CXCR5 Signals Fine-Tune Dendritic Cell Transcription and Regulate TH2 Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
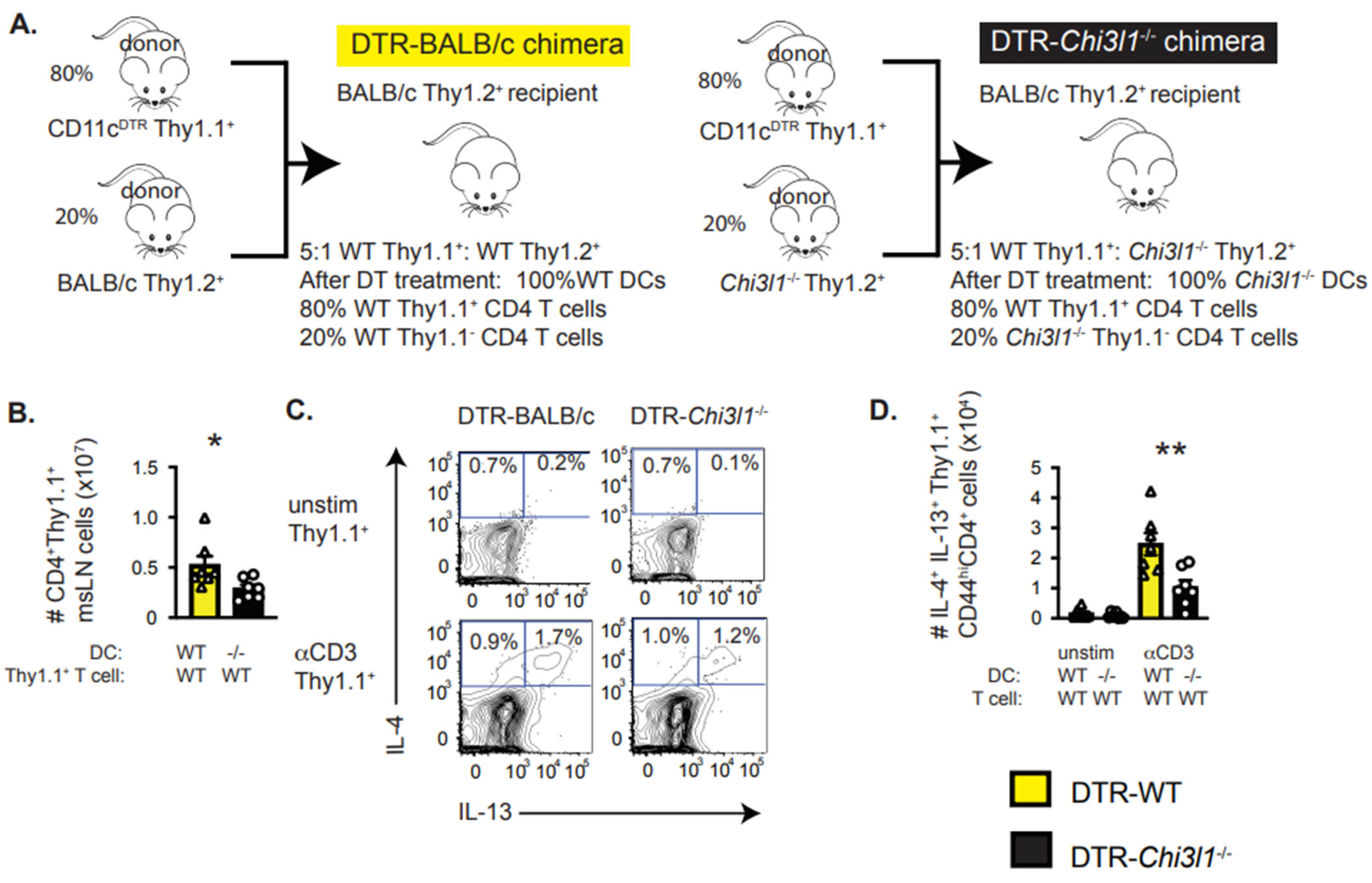
2.2. Bone Marrow Chimeras
2.3. Infections and Diphtheria Toxin Treatment
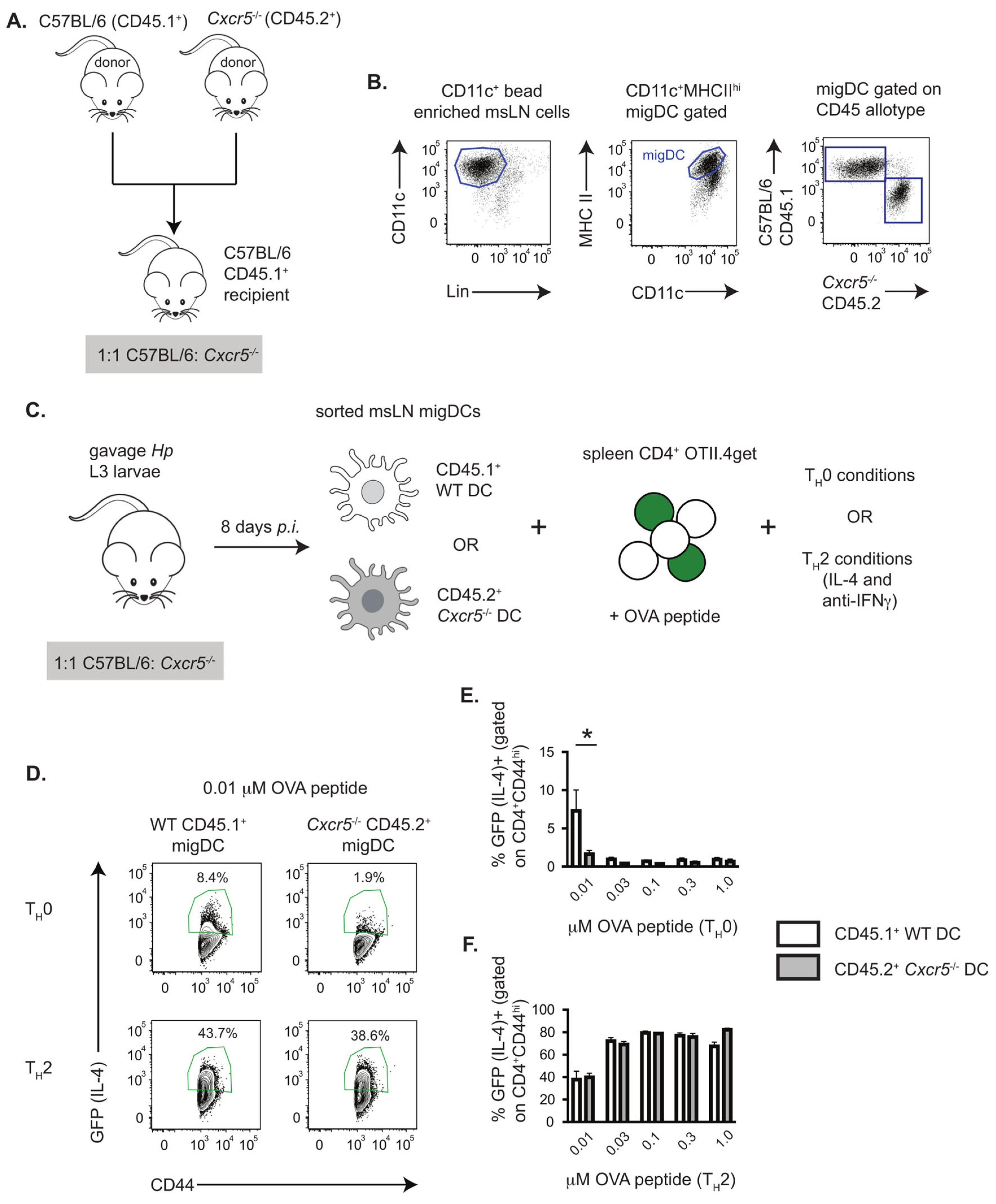
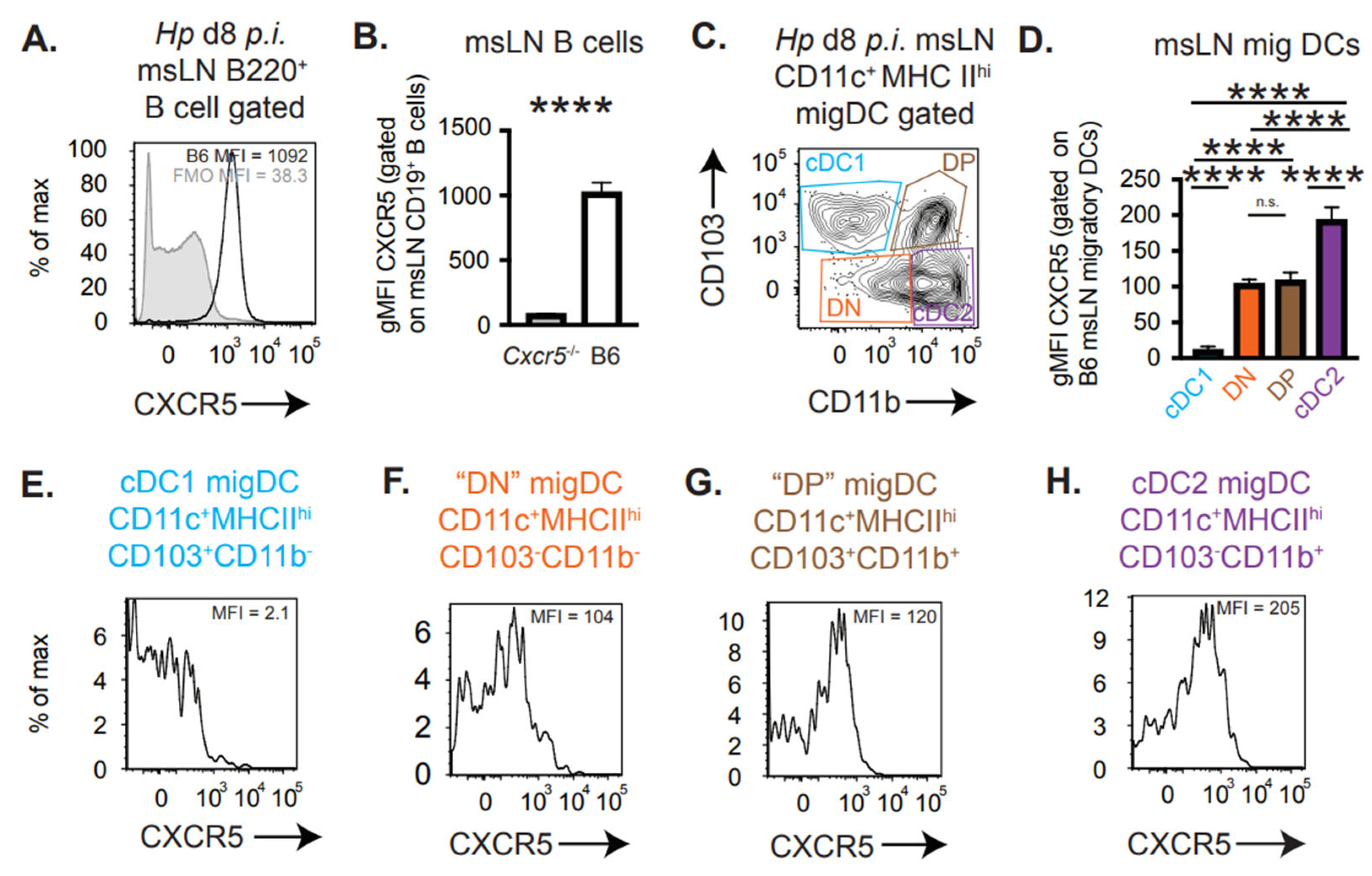
2.4. Flow Cytometry
2.5. DC Enrichment and Sort Purification
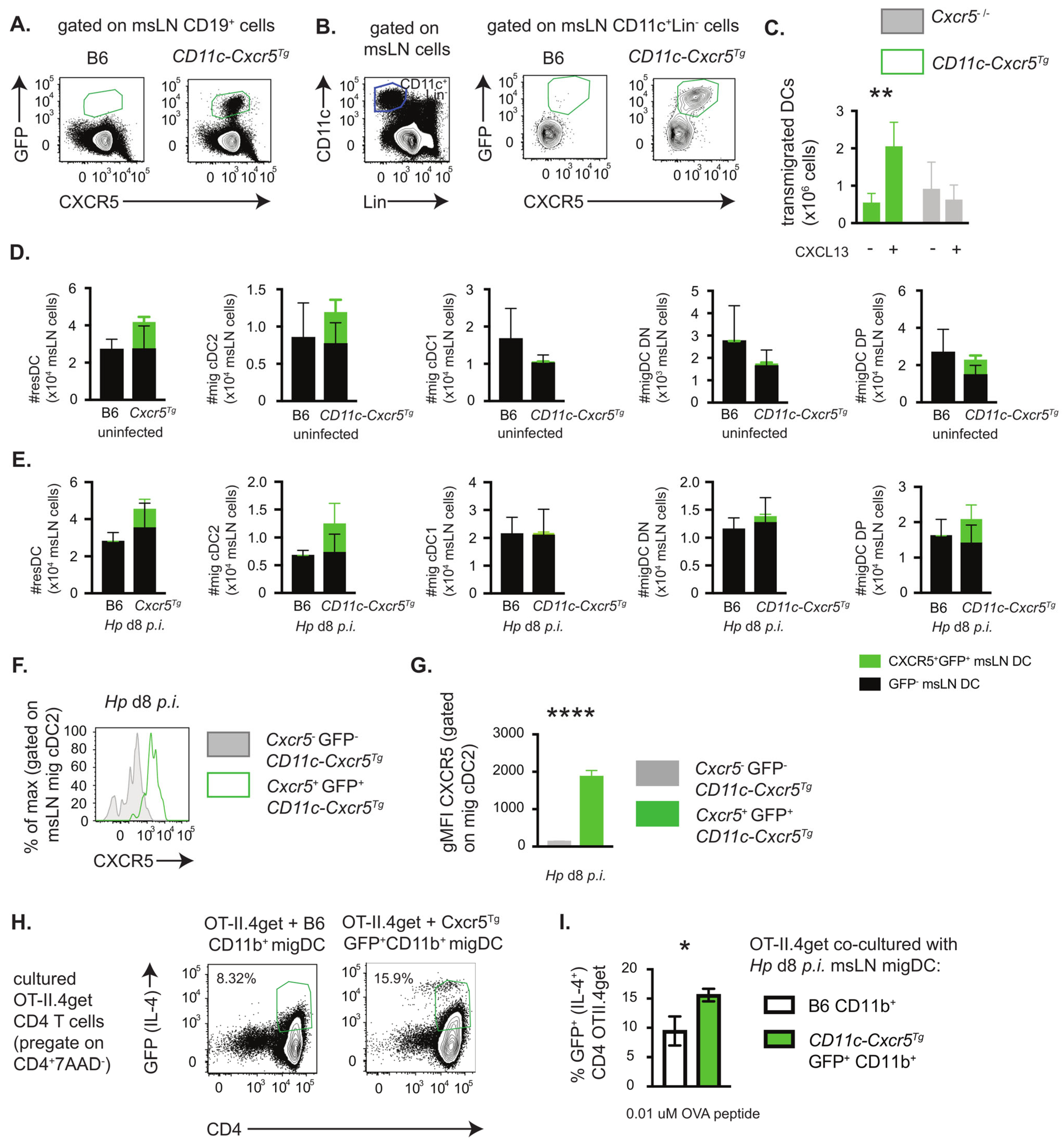
2.6. In Vitro TH2 Cell Priming Assays
2.7. DC Chemotaxis Assay
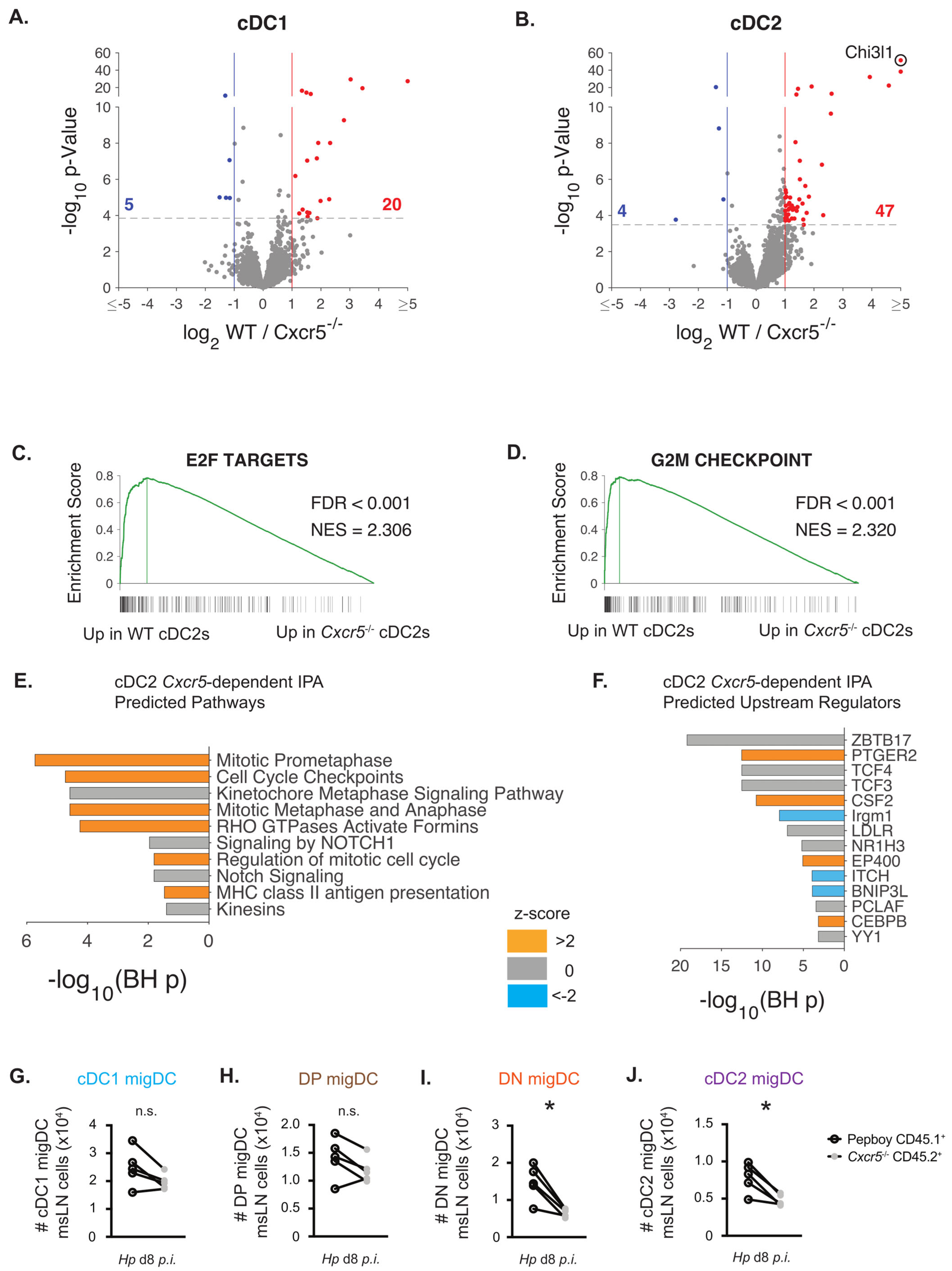
2.8. Bulk RNA-Seq Analysis
2.9. Statistical Analysis
3. Results
3.1. TH2 Priming In Vitro by Hp-Induced DCs Requires DC Intrinsic Expression of CXCR5
3.2. CXCR5 Is Expressed by Migratory cDC2 and Not cDC1 Cells
3.3. Enforced CXCR5 Expression by Migratory CD11b+ DCs Enhances TH2 Priming In Vitro
3.4. CXCR5 Regulates Cell Division Pathways and Modulates the Size of the Migratory cDC2 Compartment
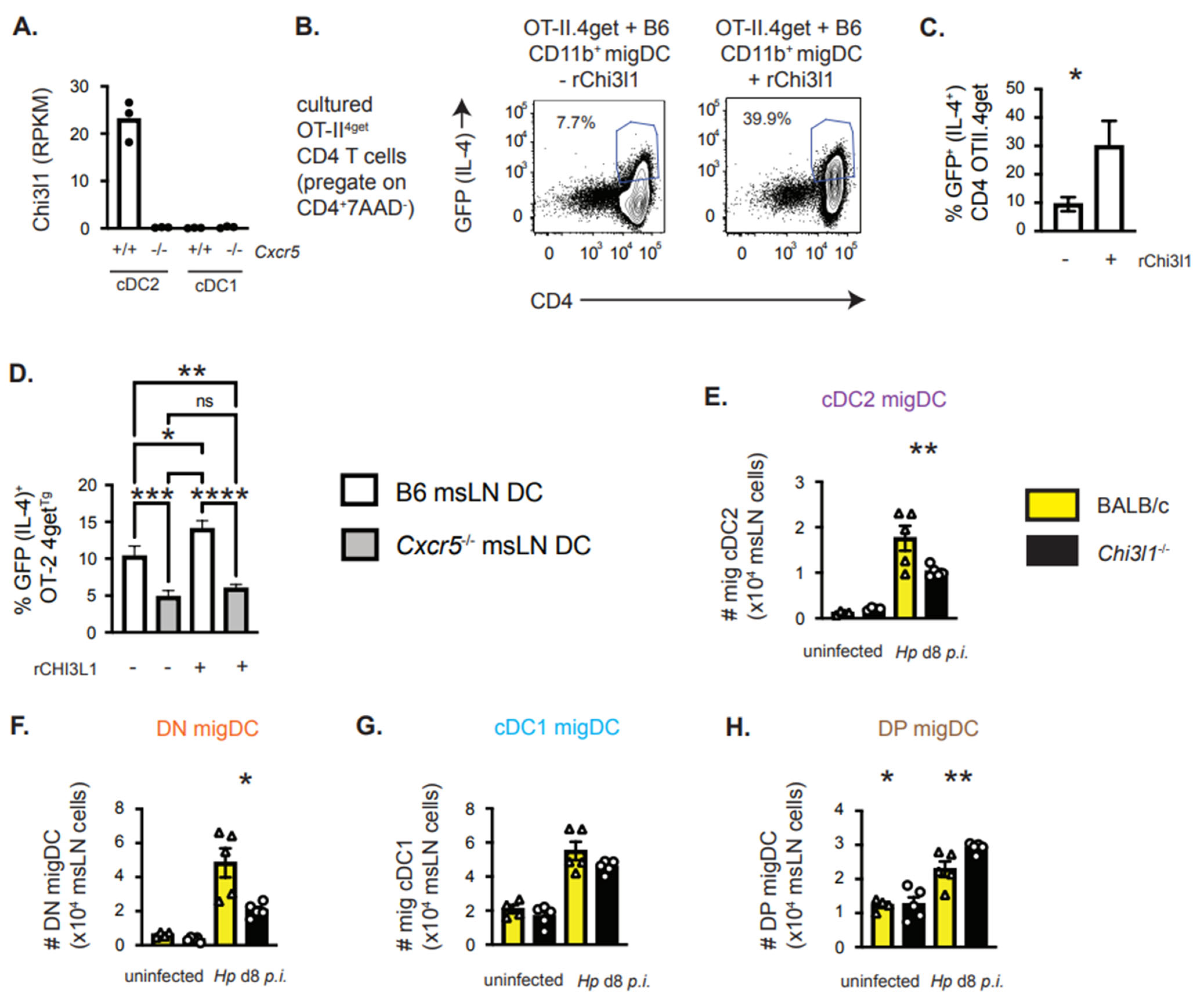
3.5. Chi3l1, a CXCR5-Regulated Gene in cDC2 Cells, Supports TH2 Priming In Vitro
3.6. Chi3l1 Regulates the Size of the msLN cDC2 Population Following Hp Infection
3.7. DC-Intrinsic Expression of Chi3l1 Regulates TH2 Responses In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BM | bone marrow |
| CCR7 | C-C motif chemokine receptor 7 |
| cDC | conventional dendritic cells |
| cDC1 | CD11b−CD103+ conventional DCs |
| cDC2 | CD11b+CD103− conventional DCs |
| Chi3l1 | chitinase 3-like-1 gene (mouse protein: Chi3l1, mouse gene Chi3l1) |
| CXCL13 | C-X-C motif chemokine ligand 13 |
| CXCR5 | C-X-C motif chemokine receptor 5 (mouse protein: CXCR5, mouse gene Cxcr5) |
| DT | diphtheria toxin |
| DCs | dendritic cells |
| DN | CD103−CD11b− conventional DCs |
| DP | CD103+CD11b+ conventional DCs |
| EGFP | enhanced green fluorescent protein |
| FRCs | fibroblastic reticular cells |
| gMFI | geometric mean fluorescence intensity |
| GSEA | gene set enrichment analysis |
| Hp | Heligmosomoides polygyrus |
| IPA | Ingenuity Pathway Analysis |
| LN | Lymph nodes |
| msLN | mesenteric lymph nodes |
| n | number of mice |
| Nb | Nippostrongylus brasiliensis |
| OVA | ovalbumin |
| TH2 cells | T helper 2 CD4 T cells |
| YKL-40 | human protein encoded by CHI3L1 gene (alternative: CHI3L1 protein) |
References
- Ronchese, F.; Webb, G.R.; Ochiai, S.; Lamiable, O.; Brewerton, M. How type-2 dendritic cells induce Th2 differentiation: Instruction, repression, or fostering T cell-T cell communication? Allergy 2025, 80, 395–407. [Google Scholar] [CrossRef]
- Cabeza-Cabrerizo, M.; Cardoso, A.; Minutti, C.M.; Pereira da Costa, M.; Reis e Sousa, C. Dendritic Cells Revisited. Annu. Rev. Immunol. 2021, 39, 131–166. [Google Scholar] [CrossRef]
- Leon, B.; Ballesteros-Tato, A.; Browning, J.L.; Dunn, R.; Randall, T.D.; Lund, F.E. Regulation of T(H)2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nat. Immunol. 2012, 13, 681–690. [Google Scholar] [CrossRef]
- Stagg, A.J. Intestinal Dendritic Cells in Health and Gut Inflammation. Front. Immunol. 2018, 9, 2883. [Google Scholar] [CrossRef] [PubMed]
- Connor, L.M.; Tang, S.C.; Cognard, E.; Ochiai, S.; Hilligan, K.L.; Old, S.I.; Pellefigues, C.; White, R.F.; Patel, D.; Smith, A.A.; et al. Th2 responses are primed by skin dendritic cells with distinct transcriptional profiles. J. Exp. Med. 2017, 214, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, I.; Kato, T.; Hemmi, H.; Fukuda-Ohta, Y.; Wakaki-Nishiyama, N.; Yamamoto, A.; Kaisho, T. Conventional Type 1 Dendritic Cells in Intestinal Immune Homeostasis. Front. Immunol. 2022, 13, 857954. [Google Scholar] [CrossRef] [PubMed]
- Tussiwand, R.; Everts, B.; Grajales-Reyes, G.E.; Kretzer, N.M.; Iwata, A.; Bagaitkar, J.; Wu, X.; Wong, R.; Anderson, D.A.; Murphy, T.L.; et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 2015, 42, 916–928. [Google Scholar] [CrossRef]
- Everts, B.; Tussiwand, R.; Dreesen, L.; Fairfax, K.C.; Huang, S.C.; Smith, A.M.; O’Neill, C.M.; Lam, W.Y.; Edelson, B.T.; Urban, J.F., Jr.; et al. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J. Exp. Med. 2016, 213, 35–51. [Google Scholar] [CrossRef]
- Akbari, M.; Honma, K.; Kimura, D.; Miyakoda, M.; Kimura, K.; Matsuyama, T.; Yui, K. IRF4 in dendritic cells inhibits IL-12 production and controls Th1 immune responses against Leishmania major. J. Immunol. 2014, 192, 2271–2279. [Google Scholar] [CrossRef]
- Persson, E.K.; Uronen-Hansson, H.; Semmrich, M.; Rivollier, A.; Hägerbrand, K.; Marsal, J.; Gudjonsson, S.; Håkansson, U.; Reizis, B.; Kotarsky, K.; et al. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 2013, 38, 958–969. [Google Scholar] [CrossRef]
- Mayer, J.U.; Demiri, M.; Agace, W.W.; MacDonald, A.S.; Svensson-Frej, M.; Milling, S.W. Different populations of CD11b+ dendritic cells drive Th2 responses in the small intestine and colon. Nat. Commun. 2017, 8, 15820. [Google Scholar] [CrossRef]
- Plantinga, M.; Guilliams, M.; Vanheerswynghels, M.; Deswarte, K.; Branco-Madeira, F.; Toussaint, W.; Vanhoutte, L.; Neyt, K.; Killeen, N.; Malissen, B.; et al. Conventional and Monocyte-Derived CD11b+ Dendritic Cells Initiate and Maintain T Helper 2 Cell-Mediated Immunity to House Dust Mite Allergen. Immunity 2013, 38, 322–335. [Google Scholar] [CrossRef]
- Zhang, S.; Audiger, C.; Chopin, M.; Nutt, S.L. Transcriptional regulation of dendritic cell development and function. Front. Immunol. 2023, 14, 1182553. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Nish, S.A.; Jiang, R.; Hou, L.; Licona-Limon, P.; Weinstein, J.S.; Zhao, H.; Medzhitov, R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity 2013, 39, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Grajales-Reyes, G.E.; Iwata, A.; Albring, J.; Wu, X.; Tussiwand, R.; Kc, W.; Kretzer, N.M.; Briseno, C.G.; Durai, V.; Bagadia, P.; et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat. Immunol. 2015, 16, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Luda, K.M.; Joeris, T.; Persson, E.K.; Rivollier, A.; Demiri, M.; Sitnik, K.M.; Pool, L.; Holm, J.B.; Melo-Gonzalez, F.; Richter, L.; et al. IRF8 Transcription-Factor-Dependent Classical Dendritic Cells Are Essential for Intestinal T Cell Homeostasis. Immunity 2016, 44, 860–874. [Google Scholar] [CrossRef]
- Edelson, B.T.; Kc, W.; Juang, R.; Kohyama, M.; Benoit, L.A.; Klekotka, P.A.; Moon, C.; Albring, J.C.; Ise, W.; Michael, D.G.; et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 2010, 207, 823–836. [Google Scholar] [CrossRef]
- Williams, J.W.; Tjota, M.Y.; Clay, B.S.; Vander Lugt, B.; Bandukwala, H.S.; Hrusch, C.L.; Decker, D.C.; Blaine, K.M.; Fixsen, B.R.; Singh, H.; et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 2013, 4, 2990. [Google Scholar] [CrossRef]
- Redpath, S.A.; Heieis, G.A.; Reynolds, L.A.; Fonseca, N.M.; Kim, S.S.; Perona-Wright, G. Functional specialization of intestinal dendritic cell subsets during Th2 helminth infection in mice. Eur. J. Immunol. 2018, 48, 87–98. [Google Scholar] [CrossRef]
- Connor, L.M.; Tang, S.C.; Camberis, M.; Le Gros, G.; Ronchese, F. Helminth-conditioned dendritic cells prime CD4+ T cells to IL-4 production in vivo. J. Immunol. 2014, 193, 2709–2717. [Google Scholar] [CrossRef]
- Krishnaswamy, J.K.; Gowthaman, U.; Zhang, B.; Mattsson, J.; Szeponik, L.; Liu, D.; Wu, R.; White, T.; Calabro, S.; Xu, L.; et al. Migratory CD11b+ conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci. Immunol. 2017, 2, eaam9169. [Google Scholar] [CrossRef] [PubMed]
- Bradford, B.M.; Donaldson, D.S.; Forman, R.; Else, K.J.; Mabbott, N.A. Increased susceptibility to oral Trichuris muris infection in the specific absence of CXCR5+ CD11c+ cells. Parasite Immunol. 2018, 40, e12566. [Google Scholar] [CrossRef] [PubMed]
- Brandum, E.P.; Jorgensen, A.S.; Rosenkilde, M.M.; Hjorto, G.M. Dendritic Cells and CCR7 Expression: An Important Factor for Autoimmune Diseases, Chronic Inflammation, and Cancer. Int. J. Mol. Sci. 2021, 22, 8340. [Google Scholar] [CrossRef] [PubMed]
- Dubey, L.K.; Ludewig, B.; Luther, S.A.; Harris, N.L. IL-4Rα-Expressing B Cells Are Required for CXCL13 Production by Fibroblastic Reticular Cells. Cell Rep. 2019, 27, 2442–2458.e2445. [Google Scholar] [CrossRef]
- Leon, B.; Lund, F.E. Compartmentalization of dendritic cell and T-cell interactions in the lymph node: Anatomy of T-cell fate decisions. Immunol. Rev. 2019, 289, 84–100. [Google Scholar] [CrossRef]
- Lee, C.G.; Hartl, D.; Lee, G.R.; Koller, B.; Matsuura, H.; Da Silva, C.A.; Sohn, M.H.; Cohn, L.; Homer, R.J.; Kozhich, A.A.; et al. Role of breast regression protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13-induced tissue responses and apoptosis. J. Exp. Med. 2009, 206, 1149–1166. [Google Scholar] [CrossRef]
- Shen, F.W.; Saga, Y.; Litman, G.; Freeman, G.; Tung, J.S.; Cantor, H.; Boyse, E.A. Cloning of Ly-5 cDNA. Proc. Natl. Acad. Sci. USA 1985, 82, 7360–7363. [Google Scholar] [CrossRef]
- Shen, F.W.; Tung, J.S.; Boyse, E.A. Further definition of the Ly-5 system. Immunogenetics 1986, 24, 146–149. [Google Scholar] [CrossRef]
- Forster, R.; Mattis, A.E.; Kremmer, E.; Wolf, E.; Brem, G.; Lipp, M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell 1996, 87, 1037–1047. [Google Scholar] [CrossRef]
- Barnden, M.J.; Allison, J.; Heath, W.R.; Carbone, F.R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998, 76, 34–40. [Google Scholar] [CrossRef]
- Caton, M.L.; Smith-Raska, M.R.; Reizis, B. Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. J. Exp. Med. 2007, 204, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Unutmaz, D.; Wong, P.; Sano, G.; De los Santos, K.; Sparwasser, T.; Wu, S.; Vuthoori, S.; Ko, K.; Zavala, F.; et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity 2002, 17, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Reif, A.E.; Allen, J.M. Mouse Thymic Iso-antigens. Nature 1966, 209, 521–523. [Google Scholar] [CrossRef]
- King, I.L.; Mohrs, M. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J. Exp. Med. 2009, 206, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Peel, J.N.; Scharer, C.D.; Risley, C.A.; Chisolm, D.A.; Schultz, M.D.; Yu, B.; Ballesteros-Tato, A.; Wojciechowski, W.; Mousseau, B.; et al. T-bet Transcription Factor Promotes Antibody-Secreting Cell Differentiation by Limiting the Inflammatory Effects of IFN-gamma on B Cells. Immunity 2019, 50, 1172–1187.e1177. [Google Scholar] [CrossRef]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Lawrence, M.; Huber, W.; Pages, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mohrs, M.; Shinkai, K.; Mohrs, K.; Locksley, R.M. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity 2001, 15, 303–311. [Google Scholar] [CrossRef]
- Rubtsov, A.V.; Rubtsova, K.; Fischer, A.; Meehan, R.T.; Gillis, J.Z.; Kappler, J.W.; Marrack, P. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c⁺ B-cell population is important for the development of autoimmunity. Blood 2011, 118, 1305–1315. [Google Scholar] [CrossRef]
- Wiese, K.E.; Walz, S.; von Eyss, B.; Wolf, E.; Athineos, D.; Sansom, O.; Eilers, M. The role of MIZ-1 in MYC-dependent tumorigenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a014290. [Google Scholar] [CrossRef]
- Ryu, S.H.; Shin, H.S.; Eum, H.H.; Park, J.S.; Choi, W.; Na, H.Y.; In, H.; Kim, T.G.; Park, S.; Hwang, S.; et al. Granulocyte Macrophage-Colony Stimulating Factor Produces a Splenic Subset of Monocyte-Derived Dendritic Cells That Efficiently Polarize T Helper Type 2 Cells in Response to Blood-Borne Antigen. Front. Immunol. 2021, 12, 767037. [Google Scholar] [CrossRef] [PubMed]
- Curtiss, M.L.; Rosenberg, A.F.; Scharer, C.D.; Mousseau, B.; Benavides, N.A.B.; Bradley, J.E.; León, B.; Steele, C.; Randall, T.D.; Lund, F.E. Chitinase-3-like 1 regulates TH2 cells, TFH cells and IgE responses to helminth infection. Front. Immunol. 2023, 14, 1158493. [Google Scholar] [CrossRef]
- Eisenbarth, S.C. Dendritic cell subsets in T cell programming: Location dictates function. Nat. Rev. Immunol. 2019, 19, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Ansel, K.M.; Ngo, V.N.; Hyman, P.L.; Luther, S.A.; Forster, R.; Sedgwick, J.D.; Browning, J.L.; Lipp, M.; Cyster, J.G. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 2000, 406, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, N.; Bornacelly, A.; Mercado, D.; Unneberg, P.; Mittermann, I.; Valenta, R.; Kennedy, M.; Scheynius, A.; Caraballo, L. Genetic Variants in CHIA and CHI3L1 Are Associated with the IgE Response to the Ascaris Resistance Marker ABA-1 and the Birch Pollen Allergen Bet v 1. PLoS ONE 2016, 11, e0167453. [Google Scholar] [CrossRef]
- Kwon, J.W.; Kim, T.W.; Cho, S.H.; Min, K.U.; Park, H.W. Serum YKL-40 levels are correlated with symptom severity in patients with allergic rhinitis. Allergy 2011, 66, 1252–1253. [Google Scholar] [CrossRef]
- Kim, E.G.; Kim, M.N.; Hong, J.Y.; Lee, J.W.; Kim, S.Y.; Kim, K.W.; Lee, C.G.; Elias, J.A.; Song, T.W.; Sohn, M.H. Chitinase 3-Like 1 Contributes to Food Allergy via M2 Macrophage Polarization. Allergy Asthma Immunol. Res. 2020, 12, 1012–1028. [Google Scholar] [CrossRef]
- Dezman, K.; Korosec, P.; Rupnik, H.; Rijavec, M. SPINK5 is associated with early-onset and CHI3L1 with late-onset atopic dermatitis. Int. J. Immunogenet. 2017, 44, 212–218. [Google Scholar] [CrossRef]
- Lee, Y.S.; Yu, J.E.; Kim, M.J.; Ham, H.J.; Jeon, S.H.; Yun, J.; Song, S.G.; Lee, C.K.; Han, S.B.; Son, D.J.; et al. New therapeutic strategy for atopic dermatitis by targeting CHI3L1/ITGA5 axis. Clin. Transl. Med. 2022, 12, e739. [Google Scholar] [CrossRef]
- Salomon, J.; Matusiak, L.; Nowicka-Suszko, D.; Szepietowski, J.C. Chitinase-3-Like Protein 1 (YKL-40) Reflects the Severity of Symptoms in Atopic Dermatitis. J. Immunol. Res. 2017, 2017, 5746031. [Google Scholar] [CrossRef]
- Kwak, E.J.; Hong, J.Y.; Kim, M.N.; Kim, S.Y.; Kim, S.H.; Park, C.O.; Kim, K.W.; Lee, C.G.; Elias, J.A.; Jee, H.M.; et al. Chitinase 3-like 1 drives allergic skin inflammation via Th2 immunity and M2 macrophage activation. Clin. Exp. Allergy 2019, 49, 1464–1474. [Google Scholar] [CrossRef]
- Mackel, J.J.; Garth, J.M.; Jones, M.; Ellis, D.A.; Blackburn, J.P.; Yu, Z.; Matalon, S.; Curtiss, M.; Lund, F.E.; Hastie, A.T.; et al. Chitinase 3-like-1 protects airway function despite promoting type 2 inflammation during fungal-associated allergic airway inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, L615–L626. [Google Scholar] [CrossRef]
- Yu, J.E.; Jeon, S.H.; Kim, M.J.; Kim, D.H.; Koo, J.K.; Kim, T.H.; Kim, B.; Yoon, J.Y.; Lim, Y.S.; Park, S.R.; et al. Anti-chitinase-3-like 1 antibody attenuated atopic dermatitis-like skin inflammation through inhibition of STAT3-dependent CXCL8 expression. Br. J. Pharmacol. 2024, 181, 3232–3245. [Google Scholar] [CrossRef]
- Zhu, X.; Dai, X.; Chen, W.; Li, Y.; Liu, Y.; Shan, C.; Wang, J.; Meng, J. Chi3l1 Knockout Mitigates Chronic Itch and Cutaneous Inflammation in Mice. J. Investig. Dermatol. 2025, 145, 975–979.e974. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, M.; Tibullo, D.; Saccone, S.; Distefano, G.; Basile, M.S.; Di Raimondo, F.; Malaguarnera, L. CHI3L1 nuclear localization in monocyte derived dendritic cells. Immunobiology 2016, 221, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Do-Umehara, H.C.; Chen, C.; Zhang, Q.; Schleimer, R.P.; Budinger, G.R.S.; Liu, J. Suppression of Allergic Asthma by Loss of Function of Miz1-mediated Th1 Skewing. Am. J. Respir. Cell Mol. Biol. 2022, 67, 346–359. [Google Scholar] [CrossRef] [PubMed]
- Kaliński, P.; Schuitemaker, J.H.; Hilkens, C.M.; Kapsenberg, M.L. Prostaglandin E2 induces the final maturation of IL-12-deficient CD1a+CD83+ dendritic cells: The levels of IL-12 are determined during the final dendritic cell maturation and are resistant to further modulation. J. Immunol. 1998, 161, 2804–2809. [Google Scholar] [CrossRef]
- Cuenca-Escalona, J.; Flórez-Grau, G.; van den Dries, K.; Cambi, A.; de Vries, I.J.M. PGE2-EP4 signaling steers cDC2 maturation toward the induction of suppressive T-cell responses. Eur. J. Immunol. 2024, 54, e2350770. [Google Scholar] [CrossRef]
- Kaliński, P.; Hilkens, C.M.; Snijders, A.; Snijdewint, F.G.; Kapsenberg, M.L. IL-12-deficient dendritic cells, generated in the presence of prostaglandin E2, promote type 2 cytokine production in maturing human naive T helper cells. J. Immunol. 1997, 159, 28–35. [Google Scholar] [CrossRef]
- Jarjour, N.N.; Bradstreet, T.R.; Schwarzkopf, E.A.; Cook, M.E.; Lai, C.-W.; Huang, S.C.-C.; Taneja, R.; Stappenbeck, T.S.; Van Dyken, S.J.; Urban, J.F., Jr.; et al. BHLHE40 Promotes TH2 Cell–Mediated Antihelminth Immunity and Reveals Cooperative CSF2RB Family Cytokines. J. Immunol. 2020, 204, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Kaisar, M.M.M.; Ritter, M.; Del Fresno, C.; Jónasdóttir, H.S.; van der Ham, A.J.; Pelgrom, L.R.; Schramm, G.; Layland, L.E.; Sancho, D.; Prazeres da Costa, C.; et al. Dectin-1/2-induced autocrine PGE2 signaling licenses dendritic cells to prime Th2 responses. PLoS Biol. 2018, 16, e2005504. [Google Scholar] [CrossRef]
- Wanzel, M.; Kleine-Kohlbrecher, D.; Herold, S.; Hock, A.; Berns, K.; Park, J.; Hemmings, B.; Eilers, M. Akt and 14-3-3eta regulate Miz1 to control cell-cycle arrest after DNA damage. Nat. Cell Biol. 2005, 7, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Peukert, K.; Staller, P.; Schneider, A.; Carmichael, G.; Hänel, F.; Eilers, M. An alternative pathway for gene regulation by Myc. Embo J. 1997, 16, 5672–5686. [Google Scholar] [CrossRef] [PubMed]
- van de Laar, L.; Coffer, P.J.; Woltman, A.M. Regulation of dendritic cell development by GM-CSF: Molecular control and implications for immune homeostasis and therapy. Blood 2012, 119, 3383–3393. [Google Scholar] [CrossRef]
- Zhan, Y.; Lew, A.M.; Chopin, M. The Pleiotropic Effects of the GM-CSF Rheostat on Myeloid Cell Differentiation and Function: More Than a Numbers Game. Front. Immunol. 2019, 10–2019, 2679. [Google Scholar] [CrossRef]
- Sinha, P.; Clements, V.K.; Fulton, A.M.; Ostrand-Rosenberg, S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007, 67, 4507–4513. [Google Scholar] [CrossRef]
- Daniel, B.; Belk, J.A.; Meier, S.L.; Chen, A.Y.; Sandor, K.; Czimmerer, Z.; Varga, Z.; Bene, K.; Buquicchio, F.A.; Qi, Y.; et al. Macrophage inflammatory and regenerative response periodicity is programmed by cell cycle and chromatin state. Mol. Cell 2023, 83, 121–138.e127. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curtiss, M.L.; Benavides, N.B.; Rosenberg, A.F.; Scharer, C.D.; León, B.; Lund, F.E. CXCR5 Signals Fine-Tune Dendritic Cell Transcription and Regulate TH2 Development. Vaccines 2025, 13, 943. https://doi.org/10.3390/vaccines13090943
Curtiss ML, Benavides NB, Rosenberg AF, Scharer CD, León B, Lund FE. CXCR5 Signals Fine-Tune Dendritic Cell Transcription and Regulate TH2 Development. Vaccines. 2025; 13(9):943. https://doi.org/10.3390/vaccines13090943
Chicago/Turabian StyleCurtiss, Miranda L., Natalia Ballesteros Benavides, Alexander F. Rosenberg, Christopher D. Scharer, Beatriz León, and Frances E. Lund. 2025. "CXCR5 Signals Fine-Tune Dendritic Cell Transcription and Regulate TH2 Development" Vaccines 13, no. 9: 943. https://doi.org/10.3390/vaccines13090943
APA StyleCurtiss, M. L., Benavides, N. B., Rosenberg, A. F., Scharer, C. D., León, B., & Lund, F. E. (2025). CXCR5 Signals Fine-Tune Dendritic Cell Transcription and Regulate TH2 Development. Vaccines, 13(9), 943. https://doi.org/10.3390/vaccines13090943





