Will Attention by Vaccine Developers to the Host’s Nuclear Hormone Levels and Immunocompetence Improve Vaccine Success?
Abstract
1. The Long Path to RSV Vaccine Licensing
2. Vaccine Development Hurdles in Human Immunodeficiency Virus (HIV) and Influenza Virus Fields
3. Will Attention to the Host’s Nuclear Hormone Levels Improve Vaccine Success?
4. Vitamins and the Immune Response
5. Sex Hormones and the Immune Response
6. Innate Immune Cells Are Influenced by Nuclear Hormones
7. B Cells Are Influenced by Nuclear Hormones Directly
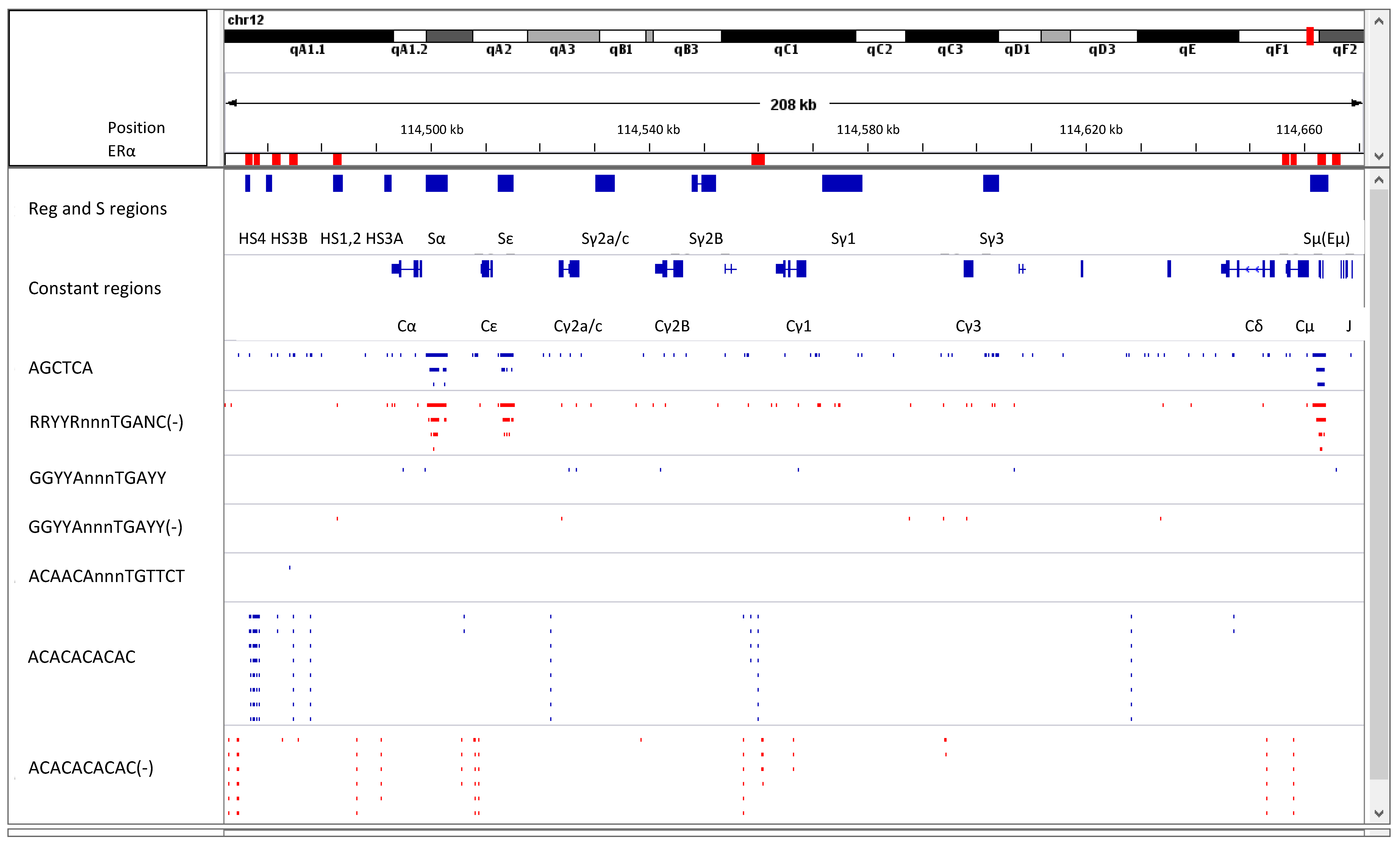
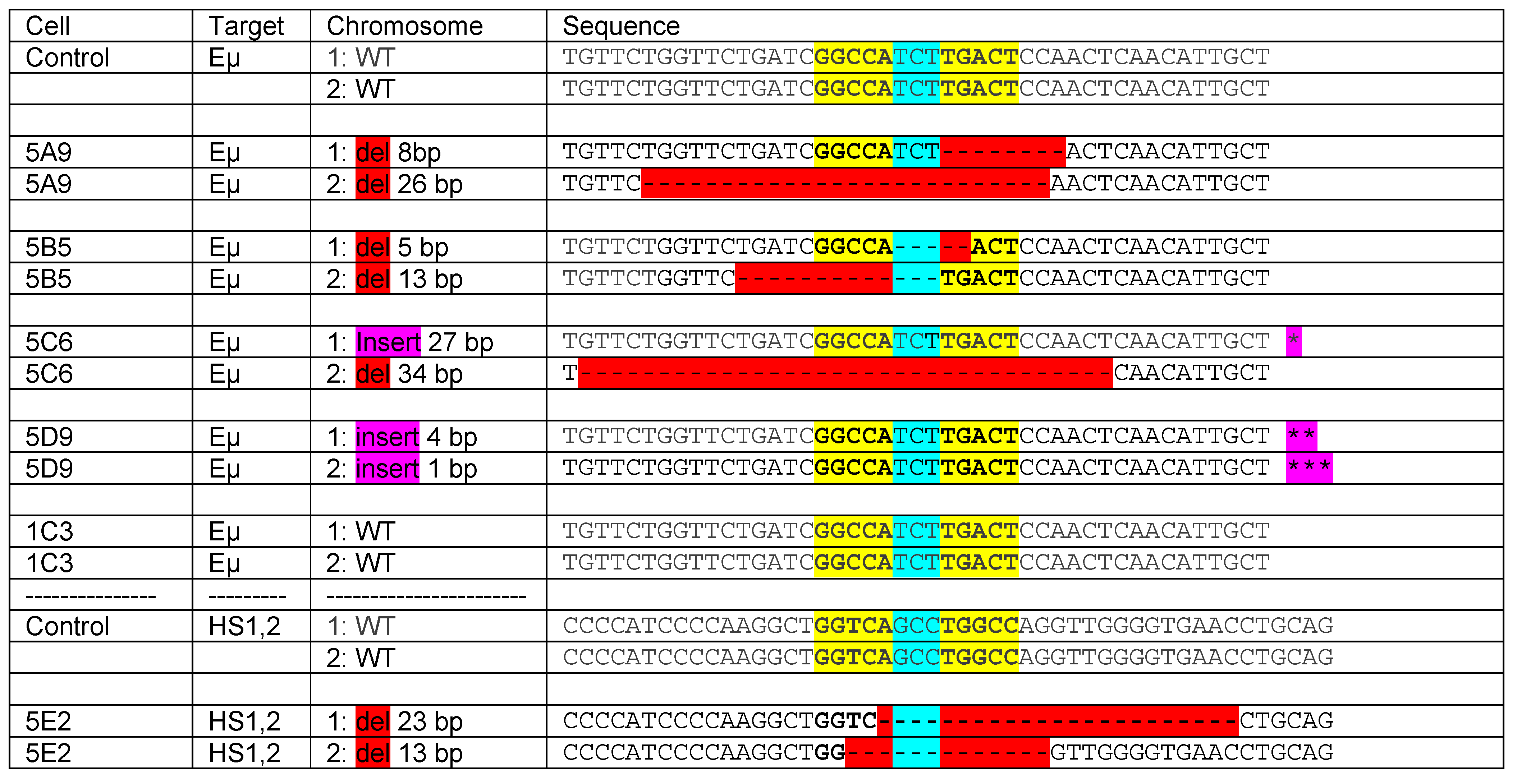
8. Estrogen Response Elements (ERE) and Adjacent Sequences Regulate Antibody Isotype Expression in B Cells
9. Additional Nuclear Hormones
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RSV | respiratory syncytial virus |
| HIV | human immunodeficiency virus |
| RAR | retinoic acid receptor |
| RARE | retinoic acid response element |
| VDR | vitamin D receptor |
| ERα | estrogen receptor α |
| ERE | estrogen response element |
| ARE | androgen response element |
| AID | activation induced deaminase |
| CSR | class switch recombination |
| 3′RR | 3′ regulatory region |
| ChIP | chromatin immunoprecipitation |
| DBD | DNA-binding domain |
| LBD | ligand-binding domain |
| FCS | fetal calf serum |
| NGS | next-generation sequencing |
References
- Belderbos, M.E.; Houben, M.L.; Wilbrink, B.; Lentjes, E.; Bloemen, E.M.; Kimpen, J.L.L.; Rovers, M.; Bont, L. Cord blood vitamin D deficiency is associated with respiratory syncytial virus bronchiolitis. Pediatrics 2011, 127, e1513–e1520. [Google Scholar] [CrossRef] [PubMed]
- Groothuis, J.R.; King, S.J.; Hogerman, D.A.; Paradiso, P.R.; Simoes, E.A. Safety and immunogenicity of a purified F protein respiratory syncytial virus (PFP-2) vaccine in seropositive children with bronchopulmonary dysplasia. J. Infect. Dis. 1998, 177, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Russell, C.J.; Simoes, E.A.F.; Hurwitz, J.L. Vaccines for the Paramyxoviruses and Pneumoviruses: Successes, Candidates, and Hurdles. Viral Immunol. 2018, 31, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Simoes, E.A.; Tan, D.H.; Ohlsson, A.; Sales, V.; Wang, E.E. Respiratory syncytial virus vaccine: A systematic overview with emphasis on respiratory syncytial virus subunit vaccines. Vaccine 2001, 20, 954–960. [Google Scholar] [CrossRef]
- Boukhvalova, M.S.; Blanco, J.C. The cotton rat Sigmodon hispidus model of respiratory syncytial virus infection. Curr. Top. Microbiol. Immunol. 2013, 372, 347–358. [Google Scholar] [PubMed]
- Wright, P.F.; Karron, R.A.; Belshe, R.B.; Thompson, J.; Crowe, J.E., Jr.; Boyce, T.G.; Halburnt, L.L.; Reed, G.W.; Whitehead, S.S.; Anderson, E.L. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J. Infect. Dis. 2000, 182, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, I.M.; Karron, R.A.; Eichelberger, M.; Walsh, E.E.; Delagarza, V.W.; Bennett, R.; Chanock, R.M.; Murphy, B.R.; Clements-Mann, M.L.; Falsey, A.R. Evaluation of the live attenuated cpts 248/404 RSV vaccine in combination with a subunit RSV vaccine (PFP-2) in healthy young and older adults. Vaccine 2000, 18, 1763–1772. [Google Scholar] [CrossRef]
- Karron, R.A.; Buchholz, U.J.; Collins, P.L. Live-attenuated respiratory syncytial virus vaccines. Curr. Top. Microbiol. Immunol. 2013, 372, 259–284. [Google Scholar] [PubMed]
- Karron, R.A.; Thumar, B.; Schappell, E.; Surman, S.; Murphy, B.R.; Collins, P.L.; Schmidt, A.C. Evaluation of two chimeric bovine-human parainfluenza virus type 3 vaccines in infants and young children. Vaccine 2012, 30, 3975–3981. [Google Scholar] [CrossRef] [PubMed]
- Fulginiti, V.A.; Eller, J.J.; Sieber, O.F.; Joyner, J.W.; Minamitani, M.; Meiklejohn, G. Respiratory virus immunization I. A field trial of two inactivated respiratory virus vaccines; an aqueous trivalent parainfluenza virus vaccine and an alum-precipitated respiratory syncytial virus vaccine. Am. J. Epidemiol. 1969, 89, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.; Magoffin, R.L.; Shearer, L.A.; Schieble, J.H.; Lennette, E.H. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am. J. Epidemiol. 1969, 89, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Glenn, G.M.; Fries, L.F.; Thomas, D.N.; Smith, G.; Kpamegan, E.; Lu, H.; Flyer, D.; Jani, D.; Hickman, S.P.; Piedra, P.A. A randomized, blinded, controlled, dose-ranging study of a respiratory syncytial virus recombinant fusion (F) nanoparticle vaccine in healthy women of childbearing age. J. Infect. Dis. 2015, 213, 411–422. [Google Scholar] [CrossRef] [PubMed]
- de Waal, L.; Power, U.F.; Yuksel, S.; van Amerongen, G.; Nguyen, T.N.; Niesters, H.G.; de Swart, R.L.; Osterhaus, A.D. Evaluation of BBG2Na in infant macaques: Specific immune responses after vaccination and RSV challenge. Vaccine 2004, 22, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.S.; Hurwitz, J.L.; Simoes, E.A.F.; Piedra, P.A. Establishing Correlates of Protection for Vaccine Development: Considerations for the Respiratory Syncytial Virus Vaccine Field. Viral Immunol. 2018, 31, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R.; Walsh, E.E.; Capellan, J.; Gravenstein, S.; Zambon, M.; Yau, E.; Gorse, G.J.; Edelman, R.; Hayden, F.G.; McElhaney, J.E.; et al. Comparison of the safety and immunogenicity of 2 respiratory syncytial virus (rsv) vaccines—Nonadjuvanted vaccine or vaccine adjuvanted with alum—Given concomitantly with influenza vaccine to high-risk elderly individuals. J. Infect. Dis. 2008, 198, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Langley, J.M.; Sales, V.; McGeer, A.; Guasparini, R.; Predy, G.; Meekison, W.; Li, M.; Capellan, J.; Wang, E. A dose-ranging study of a subunit Respiratory Syncytial Virus subtype A vaccine with and without aluminum phosphate adjuvantation in adults > or =65 years of age. Vaccine 2009, 27, 5913–5919. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.S.; Chen, M.; Chang, J.S.; Yang, Y.; Kim, A.; Graham, B.S.; Kwong, P.D. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J. Virol. 2010, 84, 12236–12244. [Google Scholar] [CrossRef] [PubMed]
- Widjojoatmodjo, M.N.; Boes, J.; van Bers, M.; van Remmerden, Y.; Roholl, P.J.; Luytjes, W. A highly attenuated recombinant human respiratory syncytial virus lacking the G protein induces long-lasting protection in cotton rats. Virol. J. 2010, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Prince, G.A.; Capiau, C.; Deschamps, M.; Fabry, L.; Garcon, N.; Gheysen, D.; Prieels, J.P.; Thiry, G.; Van Opstal, O.; Porter, D.D. Efficacy and safety studies of a recombinant chimeric respiratory syncytial virus FG glycoprotein vaccine in cotton rats. J. Virol. 2000, 74, 10287–10292. [Google Scholar] [CrossRef] [PubMed]
- Ofek, G.; Guenaga, F.J.; Schief, W.R.; Skinner, J.; Baker, D.; Wyatt, R.; Kwong, P.D. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. USA 2010, 107, 17880–17887. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.A.; Settembre, E.C.; Shaw, C.A.; Dey, A.K.; Rappuoli, R.; Mandl, C.W.; Dormitzer, P.R.; Carfi, A. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc. Natl. Acad. Sci. USA 2011, 108, 9619–9624. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.S.; Chen, M.; Leung, S.; Graepel, K.W.; Du, X.; Yang, Y.; Zhou, T.; Baxa, U.; Yasuda, E.; Beaumont, T.; et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 2013, 340, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.M.; Korban, S.S. Transgenic apple expressing an antigenic protein of the human respiratory syncytial virus. J. Plant Physiol. 2010, 167, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.H.; He, J.S.; Wang, X.B.; Zheng, X.X.; Wu, Q.; Xie, C.; Zhang, M.; Wei, W.; Tang, Q.; Song, J.D.; et al. A prime-boost vaccination strategy using attenuated Salmonella typhimurium and a replication-deficient recombinant adenovirus vector elicits protective immunity against human respiratory syncytial virus. Biochem. Biophys. Res. Commun. 2010, 395, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; He, J.S.; Zhang, M.; Xue, S.L.; Wu, Q.; Ding, X.D.; Song, W.; Yuan, Y.; Li, D.L.; Zheng, X.X.; et al. Oral respiratory syncytial virus (RSV) DNA vaccine expressing RSV F protein delivered by attenuated Salmonella typhimurium. Hum. Gene Ther. 2007, 18, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Cautivo, K.M.; Bueno, S.M.; Cortes, C.M.; Wozniak, A.; Riedel, C.A.; Kalergis, A.M. Efficient lung recruitment of respiratory syncytial virus-specific Th1 cells induced by recombinant bacillus Calmette-Guerin promotes virus clearance and protects from infection. J. Immunol. 2010, 185, 7633–7645. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.L.; Purcell, R.H.; London, W.T.; Lawrence, L.A.; Chanock, R.M.; Murphy, B.R. Evaluation in chimpanzees of vaccinia virus recombinants that express the surface glycoproteins of human respiratory syncytial virus. Vaccine 1990, 8, 164–168. [Google Scholar] [CrossRef]
- Mok, H.; Lee, S.; Utley, T.J.; Shepherd, B.E.; Polosukhin, V.V.; Collier, M.L.; Davis, N.L.; Johnston, R.E.; Crowe, J.E., Jr. Venezuelan equine encephalitis virus replicon particles encoding respiratory syncytial virus surface glycoproteins induce protective mucosal responses in mice and cotton rats. J. Virol. 2007, 81, 13710–13722. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sobrido, L.; Gitiban, N.; Fernandez-Sesma, A.; Cros, J.; Mertz, S.E.; Jewell, N.A.; Hammond, S.; Flano, E.; Durbin, R.K.; Garcia-Sastre, A.; et al. Protection against respiratory syncytial virus by a recombinant Newcastle disease virus vector. J. Virol. 2006, 80, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Hu, K.F.; Rozell, B.; Orvell, C.; Morein, B.; Liljestrom, P. Vaccination with recombinant alphavirus or immune-stimulating complex antigen against respiratory syncytial virus. J. Immunol. 2002, 169, 3208–3216. [Google Scholar] [CrossRef] [PubMed]
- Takimoto, T.; Hurwitz, J.L.; Coleclough, C.; Prouser, C.; Krishnamurthy, S.; Zhan, X.; Boyd, K.; Scroggs, R.A.; Brown, B.; Nagai, Y.; et al. Recombinant Sendai virus expressing the G glycoprotein of respiratory syncytial virus (RSV) elicits immune protection against RSV. J. Virol. 2004, 78, 6043–6047. [Google Scholar] [CrossRef] [PubMed]
- Takimoto, T.; Hurwitz, J.L.; Zhan, X.; Krishnamurthy, S.; Prouser, C.; Brown, B.; Coleclough, C.; Boyd, K.; Scroggs, R.A.; Portner, A.; et al. Recombinant Sendai virus as a novel vaccine candidate for respiratory syncytial virus. Viral Immunol. 2005, 18, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Hurwitz, J.L.; Krishnamurthy, S.; Takimoto, T.; Boyd, K.; Scroggs, R.A.; Surman, S.; Portner, A.; Slobod, K.S. Respiratory syncytial virus (RSV) fusion protein expressed by recombinant Sendai virus elicits B-cell and T-cell responses in cotton rats and confers protection against RSV subtypes A and B. Vaccine 2007, 25, 8782–8793. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Slobod, K.S.; Jones, B.G.; Sealy, R.E.; Takimoto, T.; Boyd, K.; Surman, S.; Russell, C.J.; Portner, A.; Hurwitz, J.L. Sendai virus recombinant vaccine expressing a secreted, unconstrained respiratory syncytial virus fusion protein protects against RSV in cotton rats. Int. Immunol. 2015, 27, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Slobod, K.S.; Krishnamurthy, S.; Luque, L.E.; Takimoto, T.; Jones, B.; Surman, S.; Russell, C.J.; Portner, A.; Hurwitz, J.L. Sendai virus recombinant vaccine expressing hPIV-3 HN or F elicits protective immunity and combines with a second recombinant to prevent hPIV-1, hPIV-3 and RSV infections. Vaccine 2008, 26, 3480–3488. [Google Scholar] [CrossRef] [PubMed]
- Phan, S.I.; Chen, Z.; Xu, P.; Li, Z.; Gao, X.; Foster, S.L.; Teng, M.N.; Tripp, R.A.; Sakamoto, K.; He, B. A respiratory syncytial virus (RSV) vaccine based on parainfluenza virus 5 (PIV5). Vaccine 2014, 32, 3050–3057. [Google Scholar] [CrossRef] [PubMed]
- Malkin, E.; Yogev, R.; Abughali, N.; Sliman, J.; Wang, C.K.; Zuo, F.; Yang, C.-F.; Eickhoff, M.; Esser, M.T.; Tang, R.S.; et al. Safety and immunogenicity of a live attenuated RSV vaccine in healthy RSV-seronegative children 5 to 24 months of age. PLoS ONE 2013, 8, e77104. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.C.; Schaap-Nutt, A.; Bartlett, E.J.; Schomacker, H.; Boonyaratanakornkit, J.; Karron, R.A.; Collins, P.L. Progress in the development of human parainfluenza virus vaccines. Expert Rev. Respir. Med. 2011, 5, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.G.; Sealy, R.E.; Rudraraju, R.; Traina-Dorge, V.L.; Finneyfrock, B.; Cook, A.; Takimoto, T.; Portner, A.; Hurwitz, J.L. Sendai virus-based RSV vaccine protects African green monkeys from RSV infection. Vaccine 2012, 30, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.; Huang, Y.; Langley, J.M. Prospects for defined epitope vaccines for respiratory syncytial virus. Future Microbiol. 2010, 5, 585–602. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, T.; Kamphuis, T.; Meijerhof, T.; Goud, E.; de Haan, A.; Wilschut, J. Lipopeptide-adjuvanted respiratory syncytial virus virosomes: A safe and immunogenic non-replicating vaccine formulation. Vaccine 2010, 28, 5543–5550. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sambhara, S.; Li, C.X.; Ettorre, L.; Switzer, I.; Cates, G.; James, O.; Parrington, M.; Oomen, R.; Du, R.P.; et al. Plasmid DNA encoding the respiratory syncytial virus G protein is a promising vaccine candidate. Virology 2000, 269, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Choi, Y.; Haynes, L.M.; Harcourt, J.L.; Anderson, L.J.; Jones, L.P.; Tripp, R.A. Vaccination to induce antibodies blocking the CX3C-CX3CR1 interaction of respiratory syncytial virus G protein reduces pulmonary inflammation and virus replication in mice. J. Virol. 2010, 84, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Block, S.L.; Falloon, J.; Hirschfield, J.A.; Krilov, L.R.; Dubovsky, F.; Yi, T.; Belshe, R.B. Immunogenicity and safety of a quadrivalent live attenuated influenza vaccine in children. Pediatr. Infect. Dis. J. 2012, 31, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Thondamal, M.; Witting, M.; Schmitt-Kopplin, P.; Aguilaniu, H. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat. Commun. 2014, 5, 4879. [Google Scholar] [CrossRef] [PubMed]
- Sirakov, M.; Plateroti, M. The thyroid hormones and their nuclear receptors in the gut: From developmental biology to cancer. Biochim. Biophys. Acta 2011, 1812, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Santos, R.S.; Criollo, A.; Nelson, M.D.; Palmer, B.F.; Clegg, D.J. The effects of oestrogens and their receptors on cardiometabolic health. Nat. Rev. Endocrinol. 2017, 13, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Rieger, S.; Zhao, H.; Martin, P.; Abe, K.; Lisse, T.S. The role of nuclear hormone receptors in cutaneous wound repair. Cell Biochem. Funct. 2015, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Neinast, M.D.; Frank, A.P.; Sun, K.; Park, J.; Zehr, J.A.; Vishvanath, L.; Morselli, E.; Amelotte, M.; Palmer, B.F.; et al. ERalpha upregulates Phd3 to ameliorate HIF-1 induced fibrosis and inflammation in adipose tissue. Mol. Metab. 2014, 3, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Klinge, C.M. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001, 29, 2905–2919. [Google Scholar] [CrossRef] [PubMed]
- Klinge, C.M. Role of estrogen receptor ligand and estrogen response element sequence on interaction with chicken ovalbumin upstream promoter transcription factor (COUP-TF). J. Steroid Biochem. Mol. Biol. 1999, 71, 1–19. [Google Scholar] [CrossRef]
- Mason, C.E.; Shu, F.J.; Wang, C.; Session, R.M.; Kallen, R.G.; Sidell, N.; Yu, T.; Liu, M.H.; Cheung, E.; Kallen, C.B. Location analysis for the estrogen receptor-alpha reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res. 2010, 38, 2355–2368. [Google Scholar] [CrossRef] [PubMed]
- El-Tanani, M.K.; Green, C.D. Two separate mechanisms for ligand-independent activation of the estrogen receptor. Mol. Endocrinol. 1997, 11, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.M.; Mangelsdorf, D.J. Nuclear Receptors, RXR, and the Big Bang. Cell 2014, 157, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Berrabah, W.; Aumercier, P.; Lefebvre, P.; Staels, B. Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Lett. 2011, 585, 1640–1650. [Google Scholar] [CrossRef] [PubMed]
- Sever, R.; Glass, C.K. Signaling by nuclear receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a016709. [Google Scholar] [CrossRef] [PubMed]
- Bastien, J.; Rochette-Egly, C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 2004, 328, 1–16. [Google Scholar] [CrossRef] [PubMed]
- De Bruyn, R.; Bollen, R.; Claessens, F. Identification and characterization of androgen response elements. Methods Mol. Biol. 2011, 776, 81–93. [Google Scholar] [PubMed]
- Wilson, S.; Qi, J.; Filipp, F.V. Refinement of the androgen response element based on ChIP-Seq in androgen-insensitive and androgen-responsive prostate cancer cell lines. Sci. Rep. 2016, 6, 32611. [Google Scholar] [CrossRef] [PubMed]
- Klinge, C.M. Estrogen receptor interaction with co-activators and co-repressors. Steroids 2000, 65, 227–251. [Google Scholar] [CrossRef]
- Klinge, C.M.; Peale, F.V., Jr.; Hilf, R.; Bambara, R.A.; Zain, S. Cooperative estrogen receptor interaction with consensus or variant estrogen responsive elements in vitro. Cancer Res. 1992, 52, 1073–1081. [Google Scholar] [PubMed]
- Napoli, J.L. Functions of Intracellular Retinoid Binding-Proteins. Subcell. Biochem. 2016, 81, 21–76. [Google Scholar] [PubMed]
- Jones, B.G.; Oshansky, C.M.; Bajracharya, R.; Tang, L.; Sun, Y.; Wong, S.S.; Webby, R.; Thomas, P.G.; Hurwitz, J.L. Retinol binding protein and vitamin D associations with serum antibody isotypes, serum influenza virus-specific neutralizing activities and airway cytokine profiles. Clin. Exp. Immunol. 2016, 183, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Stephens, D.; Jackson, P.L.; Gutierrez, Y. Subclinical vitamin A deficiency: A potentially unrecognized problem in the United States. Pediatr. Nurs. 1996, 22, 377–389, 456. [Google Scholar] [PubMed]
- Trasino, S.E.; Tang, X.H.; Jessurun, J.; Gudas, L.J. Obesity Leads to Tissue, but not Serum Vitamin A Deficiency. Sci. Rep. 2015, 5, 15893. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, J.L.; Jones, B.G.; Penkert, R.R.; Gansebom, S.; Sun, Y.; Tang, L.; Bramley, A.M.; Jain, S.; McCullers, J.A.; Arnold, S.R. Low Retinol-Binding Protein and Vitamin D Levels Are Associated with Severe Outcomes in Children Hospitalized with Lower Respiratory Tract Infection and Respiratory Syncytial Virus or Human Metapneumovirus Detection. J Pediatr. 2017, 187, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Rudraraju, R.; Surman, S.L.; Jones, B.G.; Sealy, R.; Woodland, D.L.; Hurwitz, J.L. Reduced frequencies and heightened CD103 expression among virus-induced CD8(+) T cells in the respiratory tract airways of vitamin A-deficient mice. Clin. Vaccine Immunol. 2012, 19, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Surman, S.L.; Jones, B.G.; Rudraraju, R.; Sealy, R.E.; Hurwitz, J.L. Intranasal administration of retinyl palmitate with a respiratory virus vaccine corrects impaired mucosal IgA response in the vitamin A-deficient host. Clin. Vaccine Immunol. 2014, 21, 598–601. [Google Scholar] [CrossRef] [PubMed]
- Surman, S.L.; Jones, B.G.; Sealy, R.E.; Rudraraju, R.; Hurwitz, J.L. Oral retinyl palmitate or retinoic acid corrects mucosal IgA responses toward an intranasal influenza virus vaccine in vitamin A deficient mice. Vaccine 2014, 32, 2521–2524. [Google Scholar] [CrossRef] [PubMed]
- Surman, S.L.; Jones, B.G.; Woodland, D.L.; Hurwitz, J.L. Enhanced CD103 Expression and Reduced Frequencies of Virus-Specific CD8(+) T Cells Among Airway Lymphocytes After Influenza Vaccination of Mice Deficient in Vitamins A + D. Viral Immunol. 2017, 30, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Surman, S.L.; Penkert, R.R.; Jones, B.G.; Sealy, R.E.; Hurwitz, J.L. Vitamin Supplementation at the Time of Immunization with a Cold-Adapted Influenza Virus Vaccine Corrects Poor Mucosal Antibody Responses in Mice Deficient for Vitamins A and D. Clin. Vaccine Immunol. 2016, 23, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Surman, S.L.; Rudraraju, R.; Sealy, R.; Jones, B.; Hurwitz, J.L. Vitamin A deficiency disrupts vaccine-induced antibody-forming cells and the balance of IgA/IgG isotypes in the upper and lower respiratory tract. Viral Immunol. 2012, 25, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Sealy, R.; Webby, R.J.; Crumpton, J.C.; Hurwitz, J.L. Differential localization and function of antibody-forming cells responsive to inactivated or live-attenuated influenza virus vaccines. Int. Immunol. 2013, 25, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Renegar, K.B.; Small, P.A., Jr.; Boykins, L.G.; Wright, P.F. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J. Immunol. 2004, 173, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- Penkert, R.R.; Iverson, A.; Rosch, J.W.; Hurwitz, J.L. Prevnar-13 vaccine failure in a mouse model for vitamin A deficiency. Vaccine 2017, 35, 6264–6268. [Google Scholar] [CrossRef] [PubMed]
- Semba, R.D.; Muhilal Scott, A.L.; Natadisastra, G.; Wirasasmita, S.; Mele, L.; Ridwan, E.; West, K.P., Jr.; Sommer, A. Depressed immune response to tetanus in children with vitamin A deficiency. J. Nutr. 1992, 122, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Villamor, E.; Fawzi, W.W. Effects of vitamin a supplementation on immune responses and correlation with clinical outcomes. Clin. Microbiol. Rev. 2005, 18, 446–464. [Google Scholar] [CrossRef] [PubMed]
- Neonatal Vitamin ASEg. Early neonatal vitamin A supplementation and infant mortality: An individual participant data meta-analysis of randomised controlled trials. Arch. Dis. Child. 2018, 104, 217–226. [Google Scholar]
- Muenchhoff, M.; Goulder, P.J. Sex differences in pediatric infectious diseases. J. Infect. Dis. 2014, 209 (Suppl. 3), S120–S126. [Google Scholar] [CrossRef] [PubMed]
- Borchers, A.T.; Chang, C.; Gershwin, M.E.; Gershwin, L.J. Respiratory syncytial virus—A comprehensive review. Clin. Rev. Allergy Immunol. 2013, 45, 331–379. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L. Sex influences immune responses to viruses, and efficacy of prophylaxis and treatments for viral diseases. Bioessays 2012, 34, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Green, M.S. The male predominance in the incidence of infectious diseases in children: A postulated explanation for disparities in the literature. Int. J. Epidemiol. 1992, 21, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Garenne, M. Sex differences in measles mortality: A world review. Int. J. Epidemiol. 1994, 23, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Segal, A.O.; Crighton, E.J.; Moineddin, R.; Mamdani, M.; Upshur, R.E. Croup hospitalizations in Ontario: A 14-year time-series analysis. Pediatrics 2005, 116, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Sue, K. The science behind “man flu”. BMJ 2017, 359, j5560. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.G.; Sealy, R.E.; Penkert, R.R.; Surman, S.L.; Maul, R.W.; Neale, G.; Xu, B.; Gearhart, P.J.; Hurwitz, J.L. Complex sex-biased antibody responses: Estrogen receptors bind estrogen response elements centered within immunoglobulin heavy chain gene enhancers. Int. Immunol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.P.; Wilhelm, C.; Yang, Q.; Hall, J.A.; Bouladoux, N.; Boyd, A.; Nutman, T.B.; Urban, J.F., Jr.; Wang, J.; Ramalingam, T.R.; et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 2014, 343, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Penkert, R.R.; Jones, B.G.; Hacker, H.; Partridge, J.F.; Hurwitz, J.L. Vitamin A differentially regulates cytokine expression in respiratory epithelial and macrophage cell lines. Cytokine 2017, 91, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Mora, J.R.; Iwata, M.; Von Andrian, U.H. Vitamin effects on the immune system: Vitamins A and D take centre stage. Nat. Rev. Immunol. 2008, 8, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Pauklin, S.; Sernandez, I.V.; Bachmann, G.; Ramiro, A.R.; Petersen-Mahrt, S.K. Estrogen directly activates AID transcription and function. J. Exp. Med. 2009, 206, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Mai, T.; Zan, H.; Zhang, J.; Hawkins, J.S.; Xu, Z.; Casali, P. Estrogen receptors bind to and activate the HOXC4/HoxC4 promoter to potentiate HoxC4-mediated activation-induced cytosine deaminase induction, immunoglobulin class switch DNA recombination, and somatic hypermutation. J. Biol. Chem. 2010, 285, 37797–37810. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, J.L.; Penkert, R.R.; Xu, B.; Fan, Y.; Partridge, J.F.; Maul, R.W.; Gearhart, P.J. Hotspots for Vitamin-Steroid-Thyroid Hormone Response Elements Within Switch Regions of Immunoglobulin Heavy Chain Loci Predict a Direct Influence of Vitamins and Hormones on B Cell Class Switch Recombination. Viral Immunol. 2016, 29, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.G.; Penkert, R.R.; Xu, B.; Fan, Y.; Neale, G.; Gearhart, P.J.; Hurwitz, J.L. Binding of estrogen receptors to switch sites and regulatory elements in the immunoglobulin heavy chain locus of activated B cells suggests a direct influence of estrogen on antibody expression. Mol. Immunol. 2016, 77, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Pan, Q.; Pardali, E.; Mills, F.C.; Bernstein, R.M.; Max, E.E.; Sideras, P.; Hammarstrom, L. Regulation of germline promoters by the two human Ig heavy chain 3′ alpha enhancers. J. Immunol. 2000, 164, 6380–6386. [Google Scholar] [CrossRef] [PubMed]
- Birshtein, B.K. Epigenetic Regulation of Individual Modules of the immunoglobulin heavy chain locus 3′ Regulatory Region. Front. Immunol. 2014, 5, 163. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, J.S.; Singh, M.; Birshtein, B.K. B cell lineage-specific activator protein (BSAP). A player at multiple stages of B cell development. J. Immunol. 1996, 156, 2349–2351. [Google Scholar] [PubMed]
- Nakamura, M.; Kondo, S.; Sugai, M.; Nazarea, M.; Imamura, S.; Honjo, T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int. Immunol. 1996, 8, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Kuliyev, E.; Gingras, S.; Guy, C.S.; Howell, S.; Vogel, P.; Pelletier, S. Overlapping Role of SCYL1 and SCYL3 in Maintaining Motor Neuron Viability. J. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, S.; Gingras, S.; Green, D.R. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity 2015, 42, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Settipane, G.A.; Pudupakkam, R.K.; McGowan, J.H. Corticosteroid effect on immunoglobulins. J. Allergy Clin. Immunol. 1978, 62, 162–166. [Google Scholar] [CrossRef]
- Oehling, A.G. Suppression of the immune system by oral glucocorticoid therapy in bronchial asthma. Allergy 1997, 52, 144–154. [Google Scholar] [CrossRef] [PubMed]
- De Vito, P.; Incerpi, S.; Pedersen, J.Z.; Luly, P.; Davis, F.B.; Davis, P.J. Thyroid hormones as modulators of immune activities at the cellular level. Thyroid 2011, 21, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Jara, E.L.; Munoz-Durango, N.; Llanos, C.; Fardella, C.; Gonzalez, P.A.; Bueno, S.M.; Kalergis, A.M.; Riedel, C.A.; et al. Modulating the function of the immune system by thyroid hormones and thyrotropin. Immunol. Lett. 2017, 184, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Winoto, A.; Littman, D.R. Nuclear hormone receptors in T lymphocytes. Cell 2002, 109, S57–S66. [Google Scholar] [CrossRef]
- Stavnezer, J.; Guikema, J.E.; Schrader, C.E. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008, 26, 261–292. [Google Scholar] [CrossRef] [PubMed]
- Maul, R.W.; Gearhart, P.J. AID and somatic hypermutation. Adv. Immunol. 2010, 105, 159–191. [Google Scholar] [PubMed]
- Hu, J.; Zhang, Y.; Zhao, L.; Meng, F.; Schatz, D.G.; Alt, F.W. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell 2015, 163, 947–959. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sealy, R.E.; Jones, B.G.; Surman, S.L.; Penkert, R.R.; Pelletier, S.; Neale, G.; Hurwitz, J.L. Will Attention by Vaccine Developers to the Host’s Nuclear Hormone Levels and Immunocompetence Improve Vaccine Success? Vaccines 2019, 7, 26. https://doi.org/10.3390/vaccines7010026
Sealy RE, Jones BG, Surman SL, Penkert RR, Pelletier S, Neale G, Hurwitz JL. Will Attention by Vaccine Developers to the Host’s Nuclear Hormone Levels and Immunocompetence Improve Vaccine Success? Vaccines. 2019; 7(1):26. https://doi.org/10.3390/vaccines7010026
Chicago/Turabian StyleSealy, Robert E., Bart G. Jones, Sherri L. Surman, Rhiannon R. Penkert, Stephane Pelletier, Geoff Neale, and Julia L. Hurwitz. 2019. "Will Attention by Vaccine Developers to the Host’s Nuclear Hormone Levels and Immunocompetence Improve Vaccine Success?" Vaccines 7, no. 1: 26. https://doi.org/10.3390/vaccines7010026
APA StyleSealy, R. E., Jones, B. G., Surman, S. L., Penkert, R. R., Pelletier, S., Neale, G., & Hurwitz, J. L. (2019). Will Attention by Vaccine Developers to the Host’s Nuclear Hormone Levels and Immunocompetence Improve Vaccine Success? Vaccines, 7(1), 26. https://doi.org/10.3390/vaccines7010026




