New Promising Targets for Synthetic Omptin-Based Peptide Vaccine against Gram-Negative Pathogens
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Design
2.2. Omptin Sequences
2.3. Prediction of Linear and Conformational B-Cell Epitopes
2.4. Peptide Library Characteristics
2.5. Predicted Antigenic and Allergenic Characteristics of the Omptin’s B-cell Epitopes
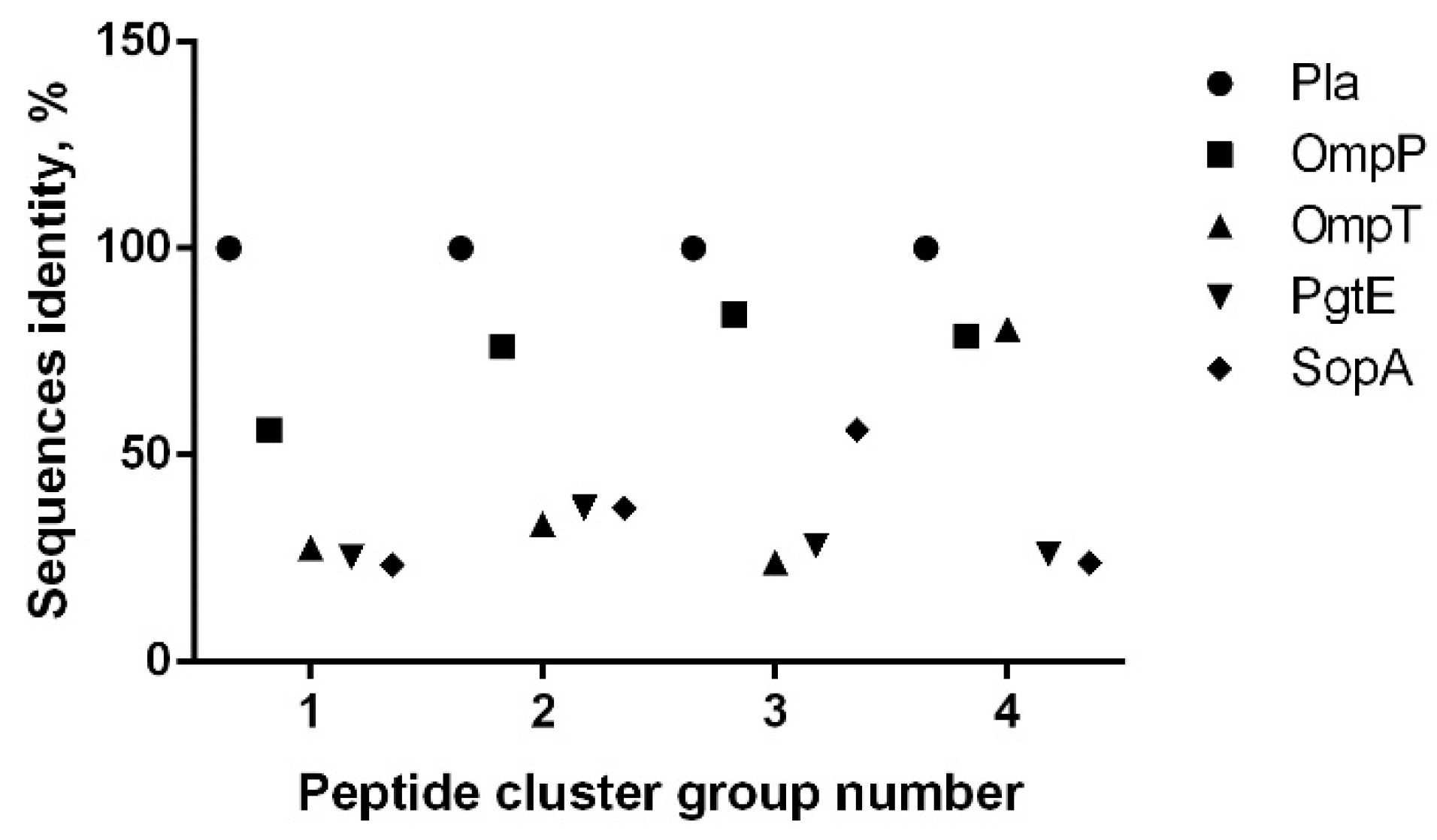
3. Results
3.1. Omptin B-Cell Epitopes Prediction
3.2. B-Cell Epitope Mapping with Human Sera
3.3. The 3D Modeling of the Predicted Location of the Omptin Epitopes
3.4. Omptins Allergenicity and Antigenicity Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Media Centre. The Top 10 Causes of Death. Available online: http://www.who.int/mediacentre/factsheets/fs310/en/ (accessed on 19 February 2019).
- Fauci, A.S.; Touchette, N.A.; Folkers, G.K. Emerging infectious diseases: A 10-year perspective from the National Institute of Allergy and Infectious Diseases. Emerg. Infect. Dis. 2005, 11, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Nii-Trebi, N.I. Emerging and Neglected Infectious Diseases. Insights, Advances, and Challenges. Biomed. Res. Int. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hays, J.N. Epidemics and Pandemics: Their Impacts on Human History; ABC-CLIO: Santa Barbara, CA, USA, 2005; p. 513. [Google Scholar]
- Bhutta, Z.A.; Sommerfeld, J.; Lassi, Z.S.; Salam, R.A.; Das, J.K. Global burden, distribution, and interventions for infectious diseases of poverty. Infect. Dis. Poverty 2014, 3, 21. [Google Scholar] [CrossRef]
- World Health Organization. Neglected Tropical Diseases. Available online: http://www.who.int/neglected_diseases/diseases/en/ (accessed on 19 February 2019).
- World Health Organization. Media Centre. Diarrhoeal Disease. Updated. May 2018. Available online: http://www.who.int/mediacentre/factsheets/fs330/en/ (accessed on 19 February 2019).
- Plotkin, S.A. Increasing Complexity of Vaccine Development. Infect. Dis. 2015, 212. [Google Scholar] [CrossRef] [PubMed]
- Cawein, A.; Emini, E.; Watson, M.; Dailey, J.; Donnelly, J.; Tresnan, D.; Evans, T.; Plotkin, S.; Gruber, W. Human capital gaps in vaccine development: An issue for global vaccine development and global health. Ann. N. Y. Acad. Sci. 2017, 1395, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Serdobova, I.; Kieny, M. Assembling a Global Vaccine Development Pipeline for Infectious Diseases in the Developing World. Am. J. Public Health 2006, 96, 1554–1559. [Google Scholar] [CrossRef]
- Feodorova, V.A.; Sayapina, L.V.; Corbel, M.J.; Motin, V.L. Russian vaccines against especially dangerous bacterial pathogens. Emerg. Microb. Infect. 2014, 3, e86. [Google Scholar] [CrossRef]
- Walker, C.L.; Aryee, M.J.; Boschi-Pinto, C.; Black, R.E. Estimating diarrhea mortality among young children in low and middle income countries. PLoS ONE 2012, 7, e29151. [Google Scholar] [CrossRef]
- Walker, C.L.F.; Rudan, I.; Liu, L.; Nair, H.; Theodoratou, E.; Bhutta, Z.A.; O’Brien, K.L.; Campbell, H.; Black, R.E. Global burden of childhood pneumonia and diarrhoea. Lancet 2013, 381, 1405–1416. [Google Scholar] [CrossRef]
- O’Ryan, M.; Vidal, R.; del Canto, F.; Salazar, J.C.; Montero, D. Vaccines for viral and bacterial pathogens causing acute gastroenteritis: Part I: Overview, vaccines for enteric viruses and Vibrio cholera. Hum. Vaccin. Immunother. 2015, 11, 584–600. [Google Scholar] [CrossRef]
- World Health Organization. Module 2: Types of Vaccine and Adverse Reactions. Available online: http://vaccine-safety-training.org/types-of-vaccine.html (accessed on 19 February 2019).
- O’Ryan, M.; Vidal, R.; del Canto, F.; Salazar, J.C.; Montero, D. Vaccines for viral and bacterial pathogens causing acute gastroenteritis: Part II: Vaccines for Shigella, Salmonella, enterotoxigenic E. coli (ETEC) enterohemorragic E. coli (EHEC) and Campylobacter jejuni. Hum. Vaccin. Immunother. 2015, 11, 601–619. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.M.C.; Pinto, M.V.; Sadarangani, M.; Plotkin, S.A. Whither vaccines? J. Infect. 2017, 74, S2–S9. [Google Scholar] [CrossRef]
- Kieny, M.P.; Excler, J.L.; Girard, M. Research and Development of New Vaccines Against Infectious Diseases. Am. J. Public Health 2004, 94, 1931–1935. [Google Scholar] [CrossRef]
- Koff, W.C.; Burton, D.R.; Johnson, P.R.; Walker, B.D.; King, C.R.; Nabel, G.J.; Ahmed, R.; Bhan, M.K.; Plotkin, S.A. Accelerating next-generation vaccine development for global disease prevention. Science 2013, 340. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; McAleer, J.P.; Lin, Y.; Paterson, D.L.; Zheng, M.; Alcorn, J.F.; Weaver, C.T.; Kolls, J.K. Th17 cells mediate clade specific, serotype-independent mucosal immunity. Immunity 2011, 35, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.; Mortensen, R.; Rosenkrands, I.; Dietrich, J.; Andersen, P. Vaccine-induced Th17 cells are established as resident memory cells in the lung and promote local IgA responses. Mucosal Immunol. 2017, 10, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Ahmada, T.A.; Eweidac, A.E.; Sheweita, S.A. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials Vaccinol. 2016, 5, 71–83. [Google Scholar] [CrossRef]
- Gayet, R.; Bioley, G.; Rochereau, N.; Paul, S.; Corthésy, B. Vaccination against Salmonella Infection: the Mucosal Way. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Kukkonen, M.; Korhonen, T.K. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 2004, 294, 7–14. [Google Scholar] [CrossRef]
- Brannon, J.R.; Thomassin, J.L.; Gruenheid, S.; Le Moual, H. Antimicrobial Peptide Conformation as a Structural Determinant of Omptin Protease Specificity. J. Bacteriol. 2015, 197, 3583–3591. [Google Scholar] [CrossRef]
- Hritonenko, V.; Stathopoulos, C. Omptin proteins: an expanding family of outer membrane proteases in Gram-negative Enterobacteriaceae. Mol. Membr. Biol. 2007, 24, 395–406. [Google Scholar] [CrossRef]
- Braciale, V.L.; Nash, M.; Sinha, N.; Zudina, I.V.; Motin, V.L. Correlates of Immunity Elicited By Live Yersinia Pestis Vaccine. In Infectious Disease; Georgiev, V.S., Western, K., McGowan, J.J., Eds.; Springer: Totowa, NJ, USA, 2008; pp. 473–480. [Google Scholar]
- Feodorova, V.; Khizhnyakova, M.; Lyapina, A.; Zaitsev, S.; Telepnev, M.; Sayapina, L.; Ulianova, O.; Ulyanov, S.; Arseneva, T.; Morozova, I.; et al. Characterization of Humoral and Cellular Immune Responses to Yersinia Pestis Pla Antigen in Humans Immunised with Live Plague Vaccine (LPV). In Proceedings of the 7th Congress of European Microbiologists (FEMS 2017), Valencia, Spain, 9–13 July 2017. [Google Scholar]
- Feodorova, V.A.; Khizhnyakova, M.A.; Lyapina, A.M.; Zaitsev, S.S.; Subbotina, I.A.; Telepnev, M.V.; Sayapina, L.V.; Ulianova, O.V.; Arseneva, T.E.; Morozova, I.V.; et al. New promising targets for synthetic Omptin-based peptide vaccine against Gram-negative pathogens. In Proceedings of the International Society for Vaccine (ISV) Annual Congress, Paris, France, 5–7 October 2017. [Google Scholar]
- Feodorova, V.A.; Lyapina, A.M.; Khizhnyakova, M.A.; Zaitsev, S.S.; Sayapina, L.V.; Arseneva, T.E.; Trukhachev, A.L.; Lebedeva, S.A.; Telepnev, M.V.; Ulianova, O.V.; et al. Humoral and Cellular Immune Responses to Yersinia pestis Pla Antigen in Humans Immunized with Live Plague Vaccine. PLoS Negl. Trop. Dis. 2018, 11, e0006511. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006, 34, W202–W209. [Google Scholar] [CrossRef]
- Cheng, J.; Randall, A.Z.; Sweredoski, M.J.; Baldi, P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids. Res. 2005, 33, W72–W76. [Google Scholar] [CrossRef]
- Butler, T. Plague gives surprises in the first decade of the 21st century in the United States and worldwide. Am. J. Trop. Med. Hyg. 2013, 89, 788–793. [Google Scholar] [CrossRef]
- Feodorova, V.A.; Motin, V.L. Plague vaccines: current developments and future perspectives. Emerg. Microbes Infect. 2012, 1, e36. [Google Scholar] [CrossRef]
- Tulchinsky, T.H.; Varavikova, E.A. Communicable Diseases. In The New Public Health, 3rd ed.; Tulchinsky, T.H., Varavikova, E.A., Eds.; Elsevier/academic Press: San Diego, CA, USA, 2014; pp. 149–236. [Google Scholar]
- Center for Disease Control and Prevention. E.coli (Escherichia coli). General Information. Available online: https://www.cdc.gov/ecoli/general/index.html (accessed on 19 February 2019).
- Haiko, J.; Kukkonen, M.; Ravantti, J.J.; Westerlund-Wikstrom, B.; Korhonen, T.K. The single substitution I259T conserved in the plasminogen activator Pla of pandemic Yersinia pestis branches enhances fibrinolytic activity. J. Bacteriol. 2009, 191, 4758–4766. [Google Scholar] [CrossRef] [PubMed]
- Zimbler, D.L.; Schroeder, J.A.; Eddy, J.L.; Lathem, W.W. Early emergence of Yersinia pestis as a severe respiratory pathogen. Nat. Commun. 2015, 6, 7487. [Google Scholar] [CrossRef] [PubMed]
- Dentovskaya, S.V.; Platonov, M.E.; Svetoch, T.E.; Kopylov, P.K.; Kombarova, T.I.; Ivanov, S.A.; Shaikhutdinova, R.Z.; Kolombet, L.V.; Chauhan, S.; Ablamunits, V.G. Two Isoforms of Yersinia pestis Plasminogen Activator Pla: Intraspecies Distribution, Intrinsic Disorder Propensity, and Contribution to Virulence. PLoS ONE 2016, 11, e0168089. [Google Scholar] [CrossRef]
- Sodeinde, O.A.; Subrahmanyam, Y.V.; Stark, K.; Quan, T.; Bao, Y.; Goguen, J.D. A surface protease and the invasive character of plague. Science 1992, 258, 1004–1007. [Google Scholar] [CrossRef]
- Lahteenmaki, K.; Edelman, S.; Korhonen, T.K. Bacterial metastasis: The host plasminogen system in bacterial invasion. Trends Microbiol. 2005, 13, 79–85. [Google Scholar] [CrossRef]
- Lathem, W.W.; Price, P.A.; Miller, V.L.; Goldman, W.E. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 2007, 315, 509–513. [Google Scholar] [CrossRef]
- Feodorova, V.A.; Devdariani, Z.L. Development, characterisation and diagnostic application of monoclonal antibodies against Yersinia pestis fibrinolysin and coagulase. J. Med. Microbiol. 2000, 49, 261–269. [Google Scholar] [CrossRef]
- Suomalainen, M.; Lobo, L.A.; Brandenburg, K.; Lindner, B.; Virkola, R.; Knirel, Y.A.; Anisimov, A.P.; Holst, O.; Korhonen, T.K. Temperature-induced changes in the lipopolysaccharide of Yersinia pestis affect plasminogen activation by the Pla surface protease. Infect. Immun. 2010, 78, 2644–2652. [Google Scholar] [CrossRef]
- Easterbrook, T.J.; Reddin, K.; Robinson, A.; Modi, N. Studies on the immunogenicity of the Pla protein from Yersinia pestis. Contrib. Microbiol. Immunol. 1995, 13, 214–215. [Google Scholar]
- Benner, G.E.; Andrews, G.P.; Byrne, W.R.; Strachan, S.D.; Sample, A.K.; Heath, D.G.; Friedlander, A.M. Immune response to Yersinia outer proteins and other Yersinia pestis antigens after experimental plague infection in mice. Infect. Immun. 1999, 67, 1922–1928. [Google Scholar]
- Holmgren, J.; Parashar, U.D.; Plotkin, S.; Louis, J.; Ng, S.P.; Desauziers, E.; Picot, V.; Saadatian-Elahi, M. Correlates of protection for enteric vaccines. Vaccine 2017, 35, 3355–3363. [Google Scholar] [CrossRef]
- Wang, S.; Heilman, D.; Liu, F.; Giehl, T.; Joshi, S.; Huang, X.; Chou, T.H.; Goguen, J.; Lu, S. A DNA vaccine producing LcrV antigen in oligomers is effective in protecting mice from lethal mucosal challenge of plague. Vaccine 2004, 22, 3348–3357. [Google Scholar] [CrossRef]
- Erova, T.E.; Rosenzweig, J.A.; Sha, J.; Suarez, G.; Sierra, J.C.; Kirtley, M.L.; van Lier, C.J.; Telepnev, M.V.; Motin, V.L.; Chopra, A.K. Evaluation of protective potential of Yersinia pestis outer membrane protein antigens as possible candidates for a new-generation recombinant plague vaccine. Clin. Vaccine Immunol. 2013, 20, 227–238. [Google Scholar] [CrossRef]
- Noster, R.; Riedel, R.; Mashreghi, M.F.; Radbruch, H.; Harms, L.; Haftmann, C.; Chang, H.D.; Radbruch, A.; Zielinski, C.E. IL-17 and GM-CSF expression are antagonistically regulated by human T helper cells. Sci. Transl. Med. 2014, 6, 241–280. [Google Scholar] [CrossRef]
- Bielinska, A.U.; O’Konek, J.J.; Janczak, K.W.; Baker, J.R., Jr. Immunomodulation of TH2 biased immunity with mucosal administration of nanoemulsion adjuvant. Vaccine 2016, 34, 4017–4024. [Google Scholar] [CrossRef] [PubMed]
- James, L.K.; Till, S.J. Potential mechanisms for IgG4 inhibition of immediate hypersensitivity reactions. Curr. Allergy Asthma Rep. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Rappuoli, R. The challenge of developing universal vaccines. F1000 Med. Rep. 2011, 3, 16. [Google Scholar] [CrossRef] [PubMed]

| No. | GenBank No. | Protein Name | No. of Amino Acids | No. of B-Cell Epitopes Predicted 2 | Allergenicity | Antigenicity | |
|---|---|---|---|---|---|---|---|
| Linear | Conformational | ||||||
| 1 | CAB53170.1 | Pla | 312 | 9 | 6 | Non-allergen | Antigenic |
| 2 | AP001918.1 | OmpP | 315 | 11 | 4 | Non-allergen | Antigenic |
| 3 | KU664810.1 | OmpT | 317 | 7 | 5 | Non-allergen | Antigenic |
| 4 | ATT45876.1 | PgtE | 312 | 10 | 5 | Non-allergen | Antigenic |
| 5 | U73461.1 | SopA | 315 | 9 | 4 | Non-allergen | Antigenic |
| Peptide Immono-Reactive Cluster No. | Omptin Epitope Predicted in ElliPro | Pla Peptide Number | Positive Reaction with Sera from Donor Group | Actual Pla Peptide Position (Library Peptide ID 1) | Pla Peptide Position for Epitope Predicted in ElliPro | |
|---|---|---|---|---|---|---|
| A | B | |||||
| 1 | 1 | 1–4 | + | + | 16–45 (4–7) | 22–36 |
| 2 | 5–7 | + | - | 41–65 (9–11) | 52–60 | |
| N/A 3 | 3 2 | N/A | - | - | 76–90 (16) | None |
| 2 | 4 | 8–10 | + | + | 86–110 (18–20) | 95–98 |
| 5 | 11–15 | + | + | 101–135 (21–25) | 107–124 | |
| 6 | 16–18 | + | + | 131–155 (27–29) | 136–150 | |
| 19 | + | - | 146–160 (30) | |||
| 7 | 20–24 | + | + | 156–190 (32–36) | 163–184 | |
| 3 | 8 | 26–27 | + | + | 221–240 (45–46) | 227–234 |
| 28 | + | - | 231–245 (47) | |||
| N/A 3 | 9 2 | N/A | - | - | 251–265 (51) | None |
| 4 | 10 | 29–32 | + | + | 266–295 (54–57) | 274–295 |
| 33–34 | + | 286–305 (58–59) | ||||
| 11 | 35 | + | 296–310 (60) | 306–312 | ||
| 36 | + | 301–312 (61) | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feodorova, V.A.; Lyapina, A.M.; Zaitsev, S.S.; Khizhnyakova, M.A.; Sayapina, L.V.; Ulianova, O.V.; Ulyanov, S.S.; Motin, V.L. New Promising Targets for Synthetic Omptin-Based Peptide Vaccine against Gram-Negative Pathogens. Vaccines 2019, 7, 36. https://doi.org/10.3390/vaccines7020036
Feodorova VA, Lyapina AM, Zaitsev SS, Khizhnyakova MA, Sayapina LV, Ulianova OV, Ulyanov SS, Motin VL. New Promising Targets for Synthetic Omptin-Based Peptide Vaccine against Gram-Negative Pathogens. Vaccines. 2019; 7(2):36. https://doi.org/10.3390/vaccines7020036
Chicago/Turabian StyleFeodorova, Valentina A., Anna M. Lyapina, Sergey S. Zaitsev, Maria A. Khizhnyakova, Lidiya V. Sayapina, Onega V. Ulianova, Sergey S. Ulyanov, and Vladimir L. Motin. 2019. "New Promising Targets for Synthetic Omptin-Based Peptide Vaccine against Gram-Negative Pathogens" Vaccines 7, no. 2: 36. https://doi.org/10.3390/vaccines7020036
APA StyleFeodorova, V. A., Lyapina, A. M., Zaitsev, S. S., Khizhnyakova, M. A., Sayapina, L. V., Ulianova, O. V., Ulyanov, S. S., & Motin, V. L. (2019). New Promising Targets for Synthetic Omptin-Based Peptide Vaccine against Gram-Negative Pathogens. Vaccines, 7(2), 36. https://doi.org/10.3390/vaccines7020036




