Stabilizing Prefusion SARS-CoV-2 Spike by Destabilizing the Postfusion Conformation
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Mutants and Designing Constructs
2.2. Recombinant Protein Expression and Purification
2.3. Estimation of Protein Yields by Capture ELISA
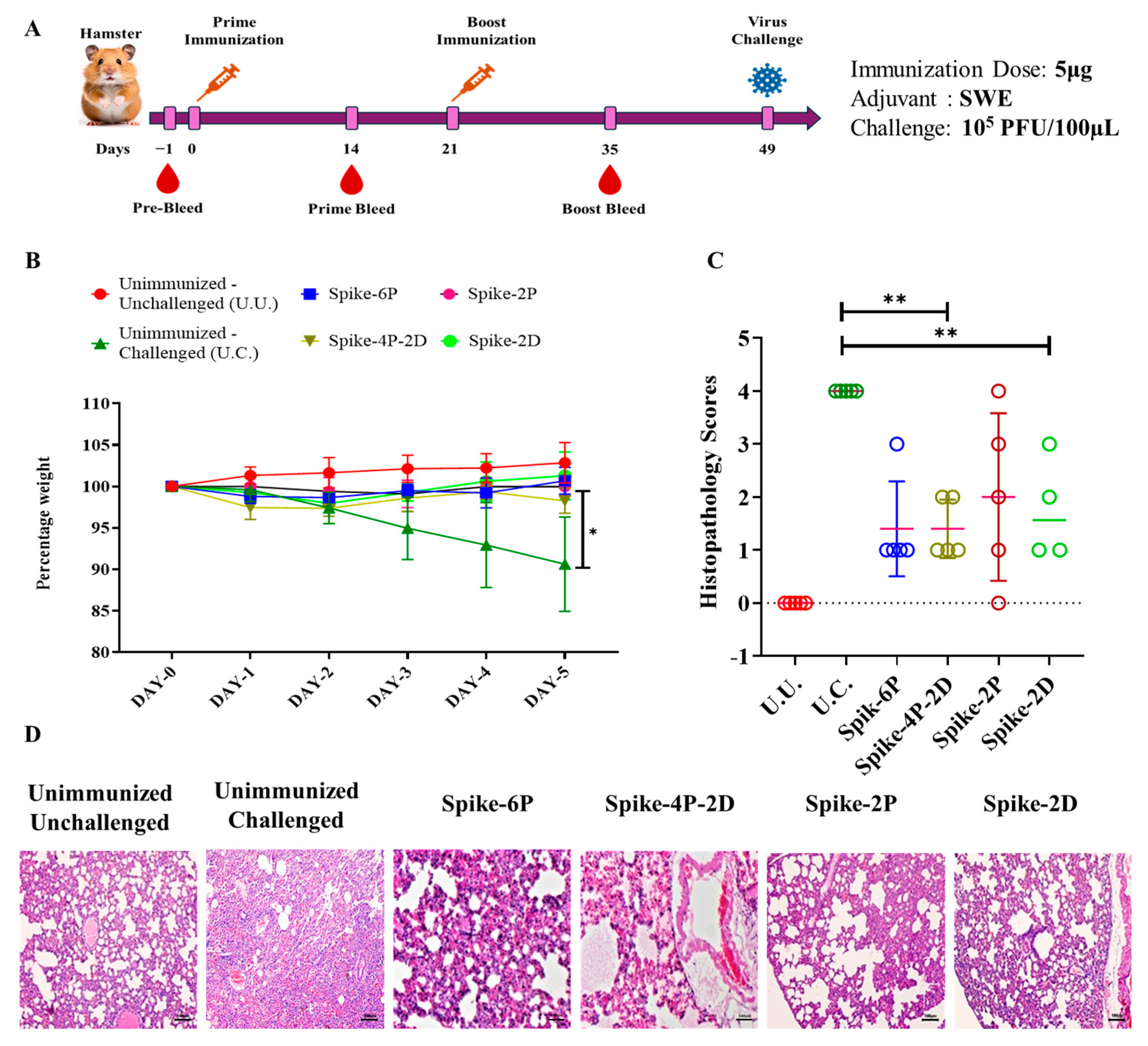
2.4. Immunization of Hamsters
2.5. ELISA for Endpoint Titer Determination
2.6. Pseudoviral Neutralization Assay
2.7. Challenge Studies of Hamsters
2.8. Histopathological Examinations
2.9. Statistical Analysis
3. Results
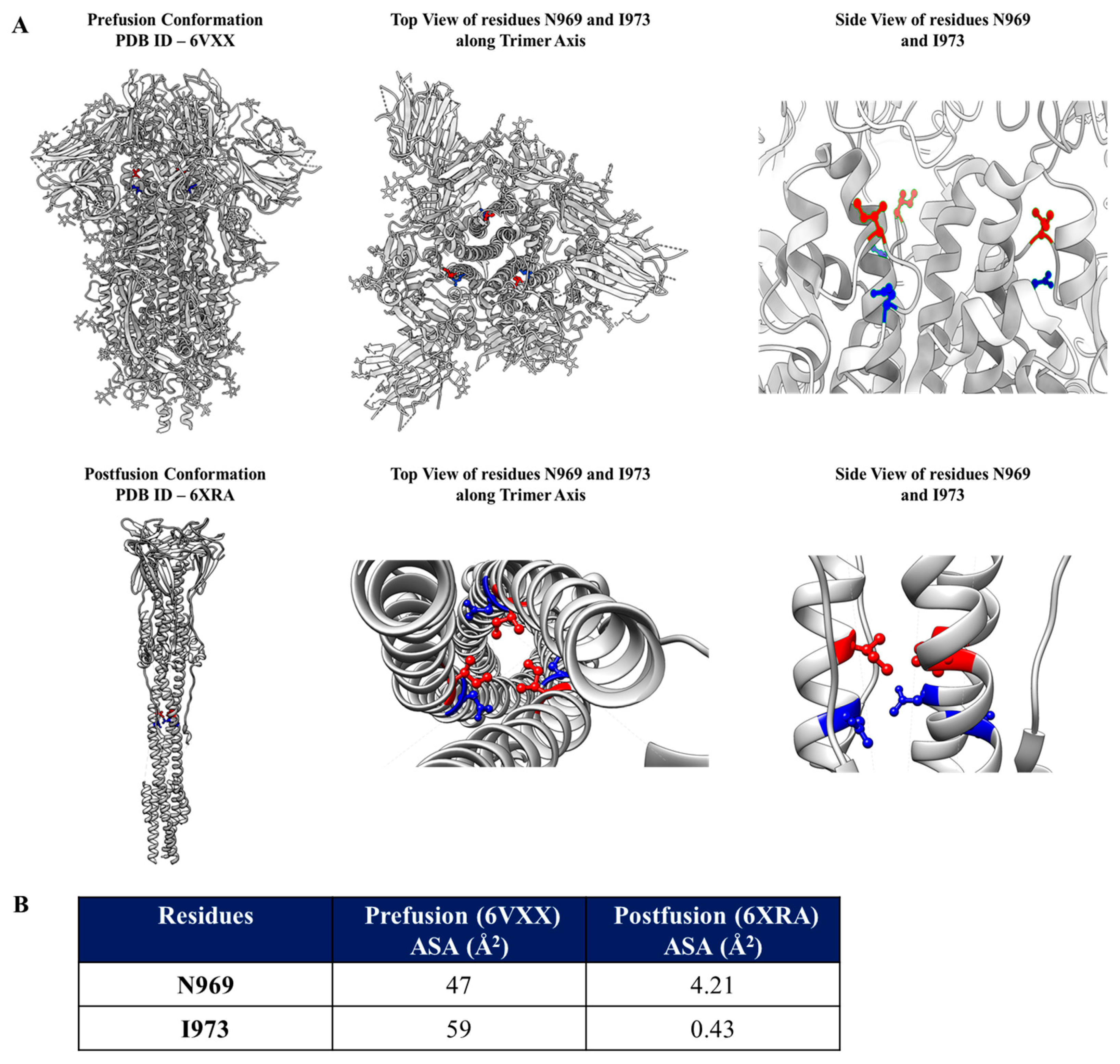
3.1. Identification of Mutations to Destabilize the Prefusion Conformation of the Full-Length Spike Protein
3.2. Introduction of the N969D and I973D Mutations into the Ectodomain of the WT Spike Increases Expression and Obviates the Need for the K986P and V987P (2P) Mutations
3.3. Spike-4P-2D Elicited a Potent Neutralizing Antibody Response in Hamsters
3.4. All Spike Derivatives Confer Protection Against a Heterologous SARS-CoV-2 (B.1.351) Live Virus Challenge in Hamsters
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RBD | Receptor-Binding Domain |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| SWE Adjuvant | Squalene-in-Water Emulsion Adjuvant |
References
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 Neutralizing Antibody Structures Inform Therapeutic Strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Malladi, S.K.; Singh, R.; Pandey, S.; Gayathri, S.; Kanjo, K.; Ahmed, S.; Khan, M.S.; Kalita, P.; Girish, N.; Upadhyaya, A.; et al. Design of a Highly Thermotolerant, Immunogenic SARS-CoV-2 Spike Fragment. J. Biol. Chem. 2021, 296, 100025. [Google Scholar] [CrossRef]
- Ahmed, S.; Khan, M.S.; Gayathri, S.; Singh, R.; Kumar, S.; Patel, U.R.; Malladi, S.K.; Rajmani, R.S.; van Vuren, P.J.; Riddell, S.; et al. A Stabilized, Monomeric, Receptor Binding Domain Elicits High-Titer Neutralizing Antibodies Against All SARS-CoV-2 Variants of Concern. Front. Immunol. 2021, 12, 765211. [Google Scholar] [CrossRef]
- Malladi, S.K.; Patel, U.R.; Rajmani, R.S.; Singh, R.; Pandey, S.; Kumar, S.; Khaleeq, S.; Van Vuren, P.J.; Riddell, S.; Goldie, S.; et al. Immunogenicity and Protective Efficacy of a Highly Thermotolerant, Trimeric SARS-CoV-2 Receptor Binding Domain Derivative. ACS Infect. Dis. 2021, 7, 2546–2564. [Google Scholar] [CrossRef]
- Mittal, N.; Kumar, S.; Rajmani, R.S.; Singh, R.; Lemoine, C.; Jakob, V.; Bj, S.; Jagannath, N.; Bhat, M.; Chakraborty, D.; et al. Enhanced Protective Efficacy of a Thermostable RBD-S2 Vaccine Formulation against SARS-CoV-2 and Its Variants. NPJ Vaccines 2023, 8, 161. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.W.; Sahi, V.; Figueroa, A.; et al. Potent Neutralizing Antibodies against Multiple Epitopes on SARS-CoV-2 Spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Gentles, L.E.; Crawford, K.H.D.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Antibodies Elicited by MRNA-1273 Vaccination Bind More Broadly to the Receptor Binding Domain than Do Those from SARS-CoV-2 Infection. Sci. Transl. Med. 2021, 13. [Google Scholar] [CrossRef]
- Barnes, C.O.; West, A.P.; Huey-Tubman, K.E.; Hoffmann, M.A.G.; Sharaf, N.G.; Hoffman, P.R.; Koranda, N.; Gristick, H.B.; Gaebler, C.; Muecksch, F.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842.e16. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 MRNA Vaccine Design Enabled by Prototype Pathogen Preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and Efficacy of the ChAdOx1 NCoV-19 Vaccine (AZD1222) Against SARS-CoV-2: An Interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef]
- Falsey, A.R.; Sobieszczyk, M.E.; Hirsch, I.; Sproule, S.; Robb, M.L.; Corey, L.; Neuzil, K.M.; Hahn, W.; Hunt, J.; Mulligan, M.J.; et al. Phase 3 Safety and Efficacy of AZD1222 (ChAdOx1 NCoV-19) COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 2348–2360. [Google Scholar] [CrossRef]
- Barrett, J.R.; Belij-Rammerstorfer, S.; Dold, C.; Ewer, K.J.; Folegatti, P.M.; Gilbride, C.; Halkerston, R.; Hill, J.; Jenkin, D.; Stockdale, L.; et al. Phase 1/2 Trial of SARS-CoV-2 Vaccine ChAdOx1 NCoV-19 with a Booster Dose Induces Multifunctional Antibody Responses. Nat. Med. 2020, 27, 279–288. [Google Scholar] [CrossRef]
- Watanabe, Y.; Mendonça, L.; Allen, E.R.; Howe, A.; Lee, M.; Allen, J.D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; et al. Native-like SARS-CoV-2 Spike Glycoprotein Expressed by ChAdOx1 NCoV-19/AZD1222 Vaccine. ACS Cent. Sci. 2021, 7, 594–602. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-Based Design of Prefusion-Stabilized SARS-CoV-2 Spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Pedenko, B.; Sulbaran, G.; Guilligay, D.; Effantin, G.; Weissenhorn, W. SARS-CoV-2 S Glycoprotein Stabilization Strategies. Viruses 2023, 15, 558. [Google Scholar] [CrossRef]
- Ellis, D.; Brunette, N.; Crawford, K.H.D.; Walls, A.C.; Pham, M.N.; Chen, C.; Herpoldt, K.L.; Fiala, B.; Murphy, M.; Pettie, D.; et al. Stabilization of the SARS-CoV-2 Spike Receptor-Binding Domain Using Deep Mutational Scanning and Structure-Based Design. Front. Immunol. 2021, 12, 710263. [Google Scholar] [CrossRef]
- Lee, J.; Stewart, C.; Schäfer, A.; Leaf, E.M.; Park, Y.J.; Asarnow, D.; Powers, J.M.; Treichel, C.; Sprouse, K.R.; Corti, D.; et al. A Broadly Generalizable Stabilization Strategy for Sarbecovirus Fusion Machinery Vaccines. Nat. Commun. 2024, 15, 5496. [Google Scholar] [CrossRef]
- Rutten, L.; Swart, M.; Koornneef, A.; Bouchier, P.; Blokland, S.; Sadi, A.; Juraszek, J.; Vijayan, A.; Schmit-Tillemans, S.; Verspuij, J.; et al. Impact of SARS-CoV-2 Spike Stability and RBD Exposure on Antigenicity and Immunogenicity. Sci. Rep. 2024, 14, 5735. [Google Scholar] [CrossRef]
- Tan, T.J.C.; Mou, Z.; Lei, R.; Ouyang, W.O.; Yuan, M.; Song, G.; Andrabi, R.; Wilson, I.A.; Kieffer, C.; Dai, X.; et al. High-Throughput Identification of Prefusion-Stabilizing Mutations in SARS-CoV-2 Spike. Nat. Commun. 2023, 14, 2003. [Google Scholar] [CrossRef]
- Olmedillas, E.; Mann, C.J.; Peng, W.; Wang, Y.-T.; Avalos, R.D.; Bedinger, D.; Valentine, K.; Shafee, N.; Schendel, S.L.; Yuan, M.; et al. Structure-Based Design of a Highly Stable, Covalently-Linked SARS-CoV-2 Spike Trimer with Improved Structural Properties and Immunogenicity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Wang, N.; Pallesen, J.; Wrapp, D.; Turner, H.L.; Cottrell, C.A.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Stabilized Coronavirus Spikes Are Resistant to Conformational Changes Induced by Receptor Recognition or Proteolysis. Sci. Rep. 2018, 8, 15701. [Google Scholar] [CrossRef]
- Bowen, J.E.; Park, Y.J.; Stewart, C.; Brown, J.T.; Sharkey, W.K.; Walls, A.C.; Joshi, A.; Sprouse, K.R.; McCallum, M.; Tortorici, M.A.; et al. SARS-CoV-2 Spike Conformation Determines Plasma Neutralizing Activity Elicited by a Wide Panel of Human Vaccines. Sci. Immunol. 2022, 7, eadf1421. [Google Scholar] [CrossRef]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and Structures of a Rationally Designed Prefusion MERS-CoV Spike Antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef]
- Henderson, R.; Edwards, R.J.; Mansouri, K.; Janowska, K.; Stalls, V.; Gobeil, S.M.C.; Kopp, M.; Li, D.; Parks, R.; Hsu, A.L.; et al. Controlling the SARS-CoV-2 Spike Glycoprotein Conformation. Nat. Struct. Mol. Biol. 2020, 27, 925–933. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Xiong, X.; Qu, K.; Ciazynska, K.A.; Hosmillo, M.; Carter, A.P.; Ebrahimi, S.; Ke, Z.; Scheres, S.H.W.; Bergamaschi, L.; Grice, G.L.; et al. A Thermostable, Closed SARS-CoV-2 Spike Protein Trimer. Nat. Struct. Mol. Biol. 2020, 27, 934–941. [Google Scholar] [CrossRef]
- Bong, Y.-S.; Brown, D.; Chung, E.; Ananthaswamy, N.; Chen, R.; Lewoczko, E.; Sabbers, W.; Patterson-Orazem, A.C.; Dorsey, Z.; Zou, Y.; et al. S6P Mutation in Delta and Omicron Variant Spike Protein Significantly Enhances the Efficacy of MRNA COVID-19 Vaccines. Front. Immunol. 2024, 15, 1495561. [Google Scholar] [CrossRef]
- Christensen, D.; Polacek, C.; Sheward, D.J.; Hanke, L.; McInerney, G.; Murrell, B.; Hartmann, K.T.; Jensen, H.E.; Zimmermann, J.; Jungersen, G.; et al. SARS-CoV-2 Spike HexaPro Formulated in Aluminium Hydroxide and Administered in an Accelerated Vaccination Schedule Partially Protects Syrian Hamsters against Viral Challenge despite Low Neutralizing Antibody Responses. Front. Immunol. 2023, 14, 941281. [Google Scholar] [CrossRef]
- Lu, M.; Chamblee, M.; Zhang, Y.; Ye, C.; Dravid, P.; Park, J.G.; Mahesh, K.C.; Trivedi, S.; Murthy, S.; Sharma, H.; et al. SARS-CoV-2 Prefusion Spike Protein Stabilized by Six Rather than Two Prolines Is More Potent for Inducing Antibodies That Neutralize Viral Variants of Concern. Proc. Natl. Acad. Sci. USA 2022, 119, e2110105119. [Google Scholar] [CrossRef]
- Bommakanti, G.; Lu, X.; Citron, M.P.; Najar, T.A.; Heidecker, G.J.; ter Meulen, J.; Varadarajan, R.; Liang, X. Design of Escherichia Coli-Expressed Stalk Domain Immunogens of H1N1 Hemagglutinin That Protect Mice from Lethal Challenge. J. Virol. 2012, 86, 13434–13444. [Google Scholar] [CrossRef] [PubMed]
- Kesavardhana, S.; Varadarajan, R. Stabilizing the Native Trimer of HIV-1 Env by Destabilizing the Heterodimeric Interface of the Gp41 Postfusion Six-Helix Bundle. J. Virol. 2014, 88, 9590–9604. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.J.; Thornton, J.M. “NACCESS” Computer Program, Department of Biochemistry and Molecular Biology, University College London, UK. 1993. Available online: http://www.bioinf.manchester.ac.uk/naccess/ (accessed on 11 May 2020).
- Lee, B.; Richards, F.M. The Interpretation of Protein Structures: Estimation of Static Accessibility. J. Mol. Biol. 1971, 55, 379–400. [Google Scholar] [CrossRef]
- Mitchison, D.A.; Wallace, J.G.; Bhatia, A.L.; Selkon, J.B.; Subbaiah, T.V.; Lancaster, M.C. A Comparison of the Virulence in Guinea-Pigs of South Indian and British Tubercle Bacilli. Tubercle 1960, 41, 1–22. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, D.; Singh, R.; Rajmani, R.S.; Kumar, S.; Ringe, R.P.; Varadarajan, R. Stabilizing Prefusion SARS-CoV-2 Spike by Destabilizing the Postfusion Conformation. Vaccines 2025, 13, 315. https://doi.org/10.3390/vaccines13030315
Chakraborty D, Singh R, Rajmani RS, Kumar S, Ringe RP, Varadarajan R. Stabilizing Prefusion SARS-CoV-2 Spike by Destabilizing the Postfusion Conformation. Vaccines. 2025; 13(3):315. https://doi.org/10.3390/vaccines13030315
Chicago/Turabian StyleChakraborty, Debajyoti, Randhir Singh, Raju S. Rajmani, Sahil Kumar, Rajesh P. Ringe, and Raghavan Varadarajan. 2025. "Stabilizing Prefusion SARS-CoV-2 Spike by Destabilizing the Postfusion Conformation" Vaccines 13, no. 3: 315. https://doi.org/10.3390/vaccines13030315
APA StyleChakraborty, D., Singh, R., Rajmani, R. S., Kumar, S., Ringe, R. P., & Varadarajan, R. (2025). Stabilizing Prefusion SARS-CoV-2 Spike by Destabilizing the Postfusion Conformation. Vaccines, 13(3), 315. https://doi.org/10.3390/vaccines13030315






