, 386, 301–321. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013, 3, 521–530. [Google Scholar] [CrossRef]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Ngai, L.; Ekiert, D.C.; Wilson, I.A.; Garcia-Sastre, A.; Moran, T.M.; Palese, P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. USA 2010, 107, 18979–18984. [Google Scholar] [CrossRef]
- Weaver, E.A.; Rubrum, A.M.; Webby, R.J.; Barry, M.A. Protection against divergent influenza H1N1 virus by a centralized influenza hemagglutinin. PLoS ONE 2011, 6, e18314. [Google Scholar] [CrossRef]
- Lingel, A.; Bullard, B.L.; Weaver, E.A. Efficacy of an Adenoviral Vectored Multivalent Centralized Influenza Vaccine. Sci. Rep. 2017, 7, 14912. [Google Scholar] [CrossRef]
- Hillaire, M.L.; van Trierum, S.E.; Kreijtz, J.H.; Bodewes, R.; Geelhoed-Mieras, M.M.; Nieuwkoop, N.J.; Fouchier, R.A.; Kuiken, T.; Osterhaus, A.D.; Rimmelzwaan, G.F. Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells. J. Gen. Virol. 2011, 92, 2339–2349. [Google Scholar] [CrossRef]
- Kreijtz, J.H.; Bodewes, R.; van Amerongen, G.; Kuiken, T.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 2007, 25, 612–620. [Google Scholar] [CrossRef]
- Schulman, J.L.; Kilbourne, E.D. Induction of partial specific heterotypic immunity in mice by a single infection with influenza A virus. J. Bacteriol. 1965, 89, 170–174. [Google Scholar]
- Savic, M.; Dembinski, J.L.; Kim, Y.; Tunheim, G.; Cox, R.J.; Oftung, F.; Peters, B.; Mjaaland, S. Epitope specific T-cell responses against influenza A in a healthy population. Immunology 2016, 147, 165–177. [Google Scholar] [CrossRef]
- Hemann, E.A.; Kang, S.M.; Legge, K.L. Protective CD8 T cell-mediated immunity against influenza A virus infection following influenza virus-like particle vaccination. J. Immunol. 2013, 191, 2486–2494. [Google Scholar] [CrossRef]
- Grant, E.J.; Chen, L.; Quinones-Parra, S.; Pang, K.; Kedzierska, K.; Chen, W. T-cell immunity to influenza A viruses. Crit. Rev. Immunol. 2014, 34, 15–39. [Google Scholar] [CrossRef]
- Fischer, W.; Perkins, S.; Theiler, J.; Bhattacharya, T.; Yusim, K.; Funkhouser, R.; Kuiken, C.; Haynes, B.; Letvin, N.L.; Walker, B.D.; et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 2007, 13, 100–106. [Google Scholar] [CrossRef]
- Santra, S.; Muldoon, M.; Watson, S.; Buzby, A.; Balachandran, H.; Carlson, K.R.; Mach, L.; Kong, W.P.; McKee, K.; Yang, Z.Y.; et al. Breadth of cellular and humoral immune responses elicited in rhesus monkeys by multi-valent mosaic and consensus immunogens. Virology 2012, 428, 121–127. [Google Scholar] [CrossRef]
- Barouch, D.H.; Stephenson, K.E.; Borducchi, E.N.; Smith, K.; Stanley, K.; McNally, A.G.; Liu, J.; Abbink, P.; Maxfield, L.F.; Seaman, M.S.; et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 2013, 155, 531–539. [Google Scholar] [CrossRef]
- Kamlangdee, A.; Kingstad-Bakke, B.; Osorio, J.E. Mosaic H5 Hemagglutinin Provides Broad Humoral and Cellular Immune Responses against Influenza Viruses. J. Virology 2016, 90, 6771–6783. [Google Scholar] [CrossRef]
- Hou, J.; Shrivastava, S.; Fraser, C.C.; Loo, H.L.; Wong, L.H.; Ho, V.; Fink, K.; Ooi, E.E.; Chen, J. Dengue Mosaic Vaccines Enhance Cellular Immunity and Expand the Breadth of Neutralizing Antibody Against All Four Serotypes of Dengue Viruses in Mice. Front. Immunol. 2019, 10, 1429. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids. Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Cummings, M.P. PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods); Sinauer Associates: Sunderland, UK, 2014. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids. Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids. Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Los Alamos National Laboratory. Mosaic Vaccine Designer. 24 March 2016. Available online: https://www.hiv.lanl.gov/content/sequence/MOSAIC/makeVaccine.html (accessed on 10 November 2017).
- Grund, S.; Adams, O.; Wahlisch, S.; Schweiger, B. Comparison of hemagglutination inhibition assay, an ELISA-based micro-neutralization assay and colorimetric microneutralization assay to detect antibody responses to vaccination against influenza A H1N1 2009 virus. J. Virol. Methods 2011, 171, 369–373. [Google Scholar] [CrossRef]
- Jacobsen, H.; Rajendran, M.; Choi, A.; Sjursen, H.; Brokstad, K.A.; Cox, R.J.; Palese, P.; Krammer, F.; Nachbagauer, R. Influenza Virus Hemagglutinin Stalk-Specific Antibodies in Human Serum are a Surrogate Marker for In Vivo Protection in a Serum Transfer Mouse Challenge Model. mBio 2017, 8. [Google Scholar] [CrossRef]
- Kamlangdee, A.; Kingstad-Bakke, B.; Anderson, T.K.; Goldberg, T.L.; Osorio, J.E. Broad protection against avian influenza virus by using a modified vaccinia Ankara virus expressing a mosaic hemagglutinin gene. J. Virol. 2014, 88, 13300–13309. [Google Scholar] [CrossRef]
- He, X.S.; Holmes, T.H.; Zhang, C.; Mahmood, K.; Kemble, G.W.; Lewis, D.B.; Dekker, C.L.; Greenberg, H.B.; Arvin, A.M. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J. Virol. 2006, 80, 11756–11766. [Google Scholar] [CrossRef]
- Hoft, D.F.; Babusis, E.; Worku, S.; Spencer, C.T.; Lottenbach, K.; Truscott, S.M.; Abate, G.; Sakala, I.G.; Edwards, K.M.; Creech, C.B.; et al. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J. Infect. Dis. 2011, 204, 845–853. [Google Scholar] [CrossRef]
- Vemula, S.V.; Mittal, S.K. Production of adenovirus vectors and their use as a delivery system for influenza vaccines. Expert Opin. Biol. Ther. 2010, 10, 1469–1487. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corder, B.N.; Bullard, B.L.; DeBeauchamp, J.L.; Ilyushina, N.A.; Webby, R.J.; Weaver, E.A. Influenza H1 Mosaic Hemagglutinin Vaccine Induces Broad Immunity and Protection in Mice. Vaccines 2019, 7, 195. https://doi.org/10.3390/vaccines7040195
Corder BN, Bullard BL, DeBeauchamp JL, Ilyushina NA, Webby RJ, Weaver EA. Influenza H1 Mosaic Hemagglutinin Vaccine Induces Broad Immunity and Protection in Mice. Vaccines. 2019; 7(4):195. https://doi.org/10.3390/vaccines7040195
Chicago/Turabian StyleCorder, Brigette N., Brianna L. Bullard, Jennifer L. DeBeauchamp, Natalia A. Ilyushina, Richard J. Webby, and Eric A. Weaver. 2019. "Influenza H1 Mosaic Hemagglutinin Vaccine Induces Broad Immunity and Protection in Mice" Vaccines 7, no. 4: 195. https://doi.org/10.3390/vaccines7040195
APA StyleCorder, B. N., Bullard, B. L., DeBeauchamp, J. L., Ilyushina, N. A., Webby, R. J., & Weaver, E. A. (2019). Influenza H1 Mosaic Hemagglutinin Vaccine Induces Broad Immunity and Protection in Mice. Vaccines, 7(4), 195. https://doi.org/10.3390/vaccines7040195




