Geographical Differences in SARS-CoV-2 Antibody Response Dynamics and Neutralisation Profiles to Mild COVID-19: Lessons from a UK–Uganda Comparison
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Study Design
2.3. Validated Enzyme-Linked Immunosorbent Assay (ELISA)-Based Quantification of SARS-CoV-2-Specific IgG, IgM, and IgA Antibodies
2.4. Assessment of Neutralising Antibody Titres Using SARS-CoV-2 Pseudotyped Viruses
2.5. Statistical Methods
3. Results
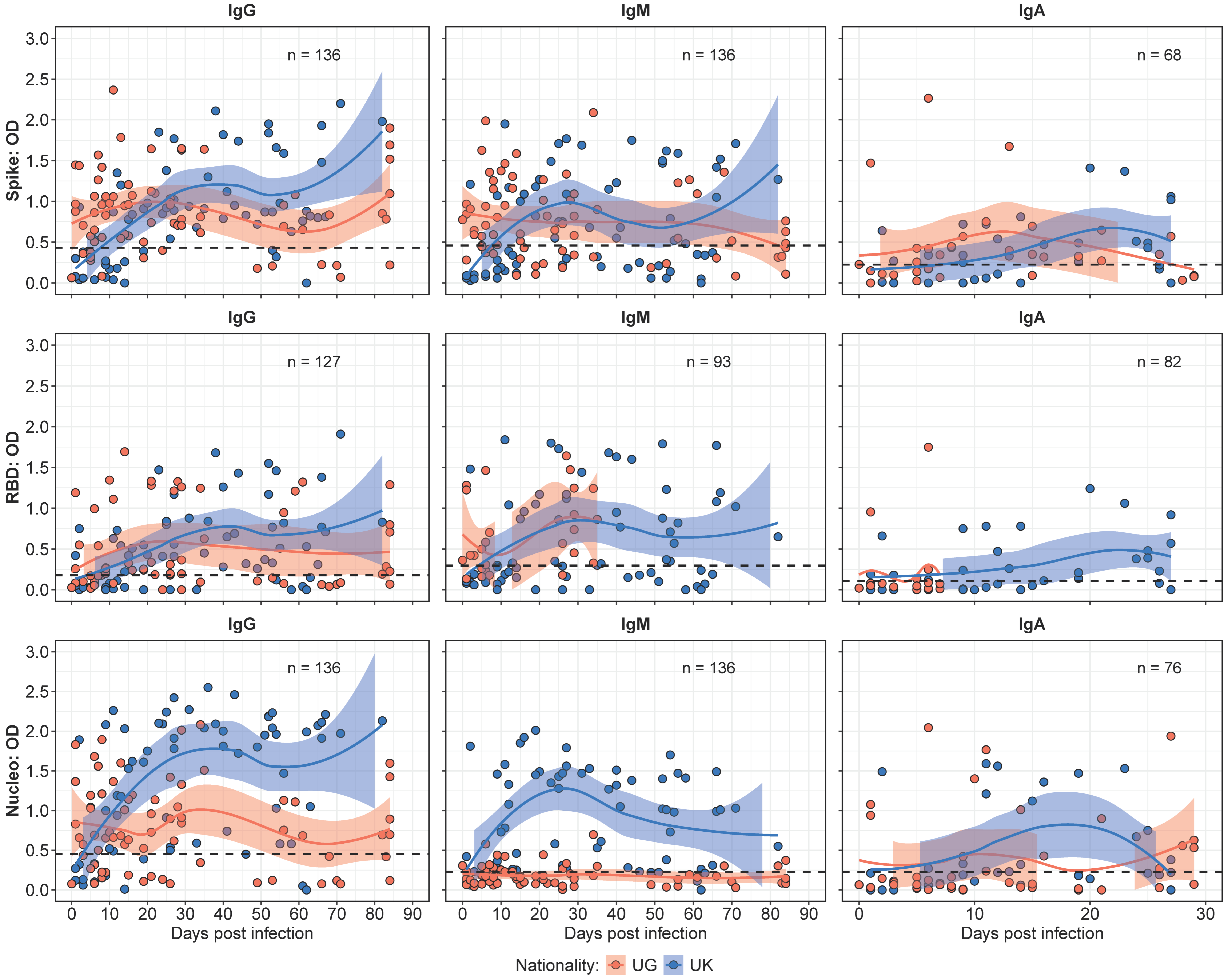
3.1. Antibody Kinetics Revealed Divergent Patterns Between Ugandan and UK Cohorts
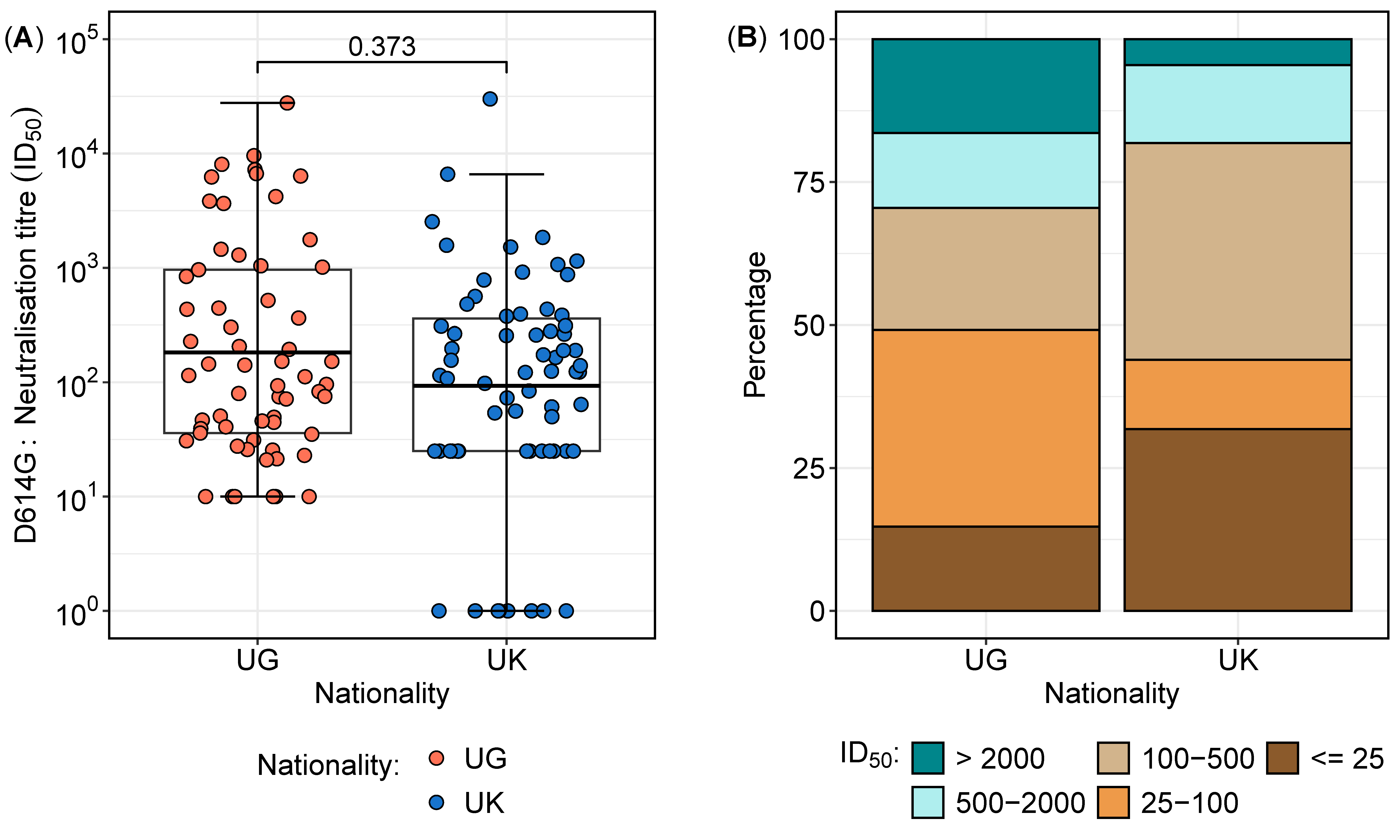
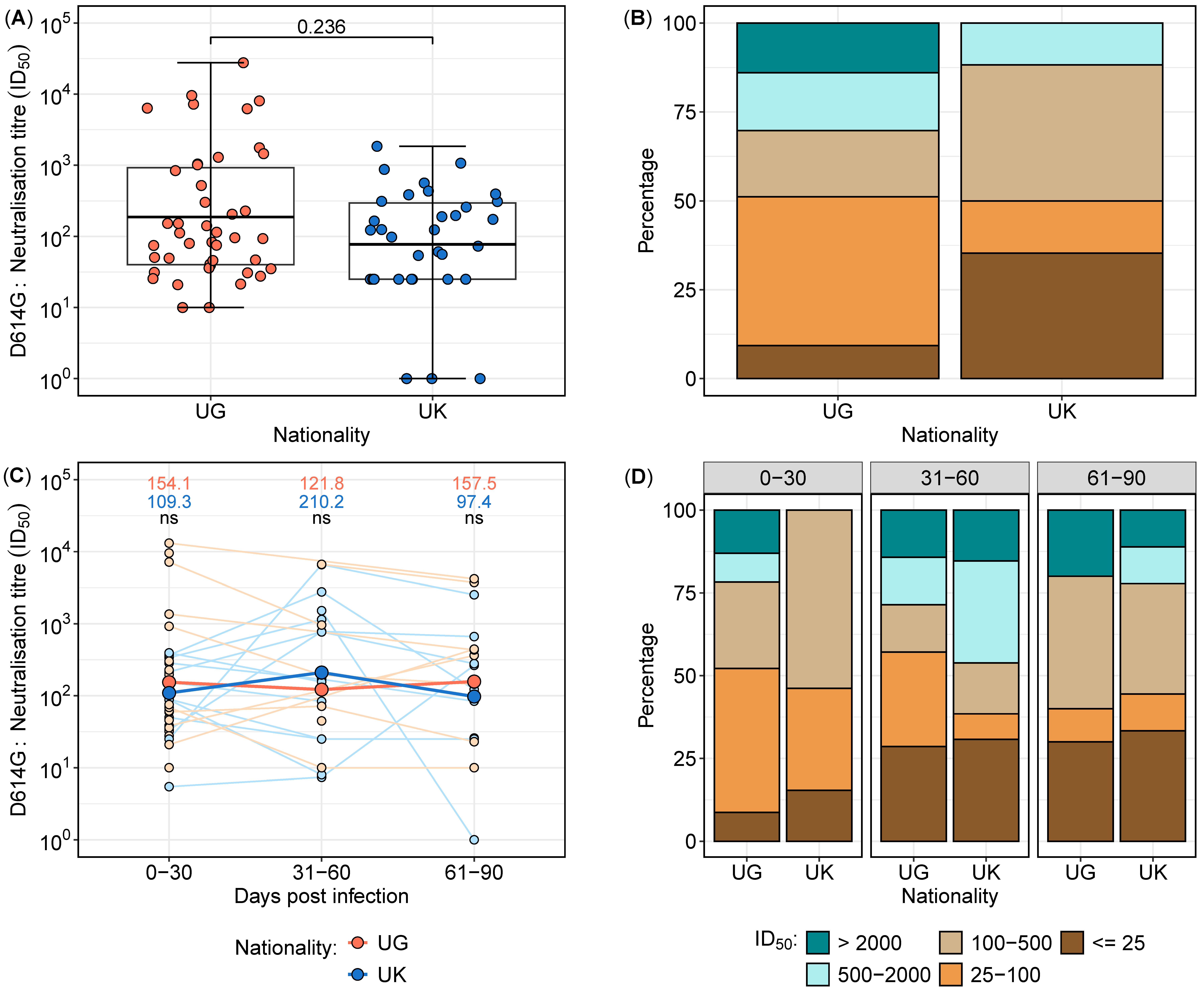
3.2. Neutralisation Titres Against the D614G Variant Reveal Regional Differences in Sensitivity and Resistance
3.3. Correlation of Neutralisation Titres with S, RBD, and N-IgG Responses Differed Between Nationalities
4. Discussion
Study Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO: World Health Organization. Clinical Management of COVID-19: Interim Guidance, 27 May 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Feng, Y.; Ling, Y.; Bai, T.; Xie, Y.; Huang, J.; Li, J.; Xiong, W.; Yang, D.; Chen, R.; Lu, F.; et al. COVID-19 with Different Severities: A Multicenter Study of Clinical Features. Am. J. Respir. Crit. Care Med. 2020, 201, 1380–1388. [Google Scholar] [CrossRef]
- Çelik, I.; Öztürk, R. From asymptomatic to critical illness: Decoding various clinical stages of COVID-19. Turk. J. Med. Sci. 2021, 51, 3284–3300. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, J.; Wang, D.; Wang, D.; Guan, W.; Qin, J.; Xu, X.; Fang, J.; Fu, B.; Zheng, X.; et al. Profiling of the immune repertoire in COVID-19 patients with mild, severe, convalescent, or retesting-positive status. J. Autoimmun. 2021, 118, 102596. [Google Scholar] [CrossRef]
- Maecker, H.T. Immune profiling of COVID-19: Preliminary findings and implications for the pandemic. J. Immunother. Cancer 2021, 9, e002550. [Google Scholar] [CrossRef] [PubMed]
- Roslund, M.I.; Nurminen, N.; Oikarinen, S.; Puhakka, R.; Grönroos, M.; Puustinen, L.; Kummola, L.; Parajuli, A.; Cinek, O.; Laitinen, O.H.; et al. Skin exposure to soil microbiota elicits changes in cell-mediated immunity to pneumococcal vaccine. Sci. Rep. 2024, 14, 18573. [Google Scholar] [CrossRef] [PubMed]
- van Riet, E.; Retra, K.; Adegnika, A.A.; Jol-van der Zijde, C.M.; Uh, H.W.; Lell, B.; Issifou, S.; Kremsner, P.G.; Yazdanbakhsh, M.; van Tol, M.J.; et al. Cellular and humoral responses to tetanus vaccination in Gabonese children. Vaccine 2008, 26, 3690–3695. [Google Scholar] [CrossRef]
- Huda, M.N.; Lewis, Z.; Kalanetra, K.M.; Rashid, M.; Ahmad, S.M.; Raqib, R.; Qadri, F.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Stool microbiota and vaccine responses of infants. Pediatrics 2014, 134, e362–e372. [Google Scholar] [CrossRef]
- Harris, V.C.; Armah, G.; Fuentes, S.; Korpela, K.E.; Parashar, U.; Victor, J.C.; Tate, J.; de Weerth, C.; Giaquinto, C.; Wiersinga, W.J.; et al. Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J. Infect. Dis. 2017, 215, 34–41. [Google Scholar] [CrossRef]
- Brummelman, J.; Suárez-Hernández, S.; de Rond, L.; Bogaard-van Maurik, M.; Molenaar, P.; van Wijlen, E.; Oomen, D.; Beckers, L.; Rots, N.Y.; van Beek, J.; et al. Distinct T cell responsiveness to different COVID-19 vaccines and cross-reactivity to SARS-CoV-2 variants with age and CMV status. Front. Immunol. 2024, 15, 1392477. [Google Scholar] [CrossRef]
- Mario Martin, B.; Cadavid Restrepo, A.; Mayfield, H.J.; Then Paulino, C.; De St Aubin, M.; Duke, W.; Jarolim, P.; Zielinski Gutiérrez, E.; Skewes Ramm, R.; Dumas, D.; et al. Using Regional Sero-Epidemiology SARS-CoV-2 Anti-S Antibodies in the Dominican Republic to Inform Targeted Public Health Response. Trop. Med. Infect. Dis. 2023, 8, 493. [Google Scholar] [CrossRef]
- Fernandes, M.; Vasconcelos, G.S.; de Melo, A.C.L.; Matsui, T.C.; Caetano, L.F.; de Carvalho Araújo, F.M.; Fonseca, M.H.G. Influence of age, gender, previous SARS-CoV-2 infection, and pre-existing diseases in antibody response after COVID-19 vaccination: A review. Mol. Immunol. 2023, 156, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Weskamm, L.M.; Tarnow, P.; Harms, C.; Huchon, M.; Raadsen, M.P.; Friedrich, M.; Rübenacker, L.; Grüttner, C.; Garcia, M.G.; Koch, T.; et al. Dissecting humoral immune responses to an MVA-vectored MERS-CoV vaccine in humans using a systems serology approach. iScience 2024, 27, 110470. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Du, L. SARS-CoV-2 spike protein: A key target for eliciting persistent neutralizing antibodies. Signal Transduct. Target. Ther. 2021, 6, 95. [Google Scholar] [CrossRef]
- Serwanga, J.; Ankunda, V.; Sembera, J.; Kato, L.; Oluka, G.K.; Baine, C.; Odoch, G.; Kayiwa, J.; Auma, B.O.; Jjuuko, M.; et al. Rapid, early, and potent Spike-directed IgG, IgM, and IgA distinguish asymptomatic from mildly symptomatic COVID-19 in Uganda, with IgG persisting for 28 months. Front. Immunol. 2023, 14, 1152522. [Google Scholar] [CrossRef] [PubMed]
- Oluka, G.K.; Sembera, J.; Katende, J.S.; Ankunda, V.; Kato, L.; Kurshan, A.; Graham, C.; Seow, J.; Doores, K.J.; Malim, M.H.; et al. Long-Term Immune Consequences of Initial SARS-CoV-2 A.23.1 Exposure: A Longitudinal Study of Antibody Responses and Cross-Neutralisation in a Ugandan Cohort. Vaccines 2025, 13, 143. [Google Scholar] [CrossRef]
- Bugembe, D.L.; Phan, M.V.T.; Ssewanyana, I.; Semanda, P.; Nansumba, H.; Dhaala, B.; Nabadda, S.; O’Toole, Á.N.; Rambaut, A.; Kaleebu, P.; et al. Emergence and spread of a SARS-CoV-2 lineage A variant (A.23.1) with altered spike protein in Uganda. Nat. Microbiol. 2021, 6, 1094–1101. [Google Scholar] [CrossRef]
- Bbosa, N.; Ssemwanga, D.; Namagembe, H.; Kiiza, R.; Kiconco, J.; Kayiwa, J.; Lutalo, T.; Lutwama, J.; Ssekagiri, A.; Ssewanyana, I.; et al. Rapid Replacement of SARS-CoV-2 Variants by Delta and Subsequent Arrival of Omicron, Uganda, 2021. Emerg. Infect. Dis. 2022, 28, 1021–1025. [Google Scholar] [CrossRef]
- Dupont, L.; Snell, L.B.; Graham, C.; Seow, J.; Merrick, B.; Lechmere, T.; Maguire, T.J.A.; Hallett, S.R.; Pickering, S.; Charalampous, T.; et al. Neutralizing antibody activity in convalescent sera from infection in humans with SARS-CoV-2 and variants of concern. Nat. Microbiol. 2021, 6, 1433–1442. [Google Scholar] [CrossRef]
- Rambaut, A.; Loman, N.; Pybus, O.; Barclay, W.; Barrett, J.; Carabelli, A.; Connor, T.; Peacock, T.; Robertson, D.L.; Volz, E. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. 2020. Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 4 January 2025).
- Cantoni, D.; Mayora-Neto, M.; Nadesalingam, A.; Wells, D.A.; Carnell, G.W.; Ohlendorf, L.; Ferrari, M.; Palmer, P.; Chan, A.C.Y.; Smith, P.; et al. Neutralisation Hierarchy of SARS-CoV-2 Variants of Concern Using Standardised, Quantitative Neutralisation Assays Reveals a Correlation with Disease Severity; Towards Deciphering Protective Antibody Thresholds. Front. Immunol. 2022, 13, 773982. [Google Scholar] [CrossRef]
- Oluka, G.K.; Namubiru, P.; Kato, L.; Ankunda, V.; Gombe, B.; Cotten, M.; Team, C.-I.; Musenero, M.; Kaleebu, P.; Fox, J.; et al. Optimisation and Validation of a conventional ELISA and cut-offs for detecting and quantifying anti-SARS-CoV-2 Spike, RBD, and Nucleoprotein IgG, IgM, and IgA antibodies in Uganda. Front. Immunol. 2023, 14, 1113194. [Google Scholar] [CrossRef]
- Pickering, S.; Betancor, G.; Galao, R.P.; Merrick, B.; Signell, A.W.; Wilson, H.D.; Kia Ik, M.T.; Seow, J.; Graham, C.; Acors, S.; et al. Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog. 2020, 16, e1008817. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, X.E. Construction and applications of SARS-CoV-2 pseudoviruses: A mini review. Int. J. Biol. Sci. 2021, 17, 1574–1580. [Google Scholar] [CrossRef] [PubMed]
- Egwang, T.G.; Owalla, T.J.; Okurut, E.; Apungia, G.; Fox, A.; De Carlo, C.; Powell, R.L. Differential pre-pandemic breast milk IgA reactivity against SARS-CoV-2 and circulating human coronaviruses in Ugandan and American mothers. Int. J. Infect. Dis. 2021, 112, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Demers-Mathieu, V.; DaPra, C.; Mathijssen, G.B.; Medo, E. Previous viral symptoms and individual mothers influenced the leveled duration of human milk antibodies cross-reactive to S1 and S2 subunits from SARS-CoV-2, HCoV-229E, and HCoV-OC43. J. Perinatol. 2021, 41, 952–960. [Google Scholar] [CrossRef] [PubMed]
- Tso, F.Y.; Lidenge, S.J.; Peña, P.B.; Clegg, A.A.; Ngowi, J.R.; Mwaiselage, J.; Ngalamika, O.; Julius, P.; West, J.T.; Wood, C. High prevalence of pre-existing serological cross-reactivity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in sub-Saharan Africa. Int. J. Infect. Dis. 2021, 102, 577–583. [Google Scholar] [CrossRef]
- Smith, T.P.; Mishra, S.; Dorigatti, I.; Dixit, M.K.; Tristem, M.; Pearse, W.D. Differential responses of SARS-CoV-2 variants to environmental drivers during their selective sweeps. Sci. Rep. 2024, 14, 13326. [Google Scholar] [CrossRef]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B. SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. Lancet 2020, 396, 1595–1606. [Google Scholar] [CrossRef]
- Lindeboom, R.G.H.; Worlock, K.B.; Dratva, L.M.; Yoshida, M.; Scobie, D.; Wagstaffe, H.R.; Richardson, L.; Wilbrey-Clark, A.; Barnes, J.L.; Kretschmer, L.; et al. Human SARS-CoV-2 challenge uncovers local and systemic response dynamics. Nature 2024, 631, 189–198. [Google Scholar] [CrossRef]
- Ahmed, M.I.M.; Einhauser, S.; Peiter, C.; Senninger, A.; Baranov, O.; Eser, T.M.; Huth, M.; Olbrich, L.; Castelletti, N.; Rubio-Acero, R.; et al. Evolution of protective SARS-CoV-2-specific B and T cell responses upon vaccination and Omicron breakthrough infection. iScience 2024, 27, 110138. [Google Scholar] [CrossRef]
- Wagstaffe, H.R.; Thwaites, R.S.; Reynaldi, A.; Sidhu, J.K.; McKendry, R.; Ascough, S.; Papargyris, L.; Collins, A.M.; Xu, J.; Lemm, N.M.; et al. Mucosal and systemic immune correlates of viral control after SARS-CoV-2 infection challenge in seronegative adults. Sci. Immunol. 2024, 9, eadj9285. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, M.; Campillo, N.E.; Canelles, M. COVID-19 Vaccine Race: Analysis of Age-Dependent Immune Responses against SARS-CoV-2 Indicates that more than Just One Strategy May Be Needed. Curr. Med. Chem. 2021, 28, 3964–3979. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Total n (col %) | UG n (col %) | UK n (col %) | p-Value | |
|---|---|---|---|---|---|
| Total | 43 (100%) | ||||
| UG | 29 (67.4) | ||||
| UK | 14 (32.6) | ||||
| Sex | 0.384 | ||||
| Female | 16 (37.2) | 9 (31.0) | 7 (50.0) | ||
| Male | 27 (62.8) | 20 (69.0) | 7 (50.0) | ||
| Age group § | 0.0811 | ||||
| 20–34 | 19 (51.4) | 16 (55.2) | 3 (37.5) | ||
| 35–50 | 13 (35.1) | 11 (37.9) | 2 (25.0) | ||
| 50+ | 5 (13.5) | 2 (6.9) | 3 (37.5) | ||
| Admission symptoms * | 0.0317 | ||||
| No | 28 (77.8) | 14 (63.6) | 14 (100.0) | ||
| Yes | 8 (22.2) | 8 (36.4) | 0 (0.0) | ||
| Neutralisation Range | Total n (%) | UG n (%) | UK n (%) |
|---|---|---|---|
| Total | 127 (100) | 61 (48.0) | 66 (52.0) |
| >2000 | 13 (10.2) | 10 (16.9) | 3 (4.5) |
| 500–2000 | 17 (13.4) | 8 (13.1) | 9 (13.6) |
| 100–500 | 38 (29.9) | 13 (21.3) | 25 (37.9) |
| 25–100 | 29 (22.8) | 21 (34.4) | 8 (12.1) |
| ≤25 | 30 (23.6) | 9 (14.8) | 21 (31.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, L.; Sembera, J.; Oluka, G.K.; Katende, J.S.; Bemanzi, J.; Ankunda, V.; Ejou, P.; Kurshan, A.; Graham, C.; Seow, J.; et al. Geographical Differences in SARS-CoV-2 Antibody Response Dynamics and Neutralisation Profiles to Mild COVID-19: Lessons from a UK–Uganda Comparison. Vaccines 2025, 13, 336. https://doi.org/10.3390/vaccines13040336
Kato L, Sembera J, Oluka GK, Katende JS, Bemanzi J, Ankunda V, Ejou P, Kurshan A, Graham C, Seow J, et al. Geographical Differences in SARS-CoV-2 Antibody Response Dynamics and Neutralisation Profiles to Mild COVID-19: Lessons from a UK–Uganda Comparison. Vaccines. 2025; 13(4):336. https://doi.org/10.3390/vaccines13040336
Chicago/Turabian StyleKato, Laban, Jackson Sembera, Gerald Kevin Oluka, Joseph Ssebwana Katende, Juliana Bemanzi, Violet Ankunda, Peter Ejou, Ashwini Kurshan, Carl Graham, Jeffrey Seow, and et al. 2025. "Geographical Differences in SARS-CoV-2 Antibody Response Dynamics and Neutralisation Profiles to Mild COVID-19: Lessons from a UK–Uganda Comparison" Vaccines 13, no. 4: 336. https://doi.org/10.3390/vaccines13040336
APA StyleKato, L., Sembera, J., Oluka, G. K., Katende, J. S., Bemanzi, J., Ankunda, V., Ejou, P., Kurshan, A., Graham, C., Seow, J., Doores, K. J., Malim, M. H., Fox, J. M., Kaleebu, P., & Serwanga, J. (2025). Geographical Differences in SARS-CoV-2 Antibody Response Dynamics and Neutralisation Profiles to Mild COVID-19: Lessons from a UK–Uganda Comparison. Vaccines, 13(4), 336. https://doi.org/10.3390/vaccines13040336







