“Gnothi Seauton”: Leveraging the Host Response to Improve Influenza Virus Vaccine Efficacy
Abstract
1. Introduction
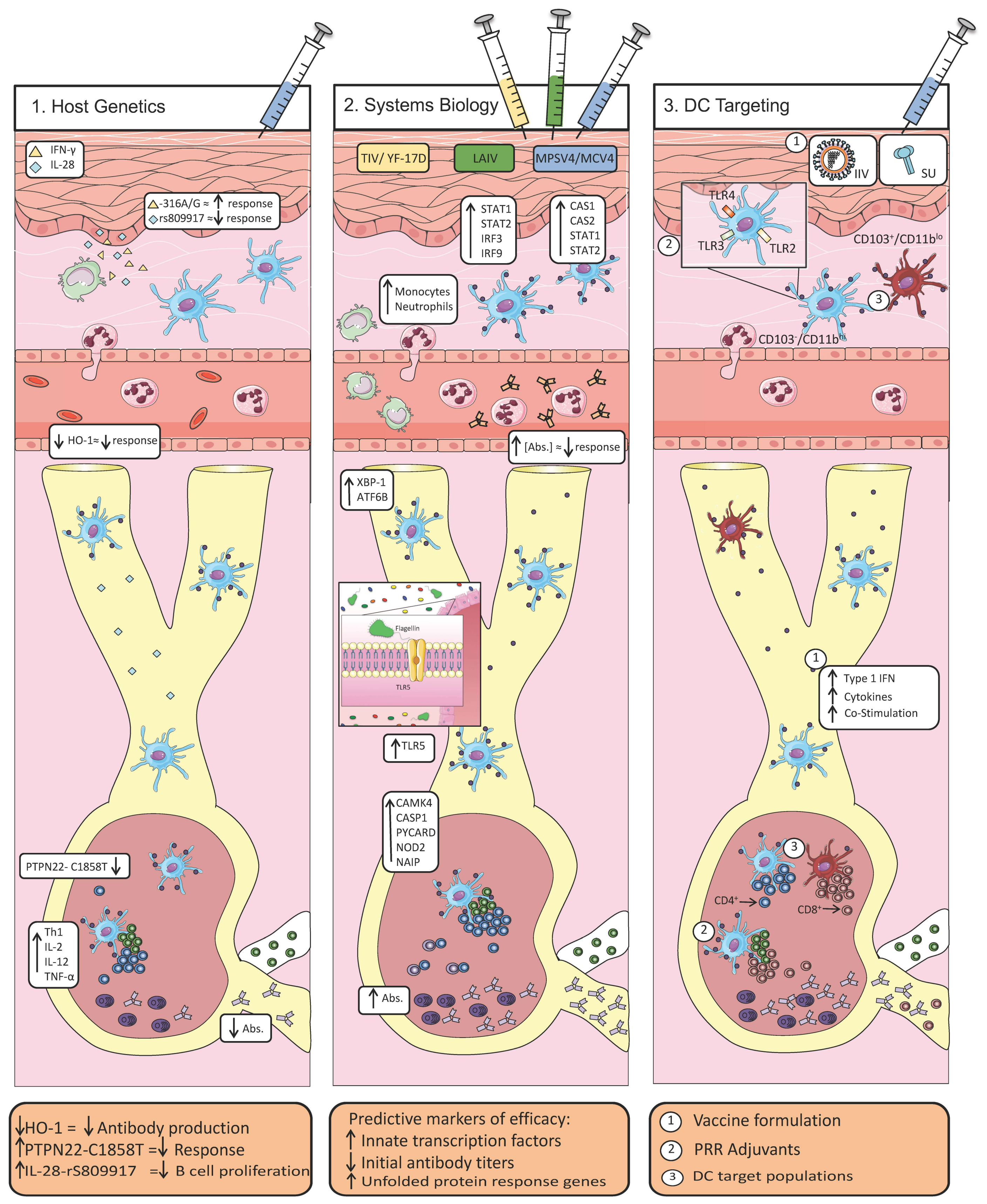
2. Host Determinants of Efficacious Vaccine Responses
2.1. Host Polymorphisms Influence the Development of Protective Immunity
2.2. A Systems-Level View of Vaccine Efficacy
2.3. Coordination of Innate and Adaptive Immunity via Dendritic Cells
3. Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Plotkin, S. History of vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 12283–12287. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S.; Palese, P. Peering into the crystal ball: Influenza pandemics and vaccine efficacy. Cell 2014, 157. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, K.; Viboud, C.; Simonsen, L. Antibody response to influenza vaccination in the elderly: A quantitative review. Vaccine 2006, 24, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, P.A.; Paich, H.A.; Handy, J.; Karlsson, E.A.; Hudgens, M.G.; Sammon, A.B.; Holland, L.A.; Weir, S.; Noah, T.L.; Beck, M.A. Obesity is associated with impaired immune response to influenza vaccination in humans. Int. J. Obes. 2012, 36, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. Novel universal influenza virus vaccine approaches. Curr. Opin. Virol. 2016, 17, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.J. Correlates of protection to influenza virus, where do we go from here? Hum. Vaccin. Immunother. 2013, 9, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Cummins, N.W.; Weaver, E.A.; May, S.M.; Croatt, A.J.; Foreman, O.; Kennedy, R.B.; Poland, G.A.; Barry, M.A.; Nath, K.A.; Badley, A.D. Heme oxygenase-1 regulates the immune response to influenza virus infection and vaccination in aged mice. FASEB J. 2012, 26, 2911–2918. [Google Scholar] [CrossRef] [PubMed]
- Linnik, J.E.; Egli, A. Impact of host genetic polymorphisms on vaccine induced antibody response. Hum. Vaccines Immunother. 2016, 12, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Ovsyannikova, I.G.; Jacobson, R.M. NIH Public Access. Vaccine 2008, 26, D35–D40. [Google Scholar] [CrossRef] [PubMed]
- Talaat, K.R.; Halsey, N.A.; Cox, A.B.; Coles, C.L.; Durbin, A.P.; Ramakrishnan, A.; Bream, J.H. Rapid changes in serum cytokines and chemokines in response to inactivated influenza vaccination. Influenza Other Respir. Viruses 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Egli, A.; Santer, D.M.; O’Shea, D.; Barakat, K.; Syedbasha, M.; Vollmer, M.; Baluch, A.; Bhat, R.; Groenendyk, J.; Joyce, M.A.; et al. IL-28B is a Key Regulator of B- and T-Cell Vaccine Responses against Influenza. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, J.N.; He, W.; Guan, W.; Flage, M.; Miller, M.S.; Peterson, E.J. Autoimmune Variant PTPN22 C1858T Is Associated with Impaired Responses to Influenza Vaccination. J. Infect. Dis. 2016, 214, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.M.; Bucasas, K.L.; Wells, J.M.; Niño, D.; Wang, X.; Zapata, G.E.; Arden, N.; Renwick, A.; Yu, P.; Quarles, J.M.; et al. Integrative genomic analysis of the human immune response to influenza vaccination. eLife 2013, 2013, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lambkin, R.; Novelli, P.; Oxford, J.; Gelder, C. Human genetics and responses to influenza vaccination: Clinical implications. Am. J. Pharmacogenom. 2004, 4, 293–298. [Google Scholar] [CrossRef]
- Gelder, C.M.; Lambkin, R.; Hart, K.W.; Fleming, D.; Williams, O.M.; Bunce, M.; Welsh, K.I.; Marshall, S.E.; Oxford, J. Associations between Human Leukocyte Antigens and Nonresponsiveness to Influenza Vaccine. J. Infect. Dis. 2002, 185, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Moss, A.J.; Gaughran, F.P.; Karasu, A.; Gilbert, A.S.; Mann, A.J.; Gelder, C.M.; Oxford, J.S.; Stephens, H.A.; Lambkin-Williams, R. Correlation between Human Leukocyte Antigen Class II Alleles and HAI Titers Detected Post-Influenza Vaccination. PLoS ONE 2013, 8, e71376. [Google Scholar] [CrossRef] [PubMed]
- Churchill, G.A.; Airey, D.C.; Allayee, H.; Angel, J.M.; Attie, A.D.; Beatty, J.; Beavis, W.D.; Belknap, J.K.; Bennett, B.; Berrettini, W.; et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004, 36, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Boon, A.; Finkelstein, D.; Zheng, M.; Liao, G. H5N1 influenza virus pathogenesis in genetically diverse mice is mediated at the level of viral load. mBio 2011, 2, e00171-11. [Google Scholar] [CrossRef] [PubMed]
- Bottomly, D.; Ferris, M.T.; Aicher, L.D.; Rosenzweig, E.; Whitmore, A.; Aylor, D.L.; Haagmans, B.L.; Gralinski, L.E.; Bradel-Tretheway, B.G.; Bryan, J.T.; et al. Expression Quantitative Trait Loci for Extreme Host Response to Influenza A in Pre-Collaborative Cross Mice. G3: Genes|Genomes|Genet. 2012, 2, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Ferris, M.T.; Aylor, D.L.; Bottomly, D.; Whitmore, A.C.; Aicher, L.D.; Bell, T.A.; Bradel-Tretheway, B.; Bryan, J.T.; Buus, R.J.; Gralinski, L.E.; et al. Modeling Host Genetic Regulation of Influenza Pathogenesis in the Collaborative Cross. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Leist, S.R.; Pilzner, C.; van den Brand, J.M.A.; Dengler, L.; Geffers, R.; Kuiken, T.; Balling, R.; Kollmus, H.; Schughart, K. Influenza H3N2 infection of the collaborative cross founder strains reveals highly divergent host responses and identifies a unique phenotype in CAST/EiJ mice. BMC Genom. 2016, 17, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Elbahesh, H.; Schughart, K. Genetically diverse CC-founder mouse strains replicate the human influenza gene expression signature. Sci. Rep. 2016, 6, 26437. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.B.; Swarts, J.L.; Mooney, M.; Choonoo, G.; Jeng, S.; Miller, D.R.; Ferris, M.T.; McWeeney, S.; Lund, J.M. Extensive Homeostatic T Cell Phenotypic Variation within the Collaborative Cross. Cell Rep. 2017, 21, 2313–2325. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Li, S.; Nakaya, H.I. Systems vaccinology. Immunity 2010, 33, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015, 14, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.J.; Patriarca, P.A.; Treanor, J. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respir. Viruses 2008, 2, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P. Yellow fever: An update. Lancet Infect. Dis. 2001, 1, 11–20. [Google Scholar] [CrossRef]
- Querec, T.D.; Akondy, R.S.; Lee, E.K.; Cao, W.; Nakaya, H.I.; Teuwen, D.; Pirani, A.; Gernert, K.; Deng, J.; Marzolf, B.; et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009, 10, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Khan, N.; Li, G.-M.; et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011, 12, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Reber, A.; Katz, J. Immunological assessment of influenza vaccines and immune correlates of protection. Expert Rev. Vaccines 2013, 12, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Hoek, K.L.; Samir, P.; Howard, L.M.; Niu, X.; Prasad, N.; Galassie, A.; Liu, Q.; Allos, T.M.; Floyd, K.A.; Guo, Y.; et al. A cell-based systems biology assessment of human blood to monitor immune responses after influenza vaccination. PLoS ONE 2015, 10, e0118528. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Rouphael, N.; Duraisingham, S.; Romero-Steiner, S.; Presnell, S.; Davis, C.; Schmidt, D.S.; Johnson, S.E.; Milton, A.; Rajam, G.; et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014, 15, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Obermoser, G.; Presnell, S.; Domico, K.; Xu, H.; Wang, Y.; Anguiano, E.; Thompson-Snipes, L.; Ranganathan, R.; Zeitner, B.; Bjork, A.; et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 2013, 38, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Furman, D.; Hejblum, B.P.; Simon, N.; Jojic, V.; Dekker, C.L.; Thiebaut, R.; Tibshirani, R.J.; Davis, M.M. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, K.L.; Fink, A.L.; Plebanski, M.; Klein, S.L. Sex and Gender Differences in the Outcomes of Vaccination over the Life Course. Annu. Rev. Cell Dev. Biol. 2017, 33, 577–599. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, O.; Binda, E.; O’Farrell, S.; Lorenc, A.; Pradines, J.; Huang, Y.; Duffner, J.; Schulz, R.; Cason, J.; Zambon, M.; et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat. Immunol. 2016, 17, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, H.I.; Clutterbuck, E.; Kazmin, D.; Wang, L.; Cortese, M.; Bosinger, S.E.; Patel, N.B.; Zak, D.E.; Aderem, A.; Dong, T.; et al. Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc. Natl. Acad. Sci. USA 2016, 113, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Howard, L.M.; Hoek, K.L.; Goll, J.B.; Samir, P.; Galassie, A.; Allos, T.M.; Niu, X.; Gordy, L.E.; Creech, C.B.; Prasad, N.; et al. Cell-based systems biology analysis of human AS03-adjuvanted H5N1 avian influenza vaccine responses: A phase i randomized controlled trial. PLoS ONE 2017, 12, e0167488. [Google Scholar] [CrossRef]
- Tsang, J.S.; Schwartzberg, P.L.; Kotliarov, Y.; Biancotto, A.; Xie, Z.; Germain, R.N.; Wang, E.; Olnes, M.J.; Narayanan, M.; Golding, H.; et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 2014, 157, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.F.; Kaur, K.; Pauli, N.T.; Huang, M.; Huang, Y.; Wilson, P.C. High Preexisting Serological Antibody Levels Correlate with Diversification of the Influenza Vaccine Response. J. Virol. 2015, 89, 3308–3317. [Google Scholar] [CrossRef] [PubMed]
- Skowronski, D.M.; Chambers, C.; De Serres, G.; Sabaiduc, S.; Winter, A.L.; Dickinson, J.A.; Gubbay, J.B.; Fonseca, K.; Drews, S.J.; Charest, H.; et al. Serial vaccination and the antigenic distance hypothesis: Effects on influenza vaccine effectiveness during A(H3N2) epidemics in Canada, 2010-2011 to 2014-2015. J. Infect. Dis. 2017, 215, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S.; Gardner, T.J.; Krammer, F.; Aguado, L.C.; Tortorella, D.; Basler, C.F.; Palese, P. Neutralizing antibodies against previously encountered influenza virus strains increase over time: A longitudinal analysis. Sci. Transl. Med. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Hemmi, H.; Steinman, R.M. Dendritic Cells: Translating Innate to Adaptive Immunity. Curr. Top. Immunol. 2006, 311, 17–58. [Google Scholar]
- Pasquale, A.; Preiss, S.; Silva, F.; Garçon, N. Vaccine Adjuvants: From 1920 to 2015 and Beyond. Vaccines 2015, 3, 320–343. [Google Scholar] [CrossRef] [PubMed]
- Moser, M.; Leo, O. Key concepts in immunology. Vaccine 2010, C2–C13. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Ahmed, R. Translating innate immunity into immunological memory: Implications for vaccine development. Cell 2006, 124, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Pang, I.K.; Ichinohe, T.; Iwasaki, A. IL-1R signaling in dendritic cells replaces pattern-recognition receptors in promoting CD8+ T cell responses to influenza A virus. Nat. Immunol. 2013, 14, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Briere, F.; Caux, C.; Davoust, J.; Lebecque, S.; Liu, Y.; Pulendran, B.; Palucka, K. Immunobiology of dendritic cells. Annu. Rev. Immunol. 2000, 18, 767–811. [Google Scholar] [CrossRef] [PubMed]
- Chatziandreou, N.; Farsakoglu, Y.; Palomino-Segura, M.; D’Antuono, R.; Pizzagalli, D.U.; Sallusto, F.; Lukacs-Kornek, V.; Uguccioni, M.; Corti, D.; Turley, S.J.; et al. Macrophage Death following Influenza Vaccination Initiates the Inflammatory Response that Promotes Dendritic Cell Function in the Draining Lymph Node. Cell Rep. 2017, 18, 2427–2440. [Google Scholar] [CrossRef] [PubMed]
- Lupfer, C.; Thomas, P.G.; Kanneganti, T.-D. Nucleotide Oligomerization and Binding Domain 2-Dependent Dendritic Cell Activation Is Necessary for Innate Immunity and Optimal CD8+ T Cell Responses to Influenza A Virus Infection. J. Virol. 2014, 88, 8946–8955. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.L.; Farber, D.L. Mucosal resident memory CD4 T cells in protection and immunopathology. Front. Immunol. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.N.; Mackay, L.K. Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 2016, 16, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Wakim, L.M.; Smith, J.; Caminschi, I.; Lahoud, M.H.; Villadangos, J.A. Antibody-targeted vaccination to lung dendritic cells generates tissue-resident memory CD8 T cells that are highly protective against influenza virus infection. Mucosal Immunol. 2015, 8, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Stoel, M.; Pool, J.; De Vries-Idema, J.; Zaaraoui-Boutahar, F.; Bijl, M.; Andeweg, A.C.; Wilschut, J.; Huckriede, A. Innate responses induced by whole inactivated virus or subunit influenza vaccines in cultured dendritic cells correlate with immune responses in vivo. PLoS ONE 2015, 10, e0125228. [Google Scholar] [CrossRef] [PubMed]
- Perez-Giron, J.V.; Belicha-Villanueva, A.; Hassan, E.; Gomez-Medina, S.; Cruz, J.L.G.; Ludtke, A.; Ruibal, P.; Albrecht, R.A.; Garcia-Sastre, A.; Munoz-Fontela, C. Mucosal Polyinosinic-Polycytidylic Acid Improves Protection Elicited by Replicating Influenza Vaccines via Enhanced Dendritic Cell Function and T Cell Immunity. J. Immunol. 2014, 193, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Osterholm, M.T.; Kelley, N.S.; Sommer, A.; Belongia, E.A. Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 36–44. [Google Scholar] [CrossRef]
- Wu, N.C.; Zost, S.J.; Thompson, A.J.; Oyen, D.; Nycholat, C.M.; McBride, R.; Paulson, J.C.; Hensley, S.E.; Wilson, I.A. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLOS Pathog. 2017, 13, e1006682. [Google Scholar] [CrossRef] [PubMed]
- Black, S.; Rappuoli, R. A crisis of public confidence in vaccines. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Ahmed, R. Immunological mechanisms of vaccination. Nat. Immunol. 2011, 12, 509. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.G.; Orr, M.T.; Fox, C.B. Key roles of adjuvants in modern vaccines. Nat. Med. 2013, 19, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Nguyen, M.T. Recent advances of vaccine adjuvants for infectious diseases. Immune Netw. 2015, 15, 51–57. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stacey, H.D.; Barjesteh, N.; Mapletoft, J.P.; Miller, M.S. “Gnothi Seauton”: Leveraging the Host Response to Improve Influenza Virus Vaccine Efficacy. Vaccines 2018, 6, 23. https://doi.org/10.3390/vaccines6020023
Stacey HD, Barjesteh N, Mapletoft JP, Miller MS. “Gnothi Seauton”: Leveraging the Host Response to Improve Influenza Virus Vaccine Efficacy. Vaccines. 2018; 6(2):23. https://doi.org/10.3390/vaccines6020023
Chicago/Turabian StyleStacey, Hannah D., Neda Barjesteh, Jonathan P. Mapletoft, and Matthew S. Miller. 2018. "“Gnothi Seauton”: Leveraging the Host Response to Improve Influenza Virus Vaccine Efficacy" Vaccines 6, no. 2: 23. https://doi.org/10.3390/vaccines6020023
APA StyleStacey, H. D., Barjesteh, N., Mapletoft, J. P., & Miller, M. S. (2018). “Gnothi Seauton”: Leveraging the Host Response to Improve Influenza Virus Vaccine Efficacy. Vaccines, 6(2), 23. https://doi.org/10.3390/vaccines6020023




