GMP Manufacturing and Characterization of the HIV Booster Immunogen HxB2.WT.Core-C4b for Germline Targeting Vaccine Strategies
Abstract
1. Introduction
2. Materials and Methods
2.1. Stable Clone Generation
2.2. Inoculum Preparation and Bioreactor Operation
2.3. Harvest Operation
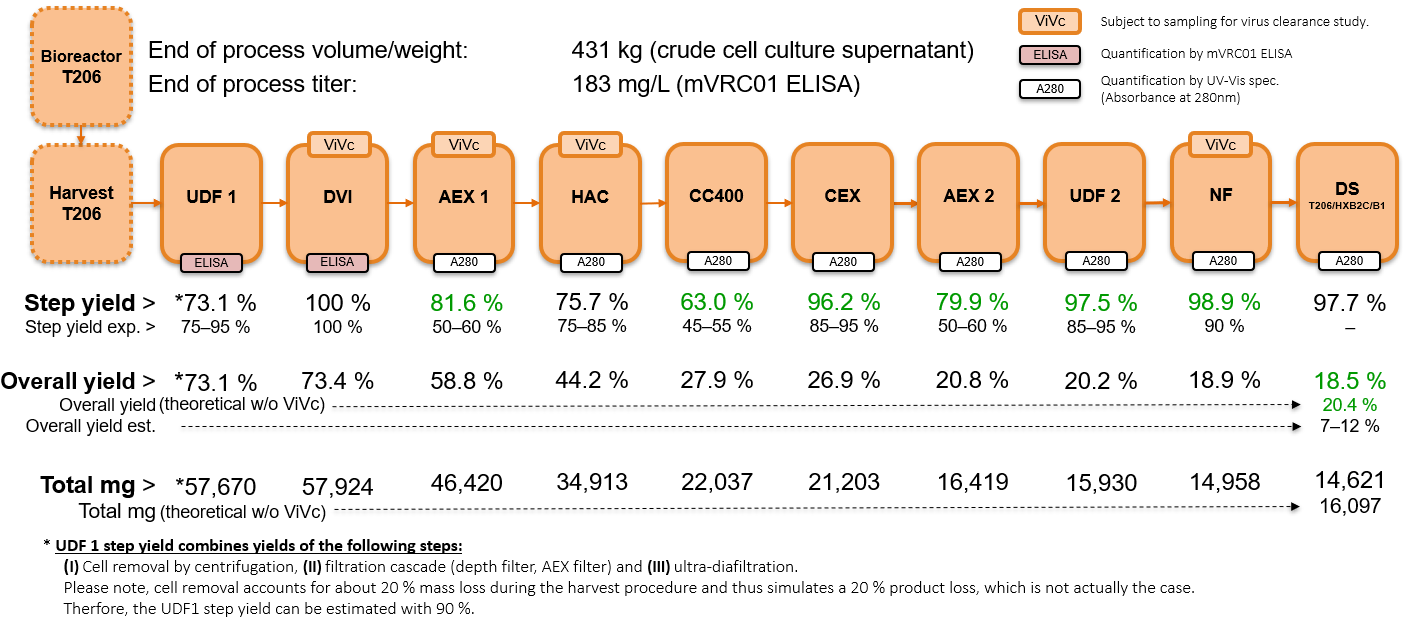
2.4. Downstream Purification of Recombinant HxB2.WT.Core-C4b Drug Substance Figure 1
2.4.1. Clarification
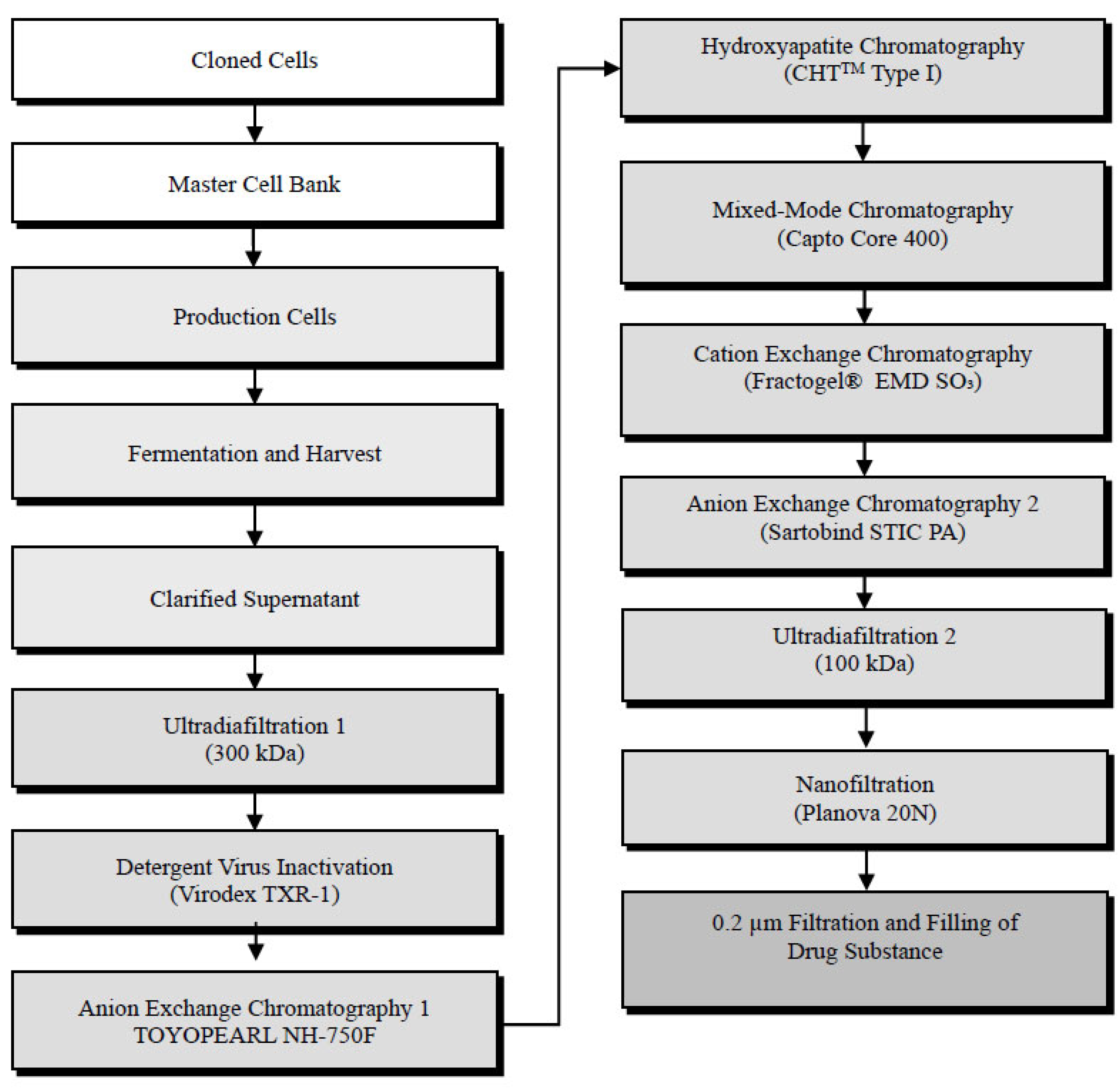
2.4.2. Ultradiafiltration
2.4.3. Virus Inactivation (Step 5)
2.4.4. Anion Exchange Chromatography 1
2.4.5. Hydroxyapatite Chromatography
2.4.6. Mixed-Mode Chromatography
2.4.7. Cation Exchange Chromatography
2.4.8. Anion Exchange Chromatography 2
2.4.9. Ultradiafiltration 2
2.4.10. Nanofiltration
2.5. Model Viruses for Viral Clearance Studies
2.5.1. Murine Leukaemia Virus (X-MuLV)
2.5.2. Murine Minute Virus (MMV)
2.6. Analytical Methods
2.6.1. Cell Concentration and Viability
2.6.2. Carbohydrates and Amino Acids
2.6.3. Product Concentration
2.6.4. Osmolality
2.6.5. Host Cell Protein Content
2.6.6. DNA Content
2.6.7. Antigenicity
2.6.8. Electron Microscopy
2.6.9. Glycosylation Analysis
3. Results
3.1. Cell Culture Process Evaluation and Optimization with the Stable Pool Cell Line
3.1.1. Effects of pH and Base Addition
3.1.2. Effect of Temperature
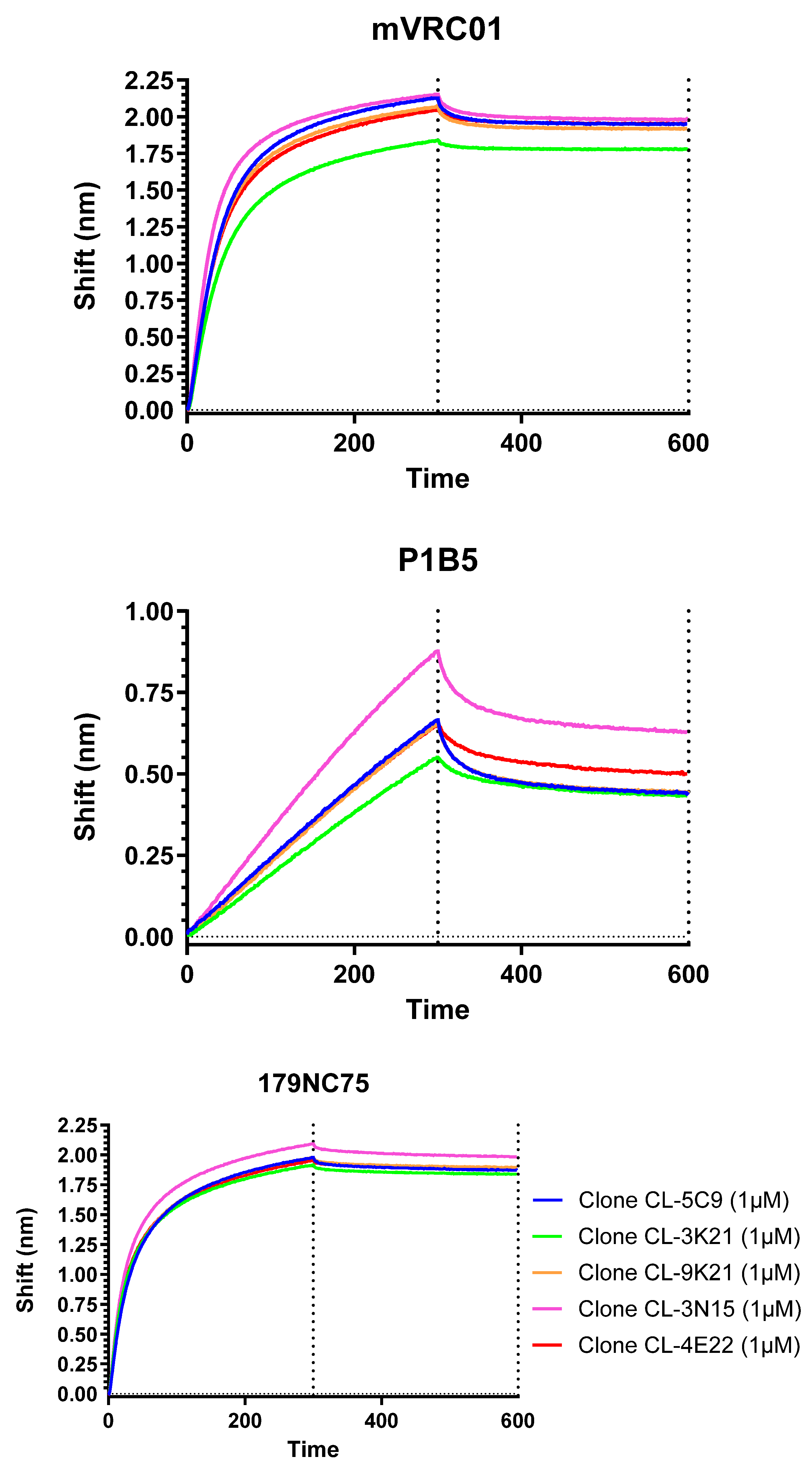
3.2. Subcloning of HxB2.WT.Core-C4b Stable Pool Cell Line and Selection of a Lead Clone
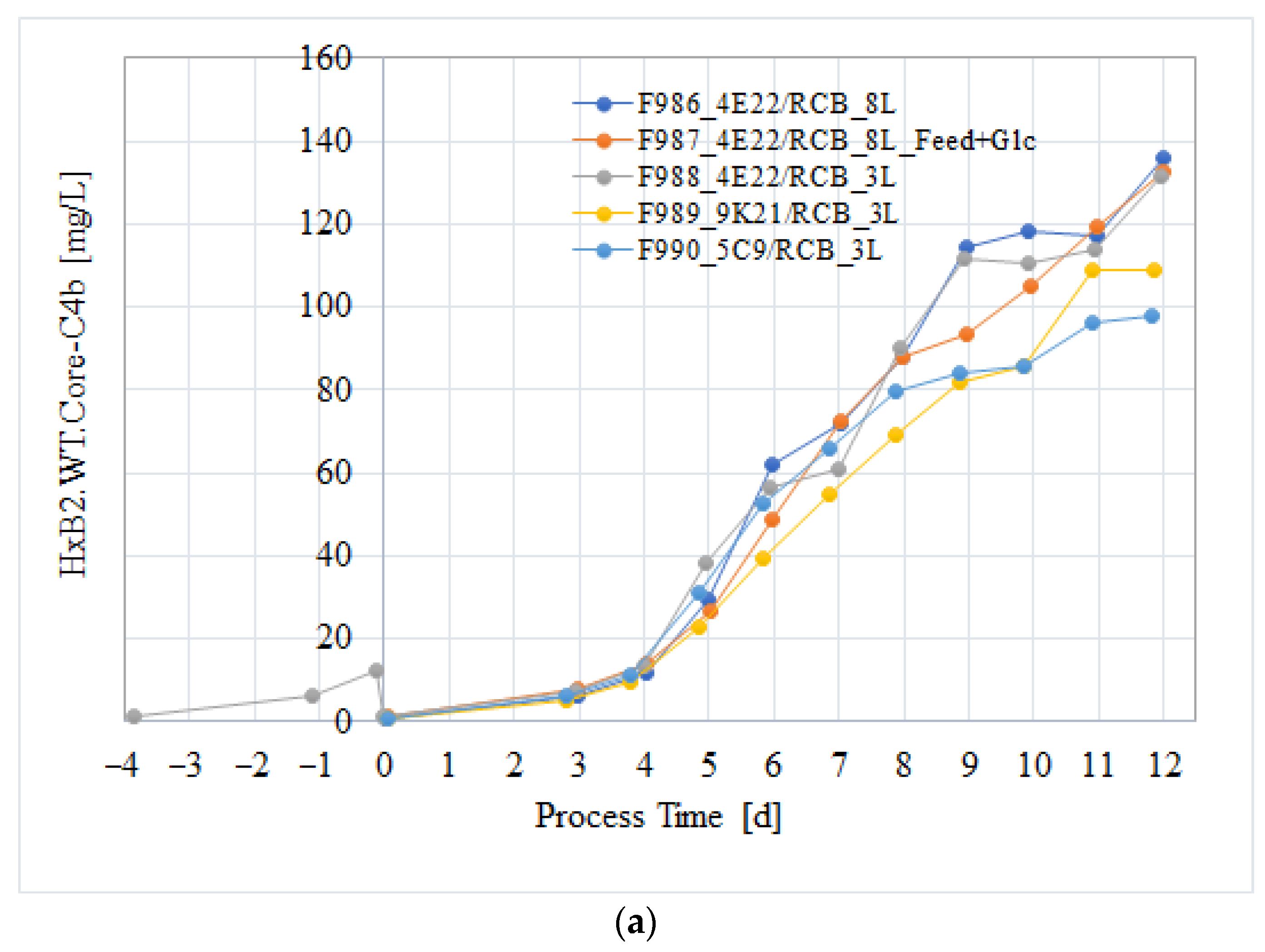
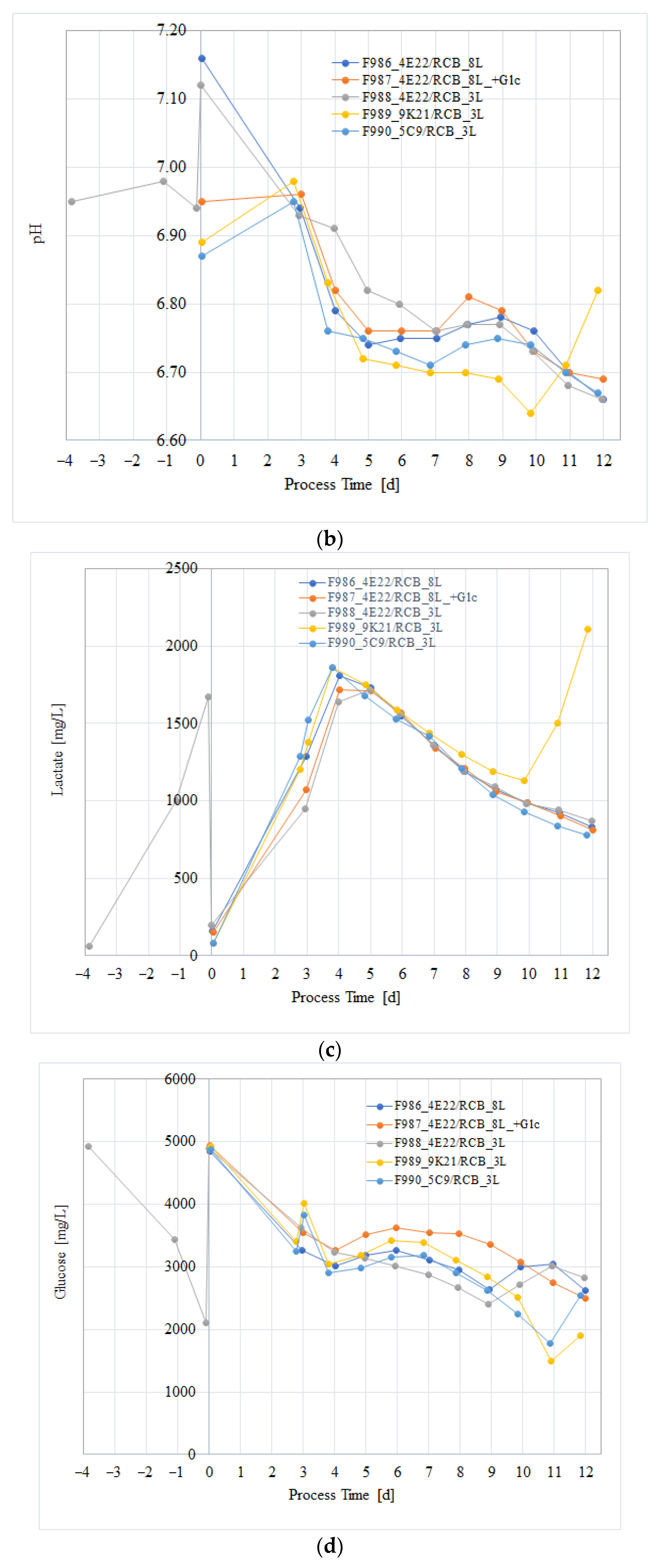
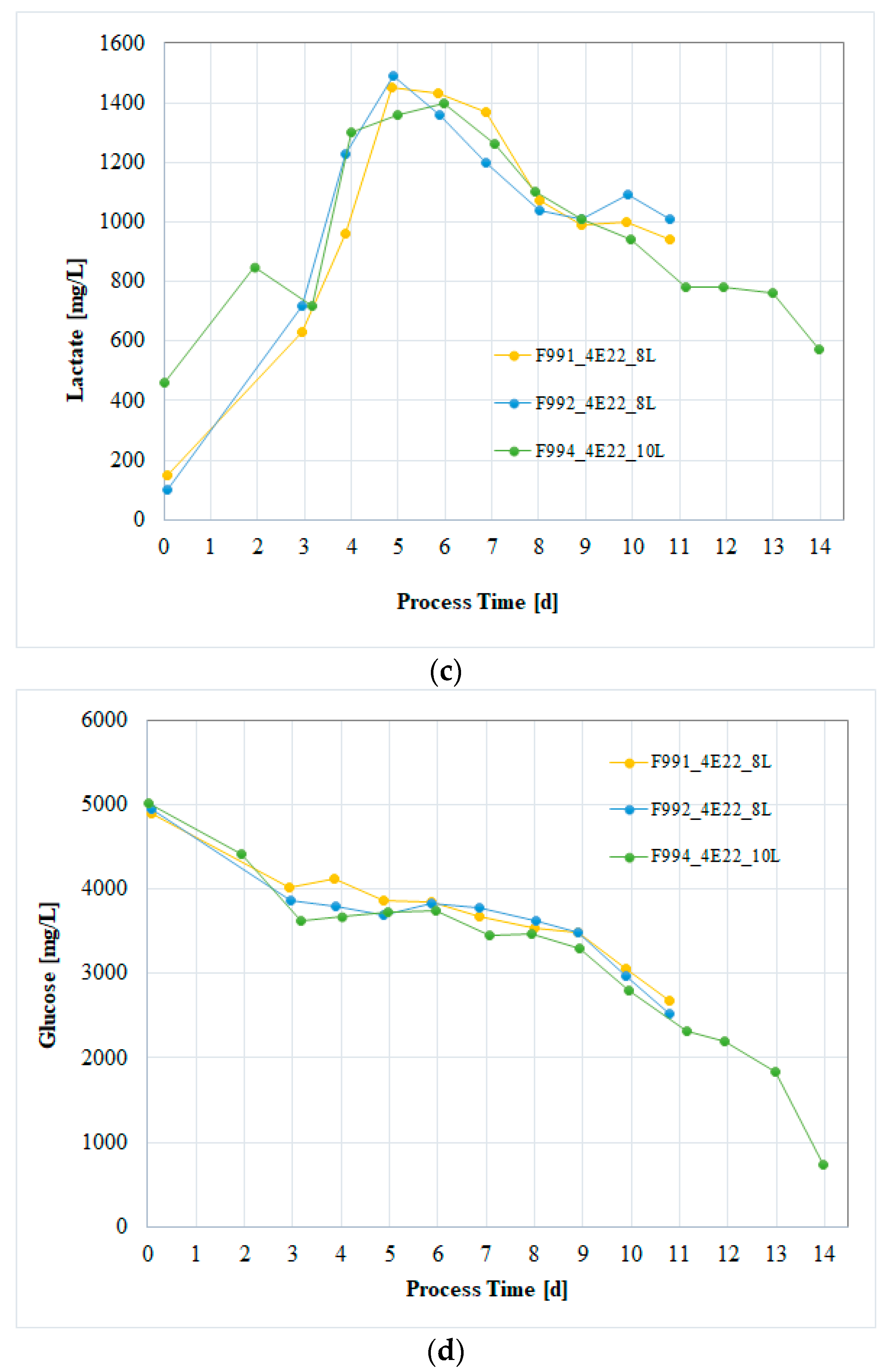
3.3. Cell Culture Process Evaluation After Subcloning
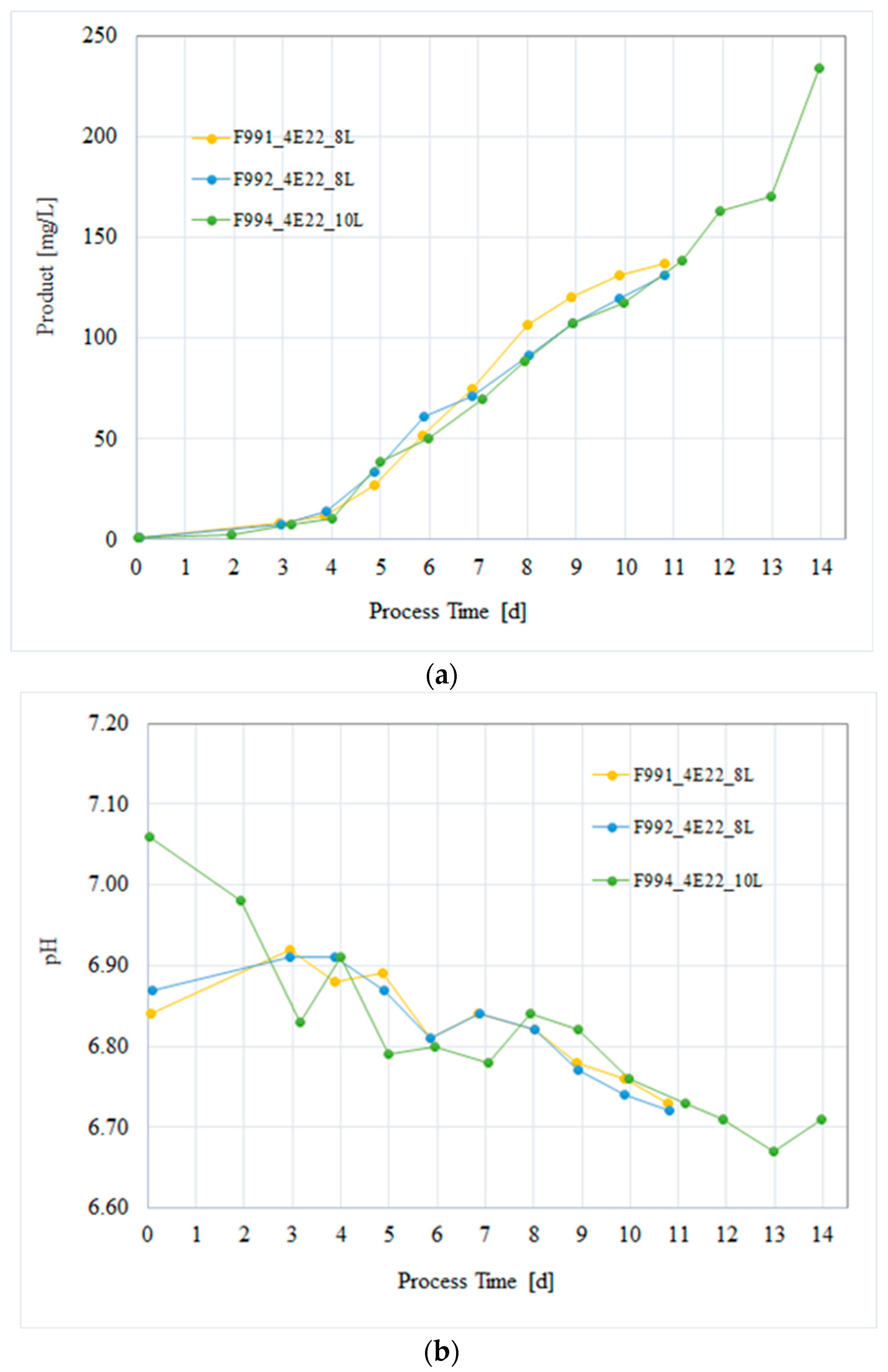
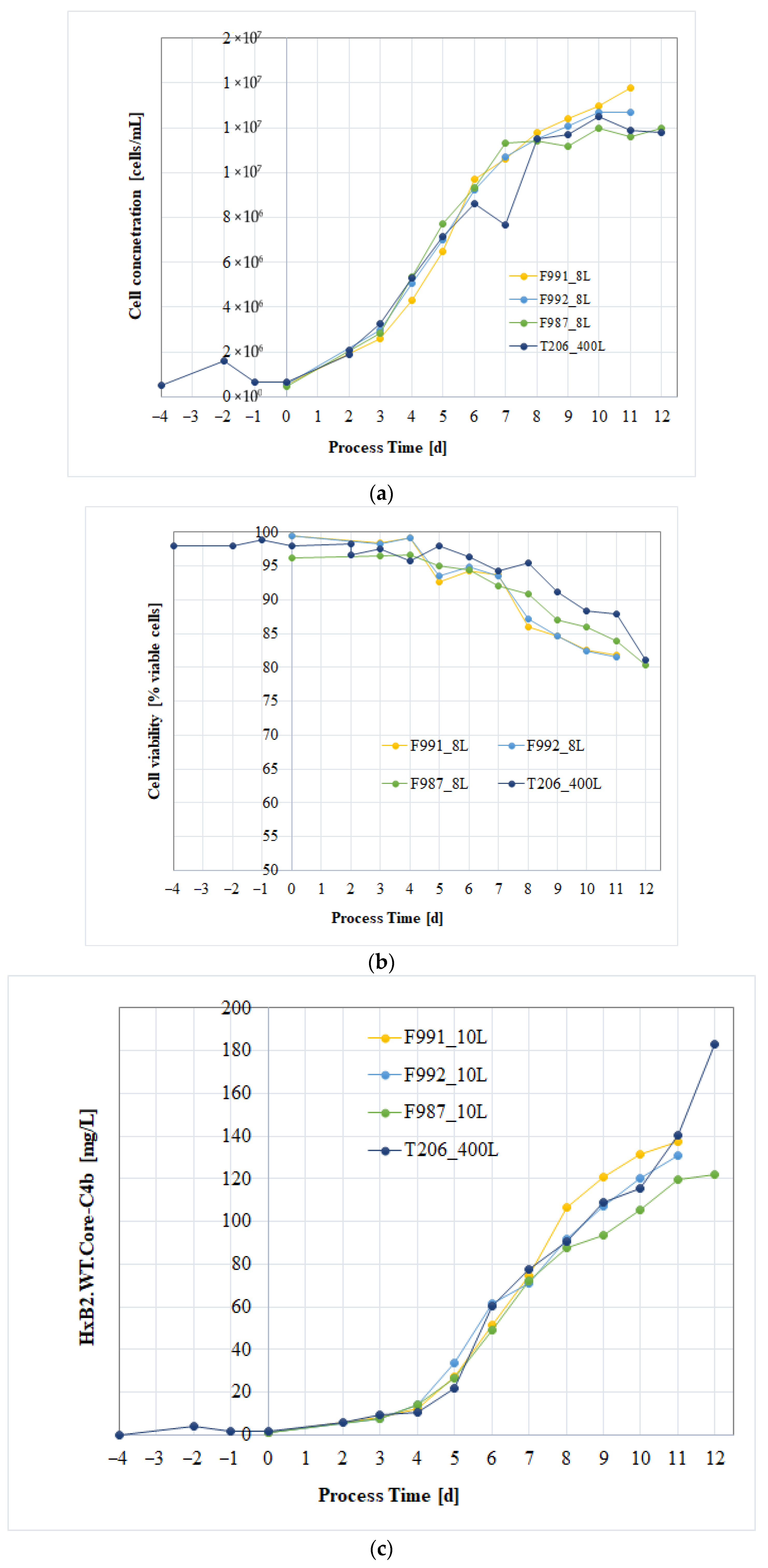
3.4. Process Verification of the CHO/HxB2C Production MCB Clone
3.5. GMP Manufacturing Process and Process Controls
3.5.1. Upstream Process: CHO Cell-Based Bioreactor Production
3.5.2. Purification and Process Yield
3.6. Viral Clearance Evaluation
3.7. Product Characterization
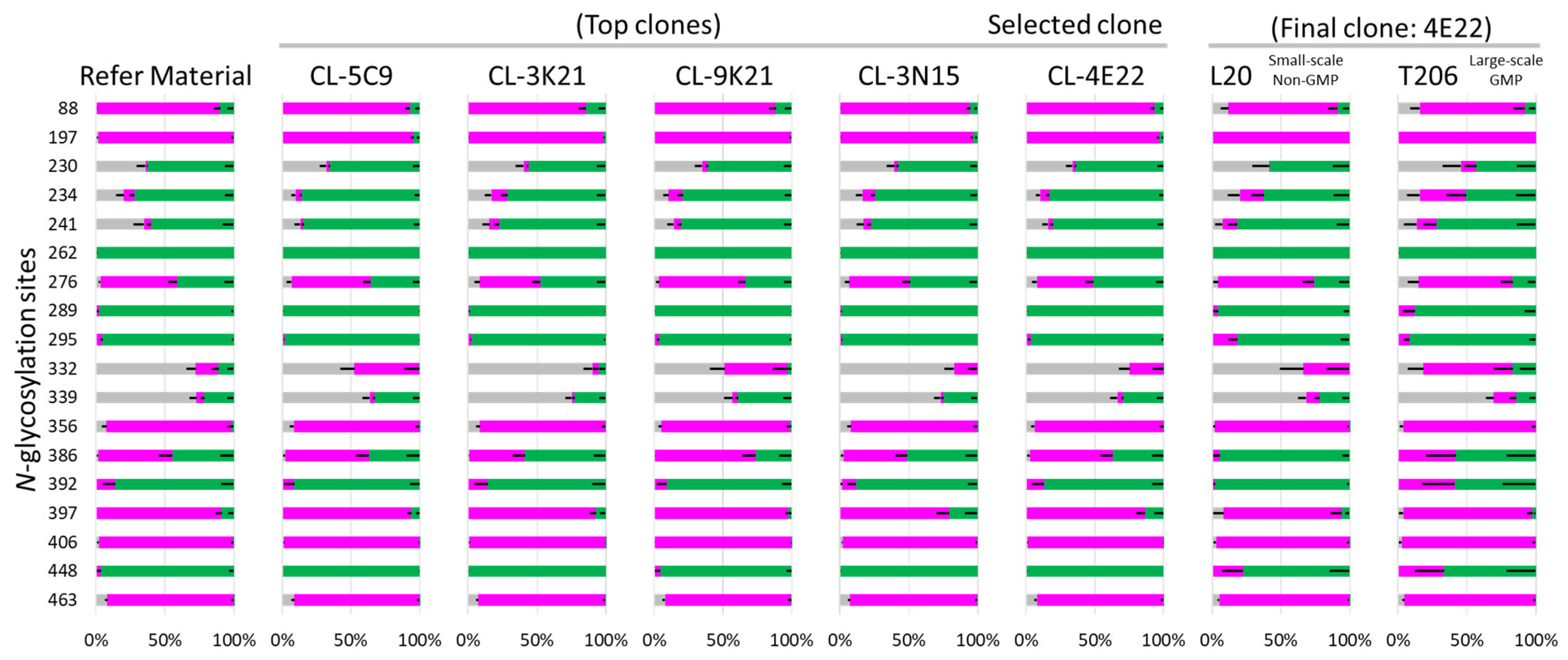
3.7.1. N-Glycosylation Profiling and Impact of Manufacturing Scale on Glycan Occupancy
3.7.2. Electron Microscopy Characterization of Nanoparticle Assembly
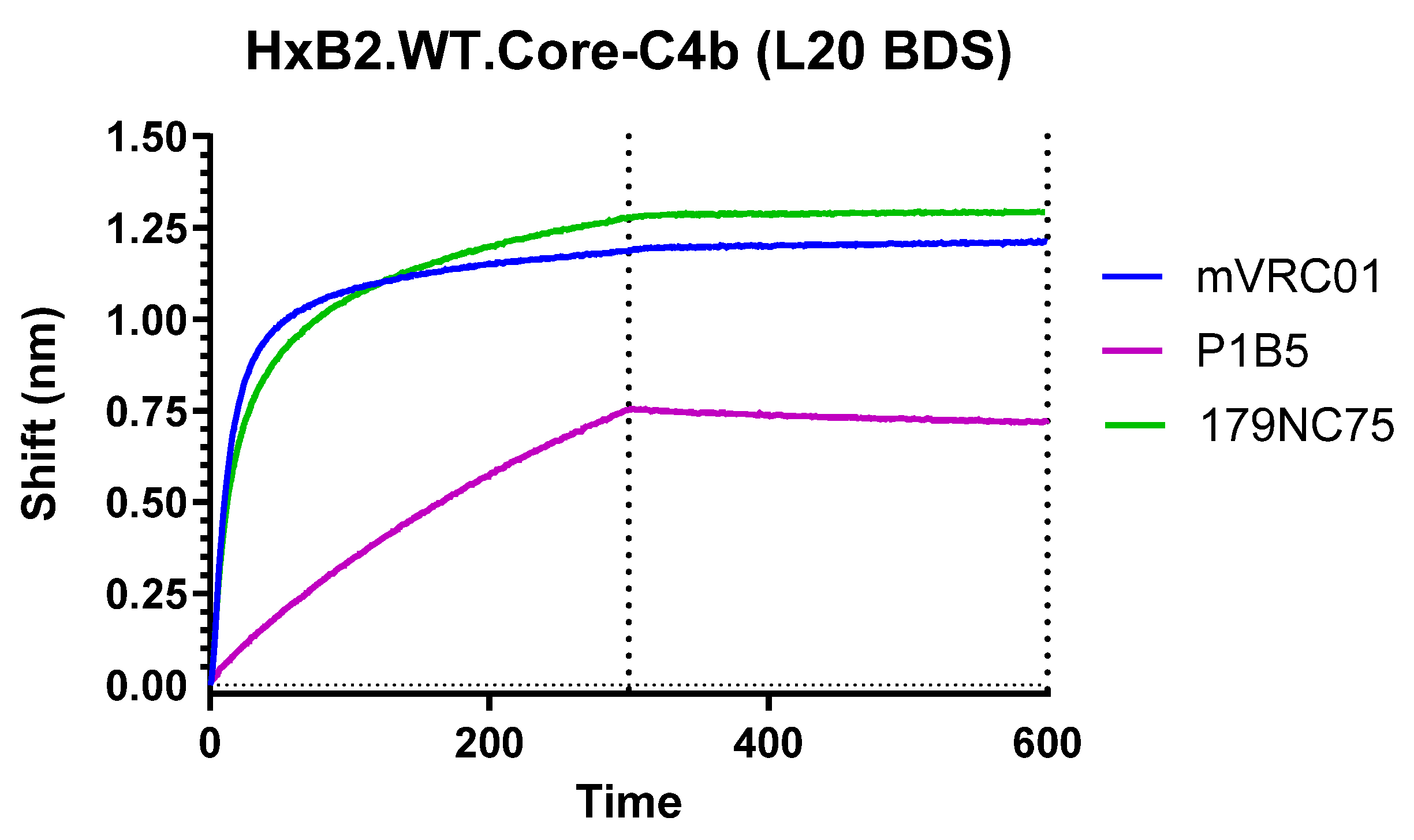
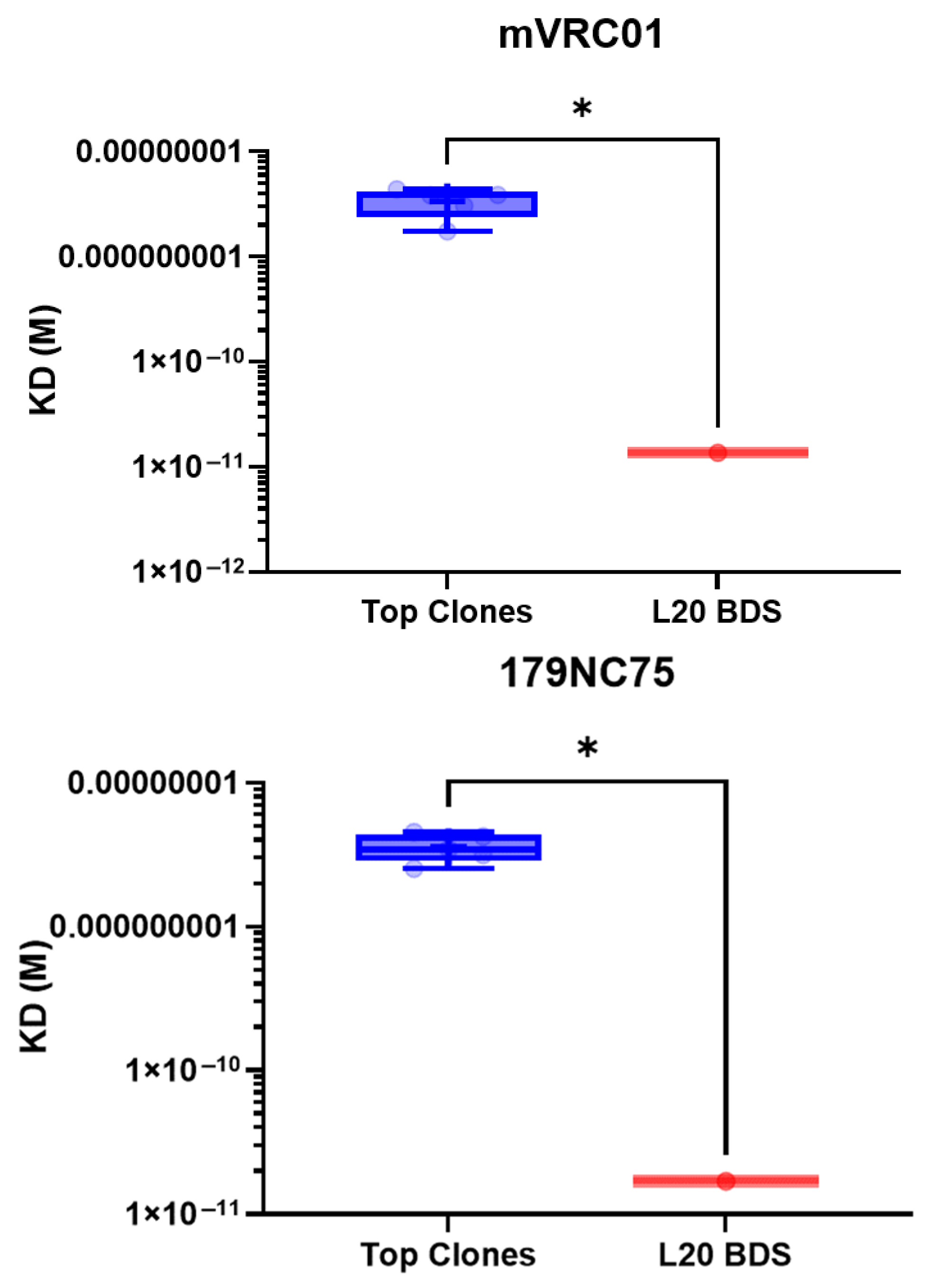
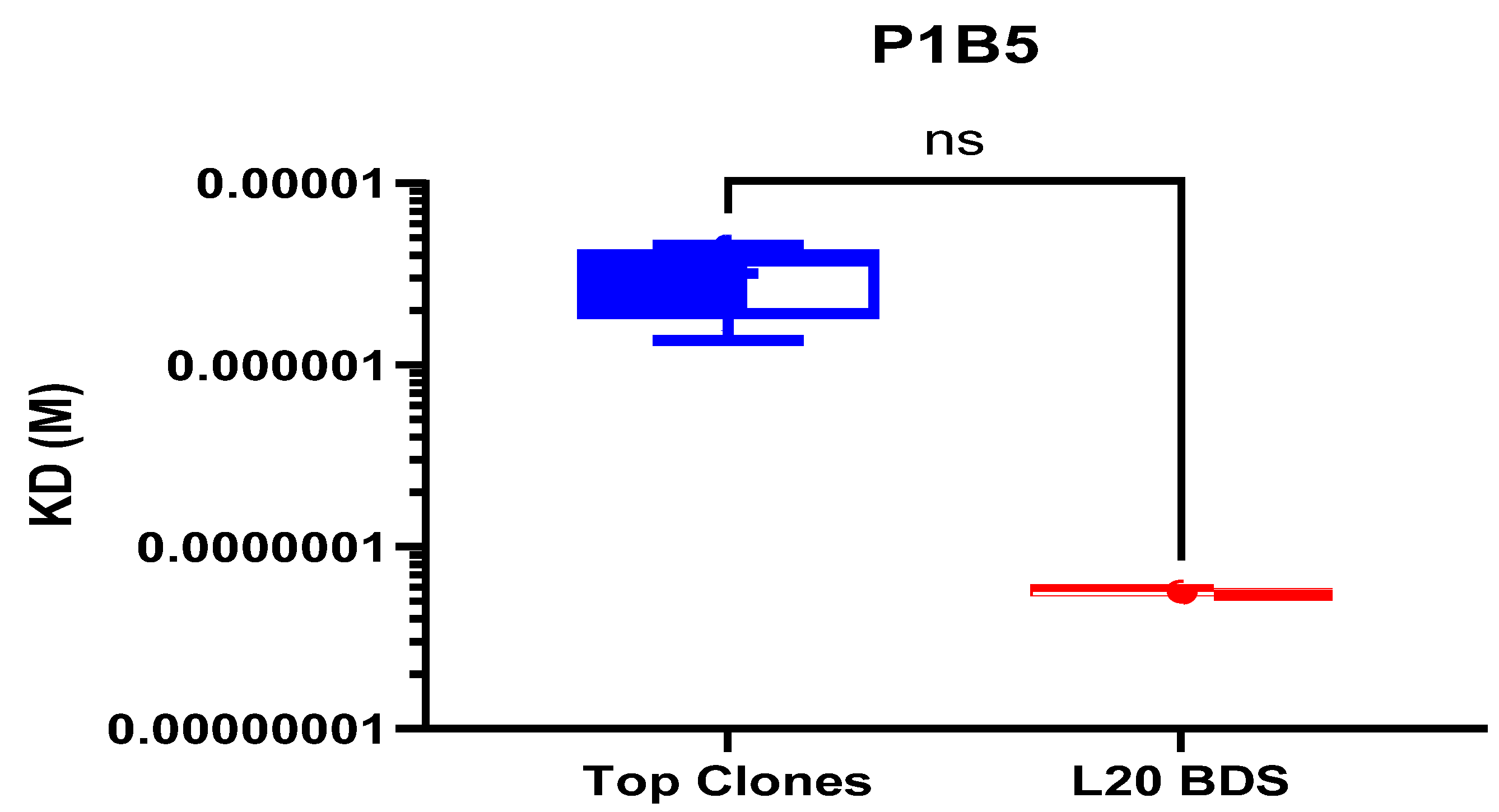
3.8. Antigenic Profile by Biolayer Interferometry
4. Discussion
4.1. Development Runs with the First Cell Line
4.2. Clone Selection and the New Production Cell Line
4.3. cGMP Production and Process Performance Comparison
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNAIDS. Global HIV & AIDS Statistics—Fact Sheet; UNAIDS: Geneva, Switzerland, 2024. [Google Scholar]
- Deeks, S.G.; Lewin, S.R.; Havlir, D.V. The end of AIDS: HIV infection as a chronic disease. Lancet 2016, 382, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Barouch, D.H. HIV vaccine development: Progress and future challenges. Immunity 2018, 48, 399–406. [Google Scholar]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Timofeeva, A.; Sedykh, S.; Nevinsky, G. Post-Immune Antibodies in HIV-1 Infection in the Context of Vaccine Development: A Variety of Biological Functions and Catalytic Activities. Vaccines 2022, 10, 384. [Google Scholar] [CrossRef]
- Jardine, J.G.; Ota, T.; Sok, D.; Pauthner, M.; Kulp, D.W.; Kalyuzhniy, O.; Skog, P.D.; Thinnes, T.C.; Bhullar, D.; Briney, B.; et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 2015, 349, 156–161. [Google Scholar] [CrossRef]
- Jardine, J.G.; Kulp, D.W.; Havenar-Daughton, C.; Sarkar, A.; Briney, B.; Sok, D.; Sesterhenn, F.; Ereño-Orbea, J.; Kalyuzhniy, O.; Deresa, I.; et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016, 351, 1458–1463. [Google Scholar] [CrossRef]
- Medina-Ramírez, M.; Garces, F.; Escolano, A.; Skog, P.; de Taeye, S.W.; Del Moral-Sanchez, I.; McGuire, A.T.; Yasmeen, A.; Behrens, A.-J.; Ozorowski, G.; et al. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 2017, 214, 2573–2590. [Google Scholar] [CrossRef]
- McGuire, A.T.; Gray, M.D.; Dosenovic, P.; Gitlin, A.D.; Freund, N.T.; Petersen, J.; Correnti, C.; Johnsen, W.; Kegel, R.; Stuart, A.B.; et al. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 2016, 7, 10618. [Google Scholar] [CrossRef] [PubMed]
- Parks, K.R.; MacCamy, A.J.; Trichka, J.; Gray, M.; Weidle, C.; Borst, A.J.; Khechaduri, A.; Takushi, B.; Agrawal, P.; Guenaga, J.; et al. Overcoming Steric Restrictions of VRC01 HIV-1 Neutralizing Antibodies through Immunization. Cell Rep. 2019, 29, 3060–3072.e7. [Google Scholar] [CrossRef]
- Knudsen, M.L.; Agrawal, P.; MacCamy, A.; Parks, K.R.; Gray, M.D.; Takushi, B.N.; Khechaduri, A.; Salladay, K.R.; Coler, R.N.; LaBranche, C.C.; et al. Adjuvants influence the maturation of VRC01-like antibodies during immunization. iScience 2022, 25, 105473. [Google Scholar] [CrossRef]
- Agrawal, P.; Knudsen, M.L.; MacCamy, A.; Hurlburt, N.K.; Khechaduri, A.; Salladay, K.R.; Kher, G.M.; Siddaramaiah, L.K.; Stuart, A.B.; Bontjer, I.; et al. Short CDRL1 in intermediate VRC01-like mAbs is not sufficient to overcome key glycan barriers on HIV-1 Env. J. Virol. 2024, 98, e0074424. [Google Scholar] [CrossRef]
- Stamatatos, L.; Pancera, M.; McGuire, A.T. Germline targeting immunogens. Immunol. Rev. 2017, 275, 203–216. [Google Scholar] [CrossRef] [PubMed Central]
- Stamatatos, L. The Germline Targeting Vaccine Concept: Overview and Updates from HIV Pre-clinical and Clinical Trials. Curr. HIV Res. 2025. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.J.; Suh, H.; Bershteyn, A.; Stephan, M.T.; Liu, H.; Huang, B.; Sohail, M.; Luo, S.; Um, S.H.; Khant, H.; et al. Interbilayer-crosslinked multilamellar vesicles as synthetic vaccines for potent humoral and cellular immune responses. Nat. Mater. 2012, 10, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Kulp, D.W.; Schief, W.R. Advances in structure-based vaccine design. Curr. Opin. Virol. 2013, 3, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Freund, N.T.; Horwitz, J.A.; Nogueira, L.; Sievers, S.A.; Scharf, L.; Scheid, J.F.; Gazumyan, A.; Liu, C.; Velinzon, K.; Goldenthal, A.; et al. A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1 In Vivo. PLoS Pathog. 2015, 11, e1005238. [Google Scholar] [CrossRef] [PubMed]
- Suloway, C.; Pulokas, J.; Fellmann, D.; Cheng, A.; Guerra, F.; Quispe, J.; Stagg, S.; Potter, C.S.; Carragher, B. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 2005, 151, 41–60. [Google Scholar] [CrossRef]
- Lander, G.C.; Stagg, S.M.; Voss, N.R.; Cheng, A.; Fellmann, D.; Pulokas, J.; Yoshioka, C.; Irving, C.; Mulder, A.; Lau, P.W.; et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009, 166, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Zivanov, J.; Nakane, T.; Forsberg, B.O.; Kimanius, D.; Hagen, W.J.H.; Lindahl, E.; Scheres, S.H.W. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 2018, 7, e42166. [Google Scholar] [CrossRef]
- Punjani , A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Baboo, S.; Diedrich, J.K.; Martínez-Bartolomé, S.; Wang, X.; Schiffner, T.; Groschel, B.; Schief, W.R.; Paulson, J.C.; Yates, J.R. DeGlyPHER: An Ultrasensitive Method for the Analysis of Viral Spike N-Glycoforms. Anal. Chem. 2021, 93, 13651–13657. [Google Scholar] [CrossRef]
- Arslan, O.; Weik, R.; Mundsperger, P.; Scientific, P.; Pallerla, S.; Craig, D.; Swoyer, R.; Blumen, T. Research Explores Alternative to EU-Banned Triton X-100. Available online: https://www.bioprocessonline.com/doc/research-explores-alternative-to-eu-banned-triton-x-0001 (accessed on 20 November 2024).
- Zhou, T.; Georgiev, I.; Wu, X.; Yang, Z.-Y.; Dai, K.; Finzi, A.; Kwon, Y.D.; Scheid, J.F.; Shi, W.; Xu, L.; et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 2010, 329, 811–817. [Google Scholar] [CrossRef]
- Walker, L.M.; Simek, M.D.; Priddy, F.; Gach, J.S.; Wagner, D.; Zwick, M.B.; Phogat, S.K.; Poignard, P.; Burton, D.R. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 2010, 6. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hofmeyer, T.; Schmelz, S.; Degiacomi, M.T.; Dal Peraro, M.; Daneschdar, M.; Scrima, A.; van den Heuvel, J.; Heinz, D.W.; Kolmar, H. Arranged sevenfold: Structural insights into the C-terminal oligomerization domain of human C4b-binding protein. J Mol Biol 2013, 425, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- ICH. ICH Q6B: Specifications: Test Procedures and Acceptance Criteria for Biotechnological/Biological Products; U.S. Food and Drug Administration (FDA): Silver Spring, MD, USA, 1999.
- FDA. Guidance for Industry: Sterile Drug Products Produced by Aseptic Processing—Current Good Manufacturing Practice; U.S. Food and Drug Administration (FDA): Silver Spring, MD, USA, 2008.
- Lee, S.L.; O’Connor, T.F.; Yang, X.; Cruz, C.N.; Chatterjee, S.; Madurawe, R.D.; Moore, C.M.V.; Yu, L.X.; Woodcock, J. Modernizing pharmaceutical manufacturing: From batch to continuous production. J. Pharm. Innov. 2015, 10, 191–199. [Google Scholar] [CrossRef]


| Process Code | Test Parameter | Process Parameters DO [%]/T [°C]/pH | Process Duration [d] | Final Viability [%] | Max. Cell Conc. [106 c/mL] | Final Yield [mg/L] |
|---|---|---|---|---|---|---|
| F977 010 | No base addition | 40/37.0/6.95 * | 12 | 86.7 | 10.6 | 81 |
| F978 010 | Standard protocol | 40/37.0/6.95 | 13 | 70.3 | 9.4 | 42 |
| F979 010 | Extended culture | 40/37.0/6.95 | 14 | 46.8 | 10.1 | 87 |
| F982 010 | No base addition | 40/37.0/6.95 * | 13 | 64.2 | 10.6 | 120 |
| F983 010 | No base addition, feed start 1 day later | 40/37.0/6.95 * | 13 | 60.3 | 10.5 | 161 |
| F984 010 | Minimum base addition | 40/37.0/6.80 | 13 | 78.8 | 8.74 | 40 |
| F981 010 (10 L) | Standard protocol | 40/37.0/6.95 | 14 | 66.0 | 13.2 | 117 |
| F985 010 (8 L) | Standard protocol, minimum base addition, including fill-up step | 40/37.0/6.80 | 3 + 13 | 77.9 | 12.0 | 107 |
| Process Code | Test Parameter | Process Parameters DO [%]/T [°C]/pH | Process Duration [d] | Final Viability [%] | Max. Cell Conc. [106 c/mL] | Final Yield [mg/L] |
|---|---|---|---|---|---|---|
| F982 020 | Temperature shift to 33 °C | 40/37.0 → 33.0/6.80 * | 14 | 74.1 | 8.97 | 64 |
| F983 020 | Temperature shift to 34 °C | 40/37.0 → 34.0/6.80 * | 14 | 72.9 | 9.96 | 84 |
| F984 020 | Temperature shift to 36 °C | 40/37.0 → 36.0/6.80 * | 14 | 76.7 | 10.6 | 65 |
| F985 010 (8 L) | Standard protocol, minimum base addition, including fill-up step | 40/37.0/6.80 * | 3 + 13 | 77.9 | 12.0 | 107 |
| Process Code | Test Parameter | Process Parameters DO [%]/T [°C]/pH | Process Duration [d] | Final Viability [%] | Max. Cell Conc. [106 c/mL] | Final Yield [mg/L] |
|---|---|---|---|---|---|---|
| F986 010 (8 L) | Clone 4E22/RCB | 40/37.0/6.80 * | 12 | 81.7 | 11.9 | 136 |
| F987 010 (8 L) | Clone 4E22/RCB Enriched feed solution (120 g/L Glc) | 40/37.0/6.80 * | 12 | 80.3 | 12.0 | 132 |
| F988 010 | Clone 4E22/RCB | 40/37.0/6.80 * | 4 + 12 | 78.6 | 11.9 | 132 |
| F989 010 | Clone 9K21/RCB | 40/37.0/6.80 * | 12 | 77.3 | 12.0 | 109 |
| F990 010 | Clone 5C9/RCB | 40/37.0/6.80 * | 12 | 79.4 | 12.6 | 98 |
| Process Code | Test Parameter | Process Parameters DO [%]/T [°C]/pH | Process Duration [d] | Final Viability [%] | Max. Cell Conc. [106 c/mL] | Final Yield [mg/L] |
|---|---|---|---|---|---|---|
| F991 010 | Verification run | 40/37.0/6.95 * | 11 | 81.8 | 13.8 | 137 |
| F992 010 | Verification run | 40/37.0/6.95 * | 11 | 81.6 | 12.7 | 131 |
| F994 010 (10 L) | Verification run, two different (late) harvest time points | 40/37.0/6.95 * | 12 (1) 14 (2) | 74.9 (1) 70.6 (2) | 12.4 | 163 233 |
| Drug Substance Specifications | Results Small Confirmation Runs | GMP Campaign | |||
|---|---|---|---|---|---|
| Test/Analysis | Acceptance Criteria | L20 | L21 | L22 | Drug Substance: T206/HXB2C/B1 |
| General | |||||
| Appearance | clear to opalescent, colorless to off-white liquid, essentially free from visible particulates | pass | pass | pass | pass |
| pH | 7.5 ± 0.5 | 7.5 | 7.6 | 7.5 | 7.4 |
| Osmolality | 220 ± 50 mOsmol/kg | 224 mOsmol/kg | 225 mOsmol/kg | 225 mOsmol/kg | 223 mOsmol/kg |
| Identity | |||||
| Binding activity (ELISA) | binding to antibodies mVRC01, P1B5, 179NC75 detected | binding to three antibodies detected | binding to three antibodies detected | binding to three antibodies detected | binding to three |
| antibodies detected | |||||
| Potency | |||||
| Antibody titration ELISA | report result of the EC50 in ng/mL for antibodies mVRC01, P1B5, 179NC75 | mVRC01: 54.3 ng/mL P1B5: 181.9 ng/mL 179NC75: 297.8 ng/mL | mVRC01: 45.7 ng/mL P1B5: 146.4 ng/mL 179NC75: 296.1 ng/mL | mVRC01: 49.6 ng/mL P1B5: 205.3 ng/mL 179NC75: 283.2 ng/mL | mVRC01: 44.2 ng/mL P1B5: 296.4 ng/mL 179NC75: 272.1 ng/mL |
| Content | |||||
| Total protein concentration (UV-Vis 280nm) | 1.0 ± 0.2 mg/mL | 0.97 mg/mL | 1.00 mg/mL | 0.95 mg/mL | 1.0 mg/mL |
| Purity | |||||
| Purity (SEC HPLC) | <10% Aggregates report result [%] for main peak area | 0.34% Aggregates 99.66% Main peak | 0.41% Aggregates 99.59% Main peak | 0.24% Aggregates 99.76% Main peak | 0% Aggregates 100% Main peak |
| Residual host cell protein (ELISA) | ≤100 ng/mg | 35 ng/mg (ppm) | 29 ng/mg (ppm) | 24 ng/mg (ppm) | 53 ng/mg (ppm) |
| Residual DNA content (qPCR) | ≤1 ng/mg | n.a. | Informing analysis by Threshold immunoassay: ≤6.3 pg/mg | Informing analysis by Threshold immunoassay: ≤6.0 pg/mg | ≤14.2 pg/mg |
| Protein purity (red. SDS-CE) | report result [%] | Main peak: 97.1% <Main peak: 1.8% >Main peak: 1.1% | Main peak: 98.2% <Main peak: 1.1% >Main peak: 0.7% | Main peak: 97.1% <Main peak: 2.0% >Main peak: 0.9% | Main peak: 98.1% <Main peak: 1.9 |
| Contaminants | |||||
| Endotoxin (rFC assay) | ≤30 EU/mg | n.a. | n.a. | n.a. | <0.16 EU/mg |
| Bioburden | ≤10 CFU/10 mL | n.a. | n.a. | n.a. | 0 CFU/10 mL |
| Test/analysis for characterization | |||||
| Test/analysis | |||||
| Nanoparticle size (Zavg) | report result | 18.3 nm | 18.5 nm | 18.3 nm | 18.8 nm |
| Nanoparticle size distribution (polydispersity index) | report result | 0.04 | 0.03 | 0.04 | 0.03 |
| MODEL VIRUS. | X-MuLV | MMV |
|---|---|---|
| Detergent treatment | ≥4.67 | - |
| Anion exchange chromatography | ≥3.59 | 2.25 |
| Hydroxy apatite chromatography | 0.75 (1) | - |
| Nanofiltration | ≥2.92 (2) | ≥5.40 (2) |
| Total LRF | ≥13.66 (2) | ≥7.65 |
| Clone # | mVRC01 (KD) | P1B5 (KD) | 179NC75 (KD) |
|---|---|---|---|
| Clone CL-5C9 | 4.37 × 10−9 | 3.75 × 10−6 | 4.53 × 10−9 |
| Clone CL-3K21 | 1.74 × 10−9 | 3.98 × 10−6 | 3.16 × 10−9 |
| Clone CL-9K21 | 3.86 × 10−9 | 4.56 × 10−6 | 3.46 × 10−9 |
| Clone CL-3N15 | 3.87 × 10−9 | 1.37 × 10−6 | 4.22 × 10−9 |
| Clone CL-4E22 | 3.06 × 10−9 | 2.28 × 10−6 | 2.54 × 10−9 |
| L20 BDS | 1.36 × 10−11 | 5.65 × 10−8 | 1.69 × 10−11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pallerla, S.; Kallur Siddaramaiah, L.; Mundsperger, P.; Katinger, D.; Fauland, K.; Kreismayr, G.; Weik, R.; Arslan, O.; Shen, M.; Ozorowski, G.; et al. GMP Manufacturing and Characterization of the HIV Booster Immunogen HxB2.WT.Core-C4b for Germline Targeting Vaccine Strategies. Vaccines 2025, 13, 980. https://doi.org/10.3390/vaccines13090980
Pallerla S, Kallur Siddaramaiah L, Mundsperger P, Katinger D, Fauland K, Kreismayr G, Weik R, Arslan O, Shen M, Ozorowski G, et al. GMP Manufacturing and Characterization of the HIV Booster Immunogen HxB2.WT.Core-C4b for Germline Targeting Vaccine Strategies. Vaccines. 2025; 13(9):980. https://doi.org/10.3390/vaccines13090980
Chicago/Turabian StylePallerla, Sammaiah, Latha Kallur Siddaramaiah, Philipp Mundsperger, Dietmar Katinger, Katharina Fauland, Günter Kreismayr, Robert Weik, Onur Arslan, Mingchao Shen, Gabriel Ozorowski, and et al. 2025. "GMP Manufacturing and Characterization of the HIV Booster Immunogen HxB2.WT.Core-C4b for Germline Targeting Vaccine Strategies" Vaccines 13, no. 9: 980. https://doi.org/10.3390/vaccines13090980
APA StylePallerla, S., Kallur Siddaramaiah, L., Mundsperger, P., Katinger, D., Fauland, K., Kreismayr, G., Weik, R., Arslan, O., Shen, M., Ozorowski, G., Lee, W.-H., Ward, A. B., Baboo, S., Diedrich, J. K., Yates, J. R., III, Paulson, J. C., Blumen, T., Craig, D., Swoyer, R., ... Stamatatos, L. (2025). GMP Manufacturing and Characterization of the HIV Booster Immunogen HxB2.WT.Core-C4b for Germline Targeting Vaccine Strategies. Vaccines, 13(9), 980. https://doi.org/10.3390/vaccines13090980






