Protective Antimicrobial Effect of the Potential Vaccine Created on the Basis of the Structure of the IgA1 Protease from Neisseria meningitidis
Abstract
1. Introduction
2. Materials and Methods
2.1. Calculations
2.1.1. Data on the Primary Structure of IgA1 Protease
2.1.2. Predicting the Location of T-Cell Epitopes
- Via an online server (accessed on 3 April 2023) for the amino acid sequences of IgA1 protease A28–P1004, its fragments: E135–H328, W329–H622, H835–P1004, W140–K833, W329–P1004 and potential fusion structures (W140–H328)-(W412–D604)-(Y866–P1004), (W140–Q299)-(Y866–P1004);
- Using the downloadable version (version 3.0.2) of the program—separately for each epitope located in the A28–P1004 chain.
2.1.3. Modeling Three-Dimensional Structures
2.1.4. Prediction of the Location of B-Cell Epitopes
2.2. Experiment
2.2.1. Production of Recombinant Proteins
2.2.2. Purification of Recombinant Proteins
2.2.3. Assessment of Immunogenic and Protective Activity of Truncated Recombinant Proteins
3. Results
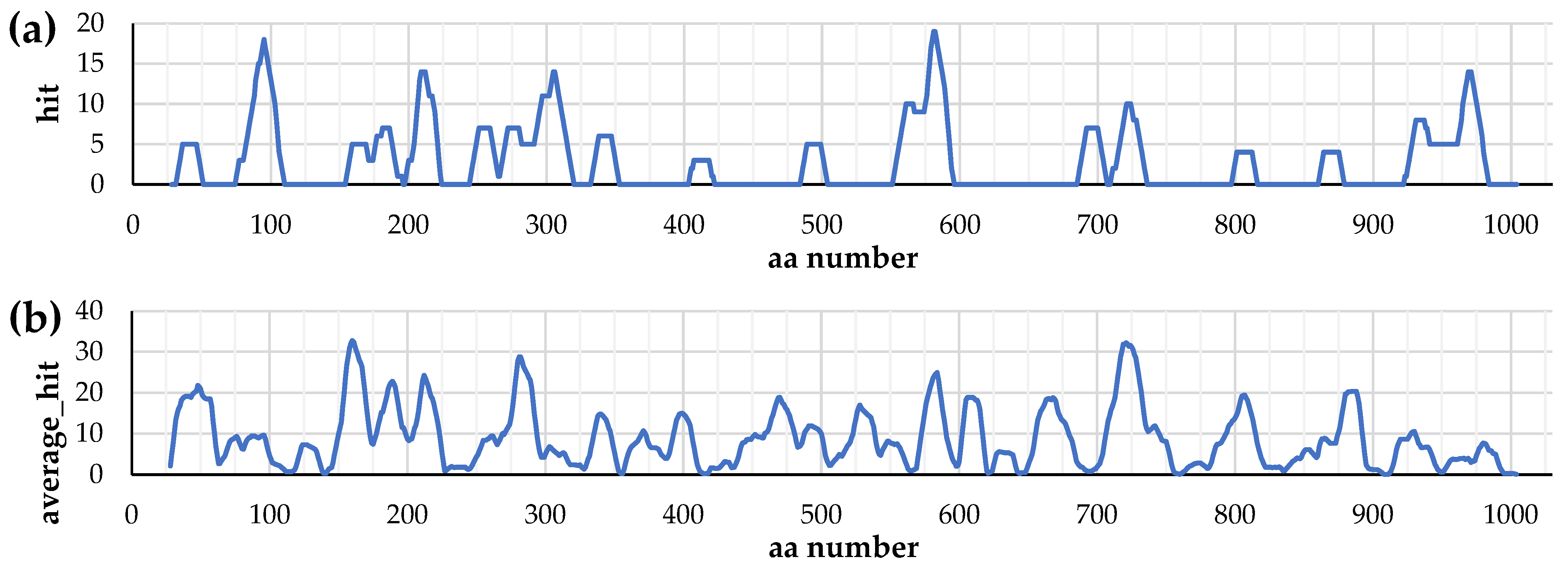
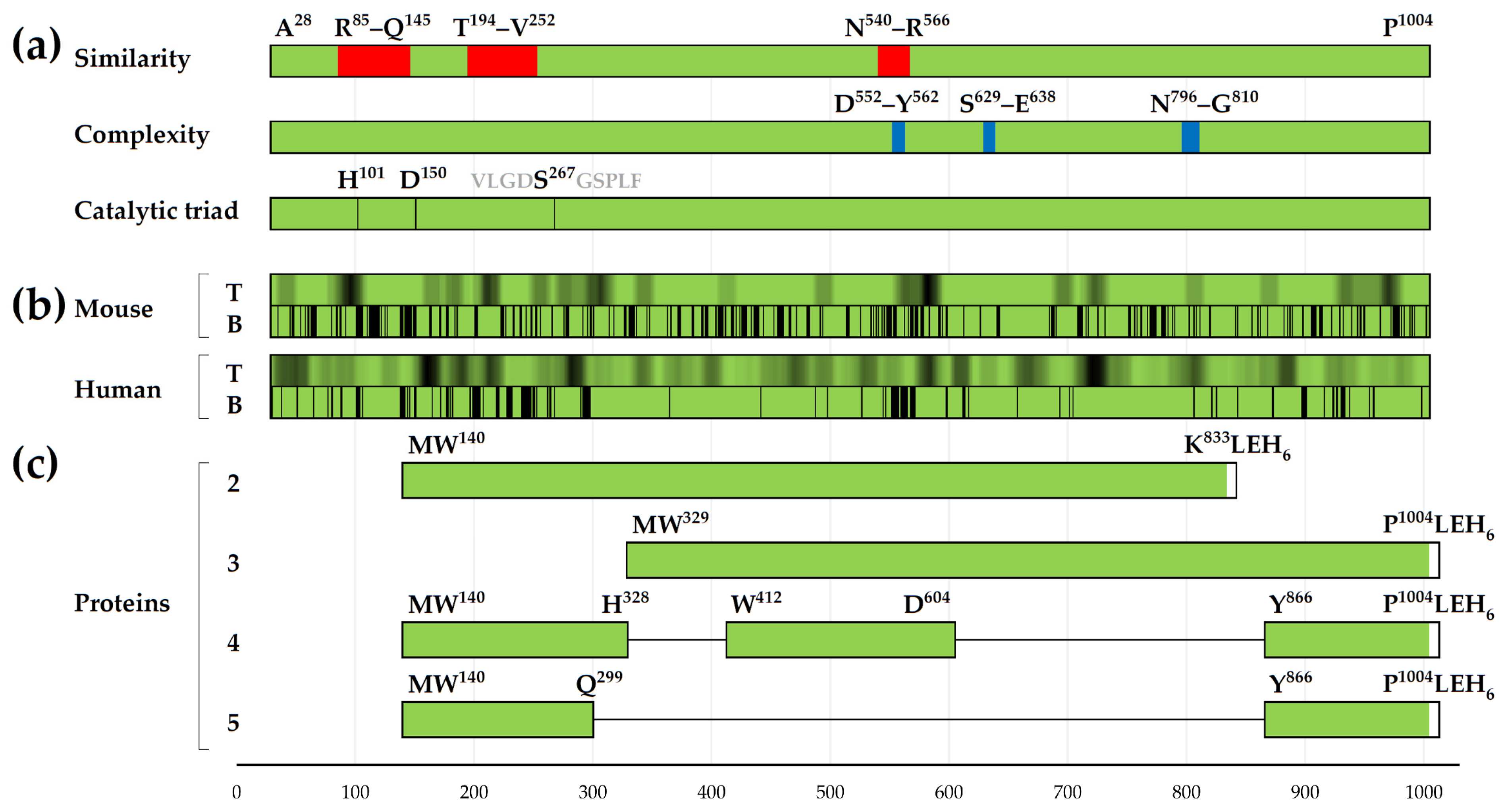
3.1. Distribution of T-Cell Epitopes in the Primary Structure of IgA1 Protease
3.2. Distribution of Conformational B-Cell Epitopes in the Tertiary Structure of IgA1 Protease
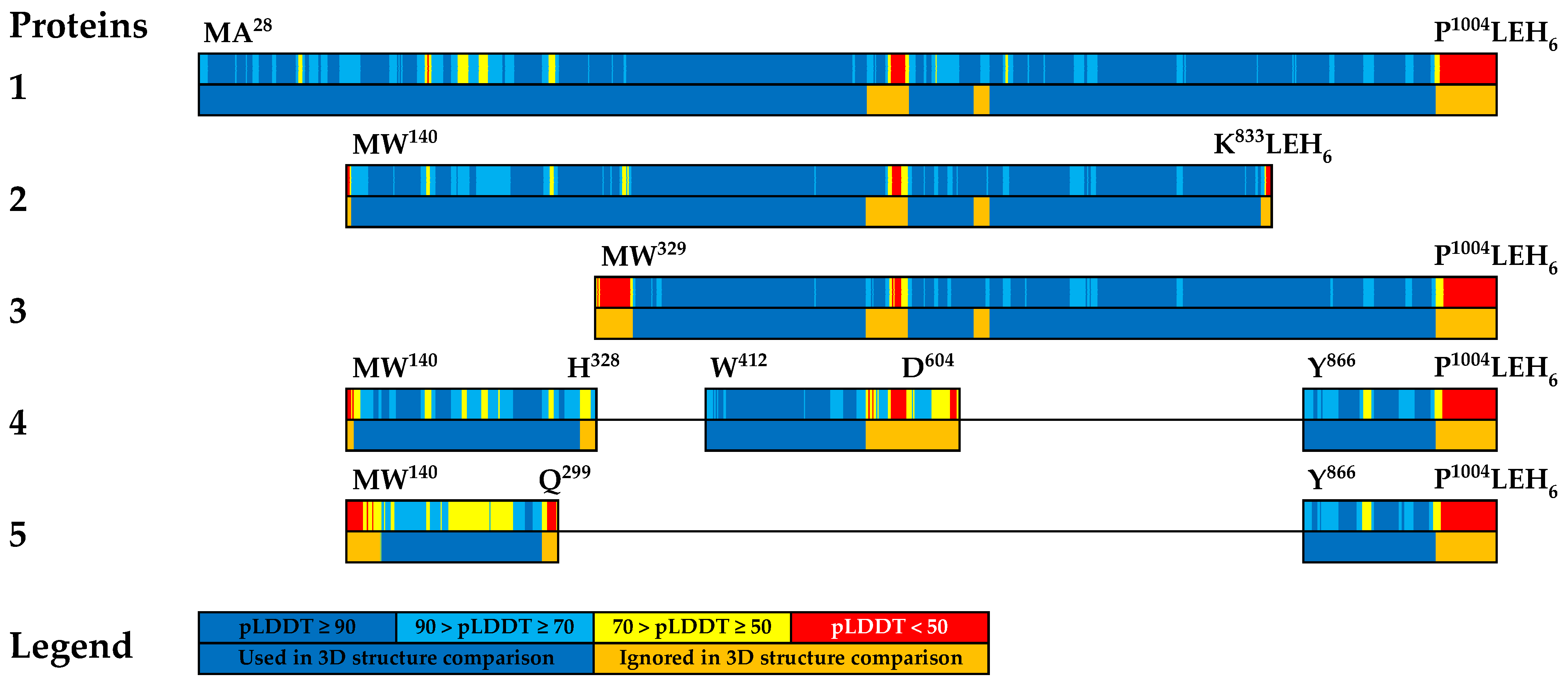
3.3. Comparative Analysis of Tertiary Structures of Created Recombinant Proteins
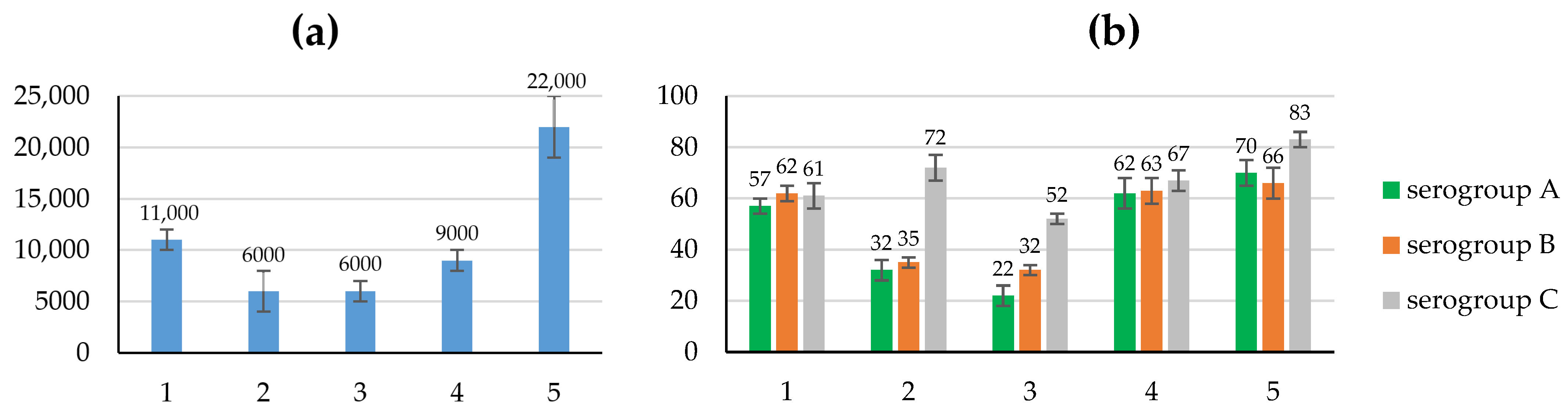
3.4. Study of the Immunogenic and Protective Properties of the Created Recombinant Proteins
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pizza, M.; Bekkat-Berkani, R.; Rappuoli, R. Vaccines against Meningococcal Diseases. Microorganisms 2020, 8, 1521. [Google Scholar] [CrossRef] [PubMed]
- Zhigis, L.S.; Kotelnikova, O.V.; Zinchenko, A.A.; Karlinsky, D.M.; Prokopenko, Y.A.; Rumsh, L.D. IgA1 Protease as a Vaccine Basis for Prevention of Bacterial Meningitis. Russ. J. Bioorg. Chem. 2021, 47, 805–814. [Google Scholar] [CrossRef]
- Rouphael, N.G.; Stephens, D.S. Neisseria meningitidis: Biology, microbiology, and epidemiology. Methods Mol. Biol. 2012, 799, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bolgiano, B.; Moran, E.; Beresford, N.J.; Gao, F.; Care, R.; Desai, T.; Nordgren, I.K.; Rudd, T.R.; Feavers, I.M.; Bore, P.; et al. Evaluation of Critical Quality Attributes of a Pentavalent (A, C, Y, W, X) Meningococcal Conjugate Vaccine for Global Use. Pathogens 2021, 10, 928. [Google Scholar] [CrossRef]
- Donald, R.G.K.; Hawkins, J.C.; Hao, L.; Liberator, P.; Jones, T.R.; Harris, S.L.; Perez, J.L.; Eiden, J.J.; Jansen, K.U.; Anderson, A.S. Meningococcal Serogroup B Vaccines: Estimating Breadth of Coverage. Hum. Vaccines Immunother. 2017, 13, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Yonts, A.B.; Gaviria-Agudelo, C.; Kimberlin, D.W.; O’Leary, S.T.; Paulsen, G.C. Fall 2023 ACIP Update on Meningococcal, RSV, COVID-19, and Other Pediatric Vaccines. Pediatrics 2023, 153, e2023064990. [Google Scholar] [CrossRef]
- Mistry, D.; Stockley, R.A. IgA1 Protease. Int. J. Biochem. Cell Biol. 2006, 38, 1244–1248. [Google Scholar] [CrossRef]
- Karlinsky, D.; Prokopenko, Y.; Zinchenko, A.; Zhigis, L.; Kotelnikova, O.; Rumsh, L.; Smirnov, I. Highly Similar Sequences of Mature IgA1 Proteases from Neisseria Meningitidis, Neisseria Gonorrhoeae and Haemophilus Influenzae. Pathogens 2022, 11, 734. [Google Scholar] [CrossRef]
- Lomholt, H.; Poulsen, K.; Kilian, M. Comparative Characterization of the Iga Gene Encoding IgA1 Protease in Neisseria meningitidis, Neisseria Gonorrhoeae and Haemophilus Influenzae. Mol. Microbiol. 1995, 15, 495–506. [Google Scholar] [CrossRef]
- Vitovski, S.; Sayers, J.R. Relaxed Cleavage Specificity of an Immunoglobulin A1 Protease from Neisseria meningitidis. Infect. Immun. 2007, 75, 2875–2885. [Google Scholar] [CrossRef]
- Spoerry, C.; Karlsson, J.; Aschtgen, M.-S.; Loh, E. Neisseria Meningitidis IgA1-Specific Serine Protease Exhibits Novel Cleavage Activity against IgG3. Virulence 2021, 12, 389–403. [Google Scholar] [CrossRef]
- Yagudaeva, E.Y.; Zhigis, L.S.; Razgulyaeva, O.A.; Zueva, V.S.; Melnikov, E.E.; Zubov, V.P.; Kozlov, L.V.; Bichucher, A.M.; Kotel’nikova, O.V.; Alliluev, A.P.; et al. Isolation and Determination of the Activity of IgA1 Protease from Neisseria Meningitidis. Russ. J. Bioorg. Chem. 2010, 36, 81–89. [Google Scholar] [CrossRef]
- Serova, O.V.; Melnikov, E.E.; Zinchenko, A.A.; Kotel’nikova, O.V.; Alliluev, A.P.; Bichucher, A.M.; Gordeeva, E.A.; Zhigis, L.S.; Zueva, V.S.; Kozlov, L.V.; et al. Recombinant IgA1 Protease from N. Meningitidis. Obtaining and Properties. Russ. J. Biopharm. 2011, 3, 42–47. [Google Scholar]
- Kotel’nikova, O.V.; Alliluev, A.P.; Drozhzhina, E.Y.; Koroleva, I.S.; Sitnikova, E.A.; Zinchenko, A.A.; Gordeeva, E.A.; Melikhova, T.D.; Nokel, E.A.; Zhigis, L.S.; et al. Protective Properties of Recombinant IgA1 Protease from Meningococcus. Biochem. Mosc. Suppl. Ser. B 2013, 7, 305–310. [Google Scholar] [CrossRef]
- Kotelnikova, O.V.; Zinchenko, A.A.; Vikhrov, A.A.; Alliluev, A.P.; Serova, O.V.; Gordeeva, E.A.; Zhigis, L.S.; Zueva, V.S.; Razgulyaeva, O.A.; Melikhova, T.D.; et al. Serological Analysis of Immunogenic Properties of Recombinant Meningococcus IgA1 Protease-Based Proteins. Bull. Exp. Biol. Med. 2016, 161, 391–394. [Google Scholar] [CrossRef]
- Kotel’nikova, O.V.; Alliluev, A.P.; Zinchenko, A.A.; Prokopenko, Y.A.; Zhigis, L.S.; Zueva, V.S.; Razgulyaeva, O.A.; Gordeeva, E.A.; Melikhova, T.D.; Nokel’, E.A.; et al. Peculiarities of the Formation of Antimeningococcus Immunity in Mice Immunized with Fragments of N. Meningitidis IgA1 Protease. Bull. Exp. Biol. Med. 2018, 165, 763–766. [Google Scholar] [CrossRef]
- Zinchenko, A.A.; Kotelnikova, O.V.; Gordeeva, E.A.; Prokopenko, Y.A.; Razgulyaeva, O.A.; Serova, O.V.; Melikhova, T.D.; Nokel, E.A.; Zhigis, L.S.; Zueva, V.S.; et al. Immunogenic and Protective Properties of Neisseria Meningitidis IgA1 Protease and of Its Truncated Fragments. Russ. J. Bioorg. Chem. 2018, 44, 64–72. [Google Scholar] [CrossRef]
- Kotelnikova, O.; Alliluev, A.; Zinchenko, A.; Zhigis, L.; Prokopenko, Y.; Nokel, E.; Razgulyaeva, O.; Zueva, V.; Tokarskaya, M.; Yastrebova, N.; et al. Protective Potency of Recombinant Meningococcal IgA1 Protease and Its Structural Derivatives upon Animal Invasion with Meningococcal and Pneumococcal Infections. Microbes Infect. 2019, 21, 336–340. [Google Scholar] [CrossRef]
- Janoff, E.N.; Rubins, J.B.; Fasching, C.; Charboneau, D.; Rahkola, J.T.; Plaut, A.G.; Weiser, J.N. Pneumococcal IgA1 Protease Subverts Specific Protection by Human IgA1. Mucosal Immunol. 2014, 7, 249–256. [Google Scholar] [CrossRef]
- Fu, L.; Zhao, J.; Lin, L.; Zhang, Q.; Xu, Z.; Han, L.; Xie, C.; Zhou, R.; Jin, M.; Zhang, A. Characterization of IgA1 Protease as a Surface Protective Antigen of Streptococcus Suis Serotype 2. Microbes Infect. 2016, 18, 285–289. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Sherry, S.T.; Yankie, L.; Karsch-Mizrachi, I. GenBank 2023 Update. Nucleic Acids Res. 2023, 51, D141–D144. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 Update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Wang, P.; Sidney, J.; Dow, C.; Mothé, B.; Sette, A.; Peters, B. A Systematic Assessment of MHC Class II Peptide Binding Predictions and Evaluation of a Consensus Approach. PLoS Comput. Biol. 2008, 4, e1000048. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sidney, J.; Kim, Y.; Sette, A.; Lund, O.; Nielsen, M.; Peters, B. Peptide Binding Predictions for HLA DR, DP and DQ Molecules. BMC Bioinform. 2010, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, M.; Karosiene, E.; Rasmussen, M.; Stryhn, A.; Buus, S.; Nielsen, M. Accurate Pan-Specific Prediction of Peptide-MHC Class II Binding Affinity with Improved Binding Core Identification. Immunogenetics 2015, 67, 641–650. [Google Scholar] [CrossRef]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved Methods for Predicting Peptide Binding Affinity to MHC Class II Molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved Predictions of MHC Antigen Presentation by Concurrent Motif Deconvolution and Integration of MS MHC Eluted Ligand Data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
- Kaabinejadian, S.; Barra, C.; Alvarez, B.; Yari, H.; Hildebrand, W.H.; Nielsen, M. Accurate MHC Motif Deconvolution of Immunopeptidomics Data Reveals a Significant Contribution of DRB3, 4 and 5 to the Total DR Immunopeptidome. Front. Immunol. 2022, 13, 835454. [Google Scholar] [CrossRef]
- Greenbaum, J.; Sidney, J.; Chung, J.; Brander, C.; Peters, B.; Sette, A. Functional Classification of Class II Human Leukocyte Antigen (HLA) Molecules Reveals Seven Different Supertypes and a Surprising Degree of Repertoire Sharing across Supertypes. Immunogenetics 2011, 63, 325–335. [Google Scholar] [CrossRef]
- Bui, H.-H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting Population Coverage of T-Cell Epitope-Based Diagnostics and Vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Clifford, J.N.; Høie, M.H.; Deleuran, S.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred -3.0: Improved B-cell Epitope Prediction Using Protein Language Models. Protein Sci. 2022, 31, e4497. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, J.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A New Structure-Based Tool for the Prediction of Antibody Epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Kringelum, J.V.; Lundegaard, C.; Lund, O.; Nielsen, M. Reliable B Cell Epitope Predictions: Impacts of Method Development and Improved Benchmarking. PLoS Comput. Biol. 2012, 8, e1002829. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Qiu, T.; Zhang, Q.; Tang, K.; Fan, Y.; Qiu, J.; Wu, D.; Zhang, W.; Chen, Y.; Gao, J.; et al. SEPPA 2.0—More Refined Server to Predict Spatial Epitope Considering Species of Immune Host and Subcellular Localization of Protein Antigen. Nucleic Acids Res. 2014, 42, W59–W63. [Google Scholar] [CrossRef]
- Zinchenko, A.A.; Serova, O.V.; Prokopenko, Y.A.; Gordeeva, E.A.; Melikhova, T.D.; Nokel, E.A.; Kaliberda, E.N.; Kotelnikova, O.V.; Zhigis, L.S.; Razgulyaeva, O.A.; et al. Recombinant protein having protective action on meningococcus (embodiments), polynucleotide coding recombinant protein, recombinant plasmid DNA containing said polynucleotide, host cell containing said recombinant plasmid DNA, method of producing recombinant protein 2019. RU 2701964 C2, 24 June 2019. [Google Scholar]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D. The Focusing Positions of Polypeptides in Immobilized pH Gradients Can Be Predicted from Their Amino Acid Sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef]
- Gupta, S.K.; Smita, S.; Sarangi, A.N.; Srivastava, M.; Akhoon, B.A.; Rahman, Q.; Gupta, S.K. In Silico CD4+ T-Cell Epitope Prediction and HLA Distribution Analysis for the Potential Proteins of Neisseria Meningitidis Serogroup B—A Clue for Vaccine Development. Vaccine 2010, 28, 7092–7097. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prokopenko, Y.; Zinchenko, A.; Karlinsky, D.; Kotelnikova, O.; Razgulyaeva, O.; Gordeeva, E.; Nokel, E.; Serova, O.; Kaliberda, E.; Zhigis, L.; et al. Protective Antimicrobial Effect of the Potential Vaccine Created on the Basis of the Structure of the IgA1 Protease from Neisseria meningitidis. Vaccines 2024, 12, 1355. https://doi.org/10.3390/vaccines12121355
Prokopenko Y, Zinchenko A, Karlinsky D, Kotelnikova O, Razgulyaeva O, Gordeeva E, Nokel E, Serova O, Kaliberda E, Zhigis L, et al. Protective Antimicrobial Effect of the Potential Vaccine Created on the Basis of the Structure of the IgA1 Protease from Neisseria meningitidis. Vaccines. 2024; 12(12):1355. https://doi.org/10.3390/vaccines12121355
Chicago/Turabian StyleProkopenko, Yuri, Alexei Zinchenko, David Karlinsky, Olga Kotelnikova, Olga Razgulyaeva, Elena Gordeeva, Elena Nokel, Oxana Serova, Elena Kaliberda, Larisa Zhigis, and et al. 2024. "Protective Antimicrobial Effect of the Potential Vaccine Created on the Basis of the Structure of the IgA1 Protease from Neisseria meningitidis" Vaccines 12, no. 12: 1355. https://doi.org/10.3390/vaccines12121355
APA StyleProkopenko, Y., Zinchenko, A., Karlinsky, D., Kotelnikova, O., Razgulyaeva, O., Gordeeva, E., Nokel, E., Serova, O., Kaliberda, E., Zhigis, L., Rumsh, L., & Smirnov, I. (2024). Protective Antimicrobial Effect of the Potential Vaccine Created on the Basis of the Structure of the IgA1 Protease from Neisseria meningitidis. Vaccines, 12(12), 1355. https://doi.org/10.3390/vaccines12121355





