Identifying the Conditions Under Which Antibodies Protect Against Infection by Equine Infectious Anemia Virus
Abstract
:1. Introduction
2. Methods
| Parameter | Definition | Value | Range | Units | Reference |
|---|---|---|---|---|---|
| rW | Virus growth rate for wild-type in the absence of antibodies | 23.60 | 0–46 | day −1 | Calculated (Appendix A) |
| rM1 | Virus growth rate for first mutant | 23.23 | 0–46 | day −1 | Calculated (Appendix A) |
| rM2 | Virus growth rate for second mutant | 23.09 | 0–46 | day −1 | Assumed |
| K | Virus carrying capacity | 1.14 × 108 | 7.47 × 108–3.82 × 108 | virus ml −1 | Calculated (Appendix A) |
| pW | Wild-type virus neutralization by antibody | 1.462 × 10−2 × m | (1.21 × 10−2–2.67 × 10−2) × m | ml mg −1 day−1 | Calculated (Appendix A) |
| pM1 | Mutant 1 virus neutralization by antibody | (varied) | — | ml mg −1 day−1 | Assumed |
| pM2 | Mutant 2 virus neutralization by antibody | (varied) | — | ml mg −1 day−1 | Assumed |
| q | Antibody decay rate | 0.0315 | 0.0277–0.0365 | day −1 | [ 32] |
| ϵ1 | Mutation rate from wild-type to first mutant (per base per cycle) | 2.7 × 10−5 | 1 × 10−5–3.4 × 10−5 | day −1 | [ 36] |
| ϵ2 | Mutation rate from wild-type to second mutant (per base per cycle) | 2.7 × 10−6 | 2 × 10−6–2.7 × 10−2 | day −1 | Assumed |
| Ai | Amount of antibody infusion | 38.4 × m | (25.6–51.2) × m | mg ml −1 | [ 10 , 11 ] |
| A0 | Antibody on Day 0 | 37.2 | 24.9–49.4 | mg ml −1 | Calculated (Appendix A) |
| VW(0) | Number of wild-type viral particles that initiated infection | 224 | 175–350 | virus ml−1 | [ 10,37] |
| VM1(0) | Number of Mutant 1 viral particles that initiated infection | 9 | — | virus ml−1 | [ 10] |
| VM2(0) | Number of Mutant 2 viral particles that initiated infection | 1 | — | virus ml−1 | Assumed |
| t1/2 | Half-life of virus due to antibody neutralization | 1.3 | 0.7–1.8 | day | [ 33–35] |
| dW , dM1, dM2 | Viral clearance rate | 23 | 9.1–36 | day−1 | [ 38] |
| m | Antibody magnification factor | {1, 10, 50} | — | — | — |
3. Results
3.1. Theoretical Results
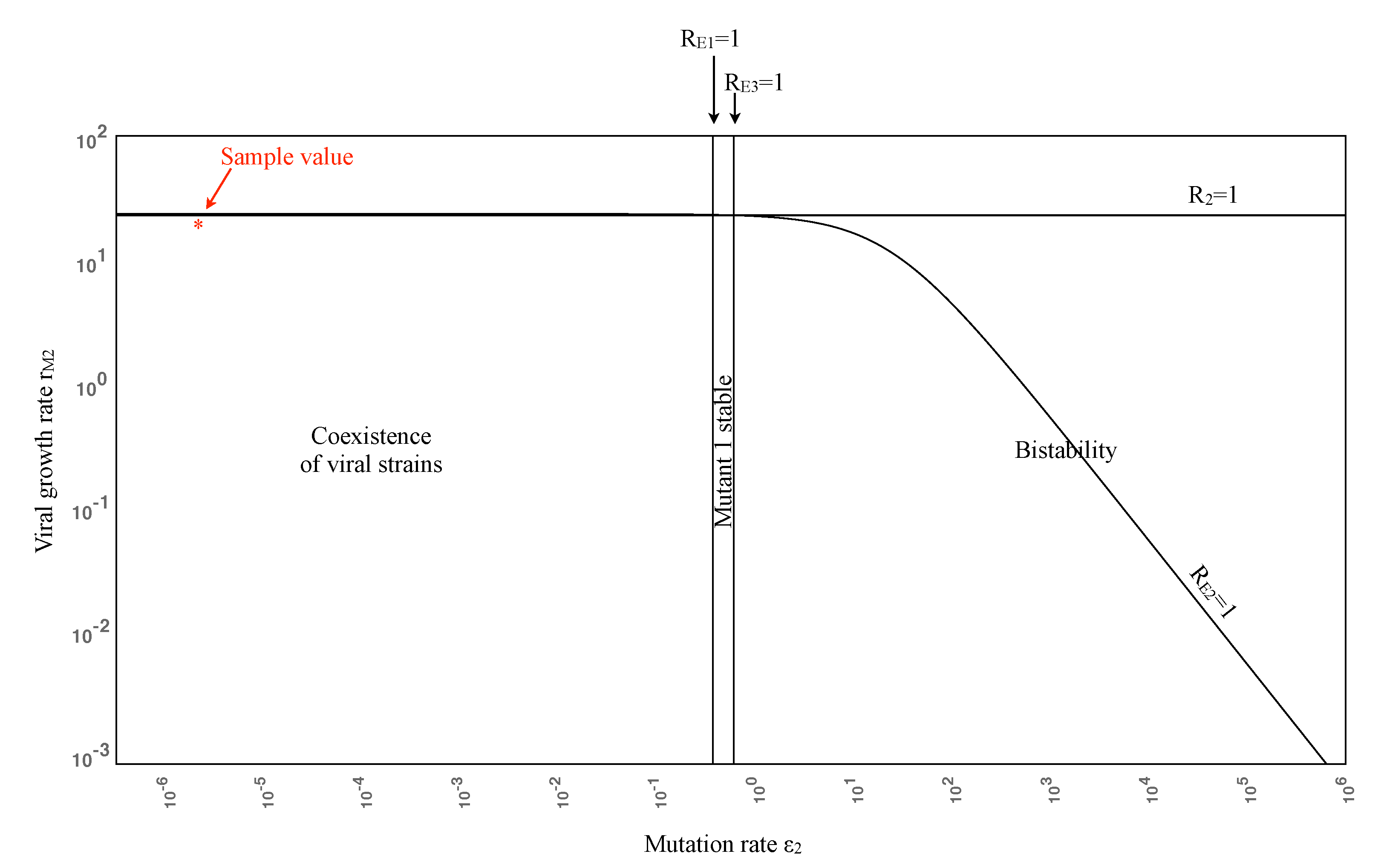
3.2. Numerical Simulations
| Relative effectiveness | Antibody magnification factor m | Figure | |||
|---|---|---|---|---|---|
| pM1 | pM2 | 1 | 10 | 50 | |
| pW | pW | Wild-type dominates (coexistence) | Eradication (wild-type last) | Eradication (exponentially fast) | 3 |
| 0.1pW | 0.01pW | Wild-type dominates (coexistence) | Mutant 2 escape (others eradicated) | Eradication (Mutant 2 last) | 4 |
| 0.01pW | 0.01pW | Wild-type dominates (coexistence) | Mutant 1 escape (Wild-type eradicated) | Mutant 1 escape or eradication | 5 |
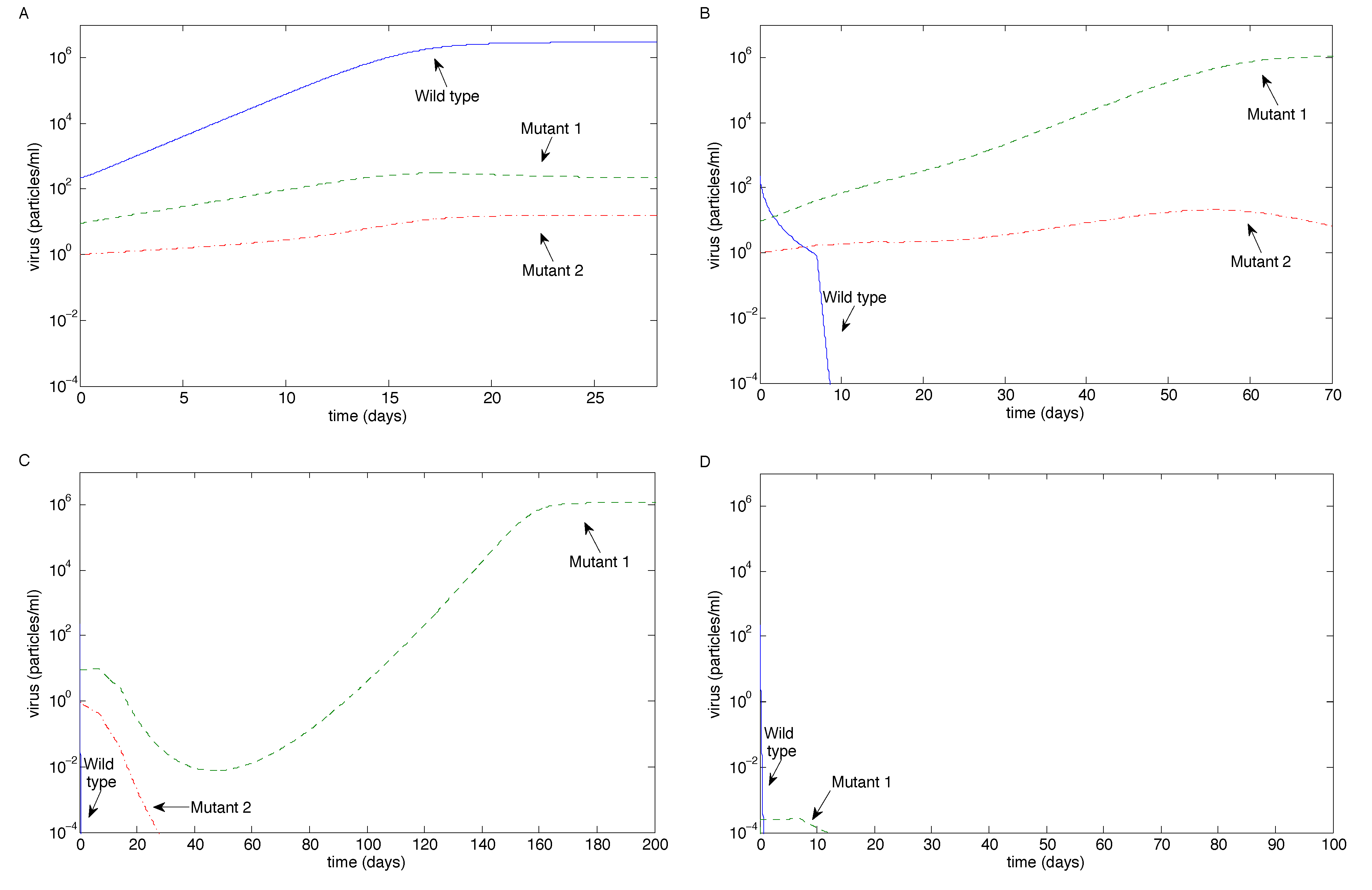

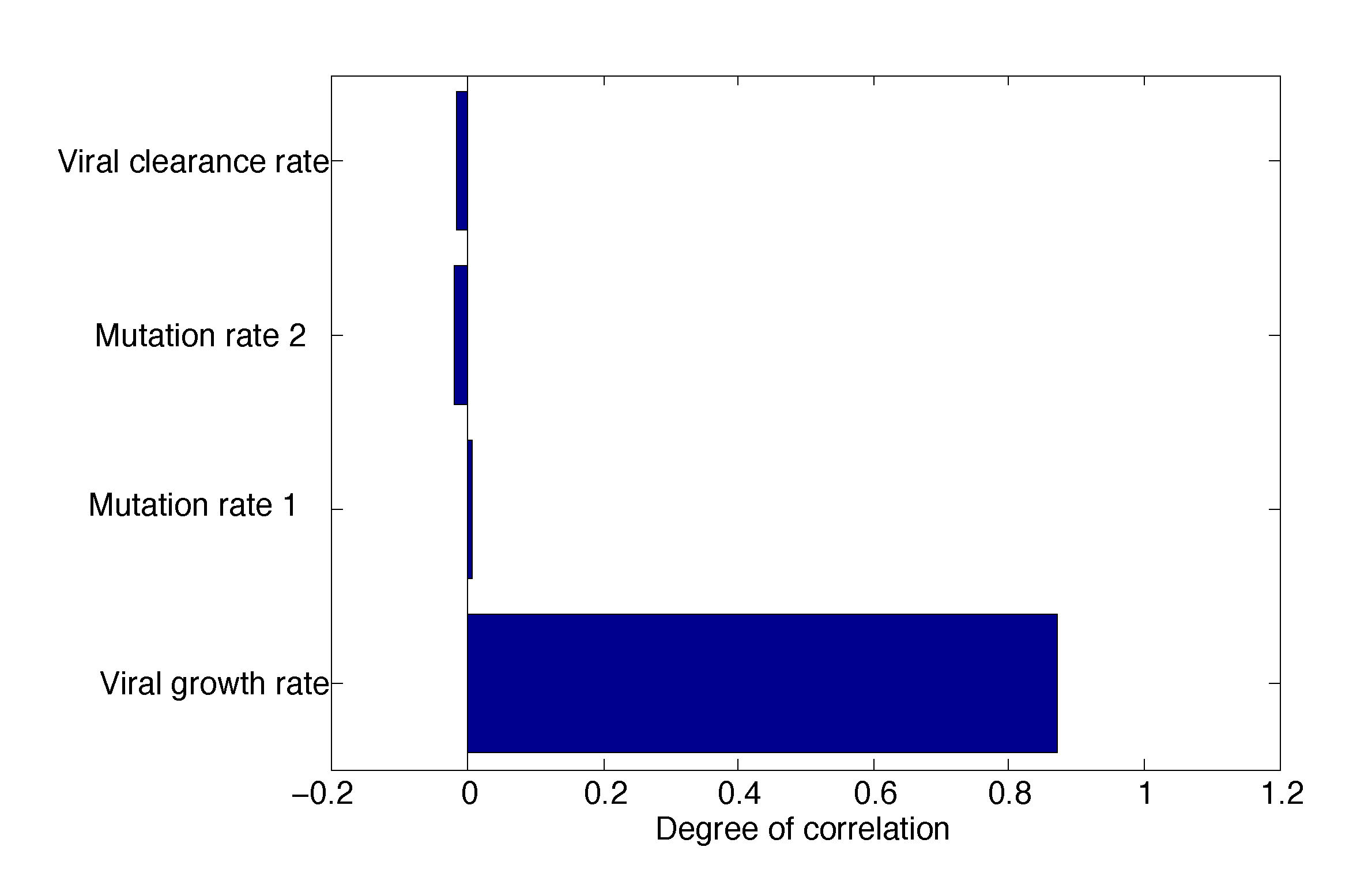
3.2.1. Sensitivity Analysis
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix
A. Calculating Parameters
A.1. Calculating p
A.2. Calculating A0
A.3. Calculating rW and K
B. The Non-Impulsive System
B.1. Equilibria
B.2. Jacobian
B.3. Calculating R0
C. Stabilizing the Mutant 1 Equilibrium
References
- Liu, P.; Overman, R.G.; Yates, N.L.; Alam, S.M.; Vandergrift, N.; Chen, Y.; Graw, F.; Freel, S.A.; Kappes, J.C.; Ochsenbauer, C.; et al. Dynamic antibody specificities and virion concentrations in circulating immune complexes in acute to chronic HIV-1 infection. J. Virol. 2011, 85, 11196–11207. [Google Scholar] [CrossRef] [PubMed]
- Tomaras, G.D.; Yates, N.L.; Liu, P.; Qin, L.; Fouda, G.G.; Chavez, L.L.; Decamp, A.C.; Parks, R.J.; Ashley, V.C.; Lucas, J.T.; et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: Virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 2008, 82, 12449–12463. [Google Scholar] [CrossRef] [PubMed]
- Leroux, C.; Cadore, J.L.; Montelaro, R.C. Equine Infectious Anemia Virus (EIAV): What has HIV’s country cousin got to tell us? Vet. Res. 2004, 35, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Maury, W.; Oaks, J.L. Equine infectious anemia virus pathogenesis and replication. In Lentiviruses and Macrophages: Molecular and Cellular Interactions; Desport, M., Ed.; Caister Academic Press: Norfolk, UK, 2010; pp. 251–276. [Google Scholar]
- Mealey, R.H.; Fraser, D.G.; Oaks, J.L.; Cantor, G.H.; McGuire, T.C. Immune reconstitution prevents continuous equine infectious anemia virus replication in an Arabian foal with severe combined immunodeficiency: Lessons for control of lentiviruses. Clin. Immunol. 2001, 101, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Perryman, L.E.; O’Rourke, K.I.; McGuire, T.C. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J. Virol. 1988, 62, 3073–3076. [Google Scholar] [PubMed]
- Carpenter, S.; Chen, W.C.; Dorman, K.S. Rev variation during persistent lentivirus infection. Viruses 2011, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mealey, R.H.; Leib, S.R.; Littke, M.H.; Wagner, B.; Horohov, D.W.; McGuire, T.C. Viral load and clinical disease enhancement associated with a lentivirus cytotoxic T lymphocyte vaccine regimen. Vaccine 2009, 27, 2453–2468. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Blythe, D.C.; Loyd, H.; Mealey, R.H.; Tallmadge, R.L.; Dorman, K.S.; Carpenter, S. Decreased infectivity of a neutralization-resistant equine infectious anemia virus variant can be overcome by efficient cell-to-cell spread. J. Virol. 2011, 85, 10421–10424. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.D.; Leib, S.R.; Carpenter, S.; Mealey, R.H. Selection of a rare neutralization-resistant variant following passive transfer of convalescent immune plasma in equine infectious anemia virus-challenged SCID horses. J. Virol. 2010, 84, 6536–6548. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.D.; Leib, S.R.; Wu, W.; Nelson, R.; Carpenter, S.; Mealey, R.H. Protective effects of broadly neutralizing immunoglobulin against homologous and heterologous equine infectious anemia virus infection in horses with severe combined immunodeficiency. J. Virol. 2011, 85, 6814–6818. [Google Scholar] [CrossRef] [PubMed]
- Mascola, J.R.; Lewis, M.G.; Stiegler, G.; Harris, D.; VanCott, T.C.; Hayes, D.; Louder, M.K.; Brown, C.R.; Sapan, C.V.; Frankel, S.S.; et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 1999, 73, 4009–4018. [Google Scholar] [PubMed]
- Ng, C.T.; Jaworski, J.P.; Jayaraman, P.; Sutton, W.F.; Delio, P.; Kuller, L.; Anderson, D.; Landucci, G.; Richardson, B.A.; Burton, D.R.; et al. Passive neutralizing antibody controls SHIV viremia and enhances B cell responses in infant macaques. Nat. Med. 2010, 16, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Igarashi, T.; Haigwood, N.L.; Sadjadpour, R.; Donau, O.K.; Buckler, C.; Plishka, R.J.; Buckler-White, A.; Martin, M.A. Transfer of neutralizing IgG to macaques 6 h but not 24 h after SHIV infection confers sterilizing protection: Implications for HIV-1 vaccine development. Proc. Natl. Acad. Sci. USA 2003, 100, 15131–15136. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, R.M.; Ferrantelli, F.; Kitabwalla, M.; Xu, W.; McClure, H.M. Antibody protection: Passive immunization of neonates against oral AIDS virus challenge. Methods Mol. Biol. 2009, 525, 559–566. [Google Scholar] [PubMed]
- Shibata, R.; Igarashi, T.; Haigwood, N.; Buckler-White, A.; Ogert, R.; Ross, W.; Willey, R.; Cho, M.W.; Martin, M.A. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 1999, 5, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.J.; Pawelek, K.A.; Harrington, K.; Cangelosi, R.; Madrid, S. Immune control of equine infectious anemia virus infection by CTLs and antibodies. Appl. Math. 2013, 4, 171–177. [Google Scholar] [CrossRef]
- Ciupe, S.; Schwartz, E.J. Understanding virus-host dynamics following EIAV infection in SCID horses. J. Theor. Biol. 2014, 343, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Smith?, R.J.; Schwartz, E.J. Predicting the potential impact of a cytotoxic T-lymphocyte HIV vaccine: How often should you vaccinate and how strong should the vaccine be? Math. Biosci. 2008, 212, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Konrad, B.P.; Vaidya, N.K.; Smith?, R.J. Modelling mutation to a cytotoxic T-lymphocyte HIV vaccine. Math. Pop. Stud. 2011, 18, 122–149. [Google Scholar]
- Bainov, D.D.; Simeonov, P.S. Systems with Impulsive Effect; Ellis Horwood Ltd.: Chichester, UK, 1989. [Google Scholar]
- Bainov, D.D.; Simeonov, P.S. Impulsive differential equations: Periodic solutions and applications; Longman Scientific and Technical; Burnt Mill: Essex, UK, 1993. [Google Scholar]
- Bainov, D.D.; Simeonov, P.S. Impulsive Differential Equations: Asymptotic Properties of the Solutions; World Scientific: Singapore City, Singapore, 1995. [Google Scholar]
- Lakshmikantham, V.; Bainov, D.D.; Simeonov, P.S. Theory of Impulsive Differential Equations; World Scientific: Singapore City, Singapore, 1989. [Google Scholar]
- Agur, Z.; Cojocaru, L.; Mazor, G.; Anderson, R.; Danon, Y. Pulse mass measles vaccination across age cohorts. Proc. Natl. Acad. Sci. USA 1993, 90, 11698–11702. [Google Scholar] [CrossRef] [PubMed]
- Stone, L.; Olinky, R.; Huppert, A. Seasonal dynamics of recurrent epidemics. Nature 2007, 446, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Miron, R.E.; Smith?, R.J. Modelling imperfect adherence to HIV induction therapy. BMC Infect. Dis. 2010, 10. [Google Scholar] [CrossRef]
- Roberts, M.G.; Kao, R.R. The dynamics of an infectious disease in a population with birth pulses. Math. Biosci. 1998, 149, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Al-arydah, M.; Smith?, R.J.; Lutscher, F. Modelling gender-structured wildlife diseases with harvesting: Chronic Wasting Disease as an example. ISRN Biomath. 2012, 2012. [Google Scholar] [CrossRef]
- Kot, M. Elements of Mathematical Ecology; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Schwartz, E.J.; Neumann, A.U.; Teixeira, A.V.; Bruggeman, L.A.; Rappaport, J.; Perelson, A.S.; Klotman, P.E. Effect of target cell availability on HIV-1 production in vitro. AIDS 2002, 16, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A.; Strober, W. Metabolism of immunoglobulins. Prog. Allergy 1969, 13, 1–110. [Google Scholar] [PubMed]
- Metzner, K.J.; Jin, X.; Lee, F.V.; Gettie, A.; Bauer, D.E.; di Mascio, M.; Perelson, A.S.; Marx, P.A.; Ho, D.D.; Kostrikis, L.G.; et al. Effects of in vivo CD8+ T cell depletion on virus replication in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Exp. Med. 2000, 191, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.A.; Lloyd, A.L.; Vasquez, G.M.; Wiltrout, T.A.; Wahl, L.M.; Bischofberger, N.; Williams, J.; Kinter, A.; Fauci, A.S.; Hirsch, V.M.; et al. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J. Virol. 1997, 71, 7518–7525. [Google Scholar] [PubMed]
- Van Rompay, K.K.; Singh, R.P.; Pahar, B.; Sodora, D.L.; Wingfield, C.; Lawson, J.R.; Marthas, M.L.; Bischofberger, N. CD8+-cell-mediated suppression of virulent simian immunod- eficiency virus during tenofovir treatment. J. Virol. 2004, 78, 5324–5337. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Temin, H.M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 1995, 69, 5087–5094. [Google Scholar] [PubMed]
- Bryan, W.R. Interpretation of host response in quantitative studies on animal viruses. Ann. NY Acad. Sci. 1957, 69, 698–728. [Google Scholar] [CrossRef] [PubMed]
- Ramratnam, B.; Bonhoeffer, S.; Binley, J.; Hurley, A.; Zhang, L.; Mittler, J.E.; Markowitz, M.; Moore, J.P.; Perelson, A.S.; Ho, D.D. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 1999, 354, 1782–1785. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, J.M.; Smith, R.J.; Wahl, L.M. Perspectives on the basic reproductive ratio. J. R. Soc. Interface 2005, 2, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.J.; Vaidya, N.K.; Dorman, K.; Carpenter, S.; Mealey, R.H. Dynamics of lentiviral infection in vivo in the absence of adaptive host immune responses. J. Virol. submitted for publication. 2014. [Google Scholar]
- Blower, S.M.; Dowlatabadi, H. Sensitivity and uncertainty analysis of complex models of disease transmission: An HIV model, as an example. Int. Stat. Rev. 1994, 2, 229–243. [Google Scholar] [CrossRef]
- Schwartz, E.J.; Blower, S. Predicting the potential individual- and population-level effects of imperfect herpes simplex virus type 2 vaccines. J. Infect. Dis. 2005, 191, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.J.; Bodine, E.N.; Blower, S. Effectiveness and efficiency of imperfect therapeutic HSV-2 vaccines. Hum. Vaccines 2007, 3, 231–238. [Google Scholar] [CrossRef]
- Smith?, R.J.; Cloutier, P.; Harrison, J.; Desforges, A. A mathematical model for the eradication of Guinea Worm Disease. In Understanding the Dynamics of Emerging and Re-Emerging Infectious Diseases Using Mathematical Models; Mushayabasa, S., Bhunu, C.P., Eds.; Transworld Research Network: Kerala, India, 2012; pp. 133–156. [Google Scholar]
- Schwartz, E.J.; Nanda, S. Antibody Escape Kinetics of EIAV Infection of Horses. J. Virol. submitted for publication. 2014. [Google Scholar]
- Tschetter, J.R.; Byrne, K.M.; Perryman, L.E.; McGuire, T.C. Control of equine infectious anemia virus is not dependent on ADCC mediating antibodies. Virology 1997, 230, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Blakeley, D.; Smith?, R.J. The Failure of R0. Comp. Math. Methods Med. 2011, 2011. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Schwartz, E.J.; Smith?, R.J. Identifying the Conditions Under Which Antibodies Protect Against Infection by Equine Infectious Anemia Virus. Vaccines 2014, 2, 397-421. https://doi.org/10.3390/vaccines2020397
Schwartz EJ, Smith? RJ. Identifying the Conditions Under Which Antibodies Protect Against Infection by Equine Infectious Anemia Virus. Vaccines. 2014; 2(2):397-421. https://doi.org/10.3390/vaccines2020397
Chicago/Turabian StyleSchwartz, Elissa J., and Robert J. Smith?. 2014. "Identifying the Conditions Under Which Antibodies Protect Against Infection by Equine Infectious Anemia Virus" Vaccines 2, no. 2: 397-421. https://doi.org/10.3390/vaccines2020397
APA StyleSchwartz, E. J., & Smith?, R. J. (2014). Identifying the Conditions Under Which Antibodies Protect Against Infection by Equine Infectious Anemia Virus. Vaccines, 2(2), 397-421. https://doi.org/10.3390/vaccines2020397




