Epitopes and Mimotopes Identification Using Phage Display for Vaccine Development against Infectious Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection, Inclusion, and Exclusion Criteria
3. Results
3.1. Techniques for Epitope Identification
3.1.1. X-ray Crystallography and Nuclear Magnetic Resonance (NMR) Spectroscopy
3.1.2. Cryo-Electron Microscopy (Cryo-EM)
3.1.3. Peptide Arrays
3.1.4. Site-Directed Mutagenesis
3.1.5. Bioinformatics and Computational Modeling
3.1.6. Phage Display
3.2. The Phage Display Library and the Samples for Epitope or Mimotope Selection
3.2.1. Types of Phage Display Libraries
3.2.2. Library Construction
3.2.3. Types of Samples
3.2.4. Antibody Purification
3.3. The selection Procedure
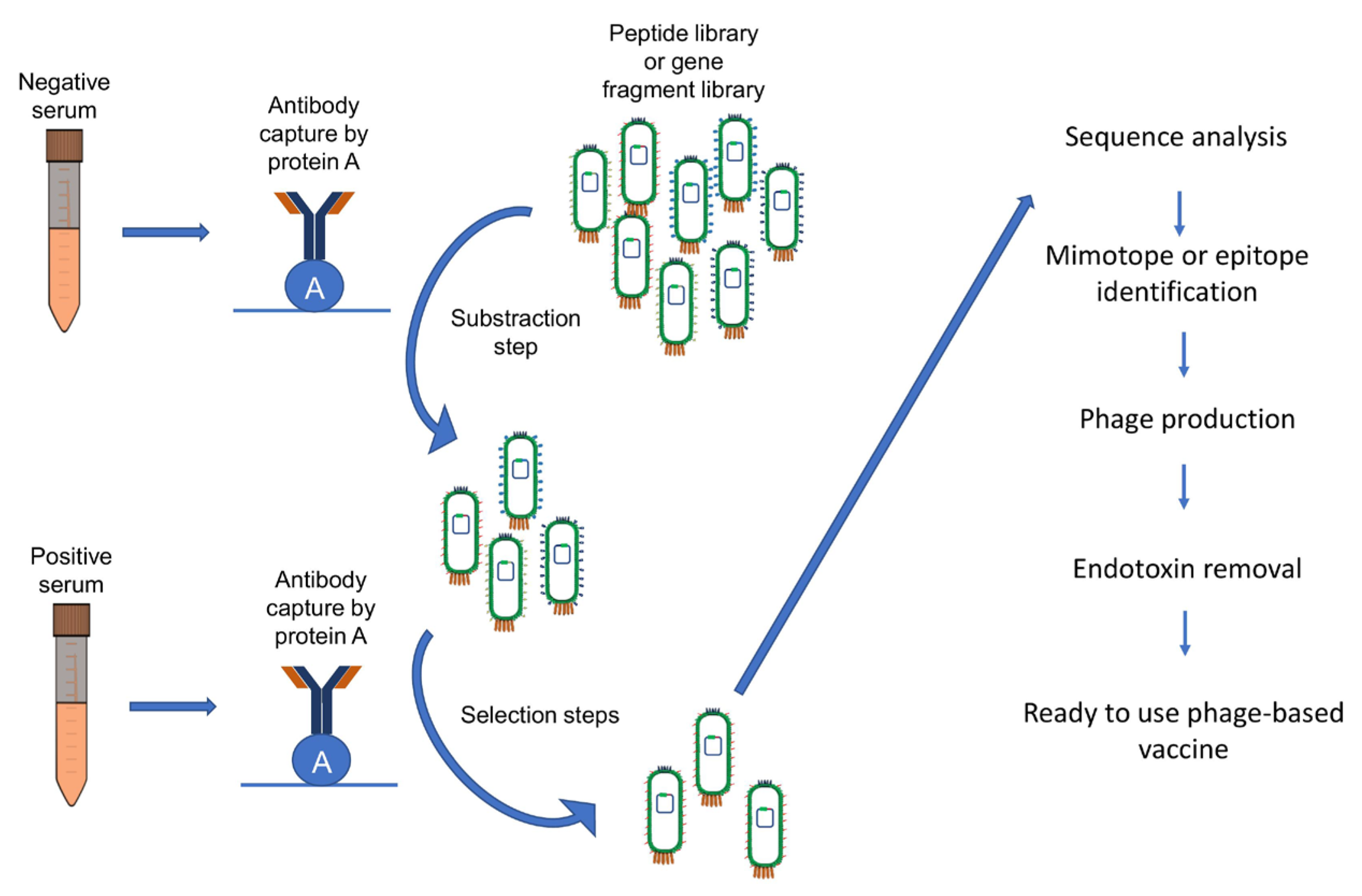
3.3.1. Biopanning
- To facilitate the selection process, it is vital to immobilize antibodies appropriately. Antibodies of interest must be purified to prevent interference from other proteins present in serum samples. Similarly, negative sera antibodies should be prepared for subsequent subtractive selection.
- To efficiently eliminate phage-specific antibodies from serum samples prior to their subsequent use, the samples can be incubated on a surface that has been coated with M13 helper phages. In the case of using a different type of bacteriophage, such as T7, for library creation, the protocol employs T7 phages lacking an insert instead of helper phages.
- To eliminate non-specific binders within the library and clones that bind to the constant region of antibodies, a subtractive selection step is essential. Thus, before proceeding with the selection process, the library is incubated with immobilized purified antibodies from negative serum.
- Subsequently, the phage solution is carefully transferred to the chosen solid support, such as immunotubes, microtiter plates, or magnetic beads, all of which are pre-coated with immobilized positive antibodies. For example, one approach involves capturing biotinylated antibodies using streptavidin-coated beads, while another option is immobilizing antibodies using protein A.
- To counteract any remaining non-specific binders within the library, negative antibodies can be added to the solution in the tube. The library is then incubated for a specific duration as required.
- The phage solution is discarded, and to ensure the removal of non-bound phages, multiple washes are conducted.
- To elute the binding phages, specific elution with purified positive antibodies or non-specific elution methods, such as pH adjustment or the use of a chaotropic agent, can be employed. When utilizing pH alteration, it is crucial to neutralize the eluted solution to prevent peptide denaturation or any negative impact on the phage particles.
- Following specific elution, the eluted phages are utilized to infect E. coli cells, facilitating the generation of a new phage stock. In instances where the library has been constructed using a phagemid, infection with a helper phage is required to enable the replication of the phage particles. If a T7 library is used, the eluted T7 phage is used to infect a suitable host strain of E. coli together with the appropriate antibiotic. The mixture is then incubated until lysis occurs. After lysis, the solution is centrifuged and subsequently transferred to a new tube.
- The selection steps are repeated in several rounds, with each round incrementally enhancing the stringency of the selection conditions. This can be accomplished by decreasing the amount of target molecules used for coating or by increasing the number of washes conducted.
3.3.2. Identification of Potential Hits with Phage ELISA
3.3.3. Optimization of Hits
3.3.4. Characterization of Hits
3.4. General Applications of Phage Display to Select Epitopes of a Pathogen
3.4.1. Diagnostic Test Development
3.4.2. Understanding Pathogen-Host Interactions
3.4.3. Developing Therapeutics
- Monoclonal antibodies represent a potent strategy for targeting pathogenic epitopes with high specificity. These antibodies are specifically designed to bind to a particular epitope, allowing for diverse therapeutic applications. By engaging with the epitopes of a pathogen, monoclonal antibodies can neutralize the pathogen, impede its interaction with host cells, or initiate an immune response against it. Notably, this approach has demonstrated promising results in anticancer treatments [58], by targeting specific molecular markers on cancer cells, leading to enhanced therapeutic outcomes and improved patient survival rates. Their ability to specifically bind to epitopes of a pathogen, block key interactions, and stimulate immune responses represents a remarkable advancement in precision medicine, offering new avenues for the development of effective and tailored treatments [59].
- Epitopes can be utilized as valuable targets for small molecule inhibitors, which are specifically designed to bind to these epitopes and disrupt the function of the pathogen. By binding to the epitope, small molecules can exert their inhibitory effects by various means, such as inhibiting enzymatic activity or blocking receptor interactions [60], serving as a potent strategy to disrupt the pathogen’s lifecycle and limit its impact on the host.
- Gene therapy presents a promising avenue where the epitopes of a pathogen can be integrated. One approach involves the incorporation of genetic material encoding the epitope into host cells. By introducing this genetic material, the host cells gain the ability to produce the epitope themselves, consequently eliciting an immune response against the pathogen [61,62,63]. This strategy offers several advantages. Firstly, it allows for the sustained production of the pathogenic epitope within the host, ensuring a prolonged immune response. Secondly, by targeting the production of the epitope directly within the host cells, gene therapy circumvents the need for external administration of epitopes, making it a potentially more convenient and controlled approach.
- The recognition of antigenic epitopes holds the utmost significance in the advancement of vaccine development [64,65,66]. These epitopes serve as the building blocks for designing vaccines that activate the immune system to generate antibodies capable of recognizing and neutralizing specific pathogens. By precisely targeting these antigenic epitopes, vaccines can achieve heightened effectiveness and specificity, while reducing the potential side effects often associated with traditional vaccines that employ whole pathogens or inactivated forms of the pathogen.
3.5. Applications of Phage Display to Select Epitopes in Infectious Diseases
3.5.1. SARS-CoV-2
3.5.2. Mycobacterium tuberculosis
3.5.3. Hepatitis Virus
3.5.4. Influenza Virus
3.5.5. HIV
3.5.6. Human T-Lymphotropic Virus 1
3.5.7. Plasmodium falciparum
3.5.8. Trypanosoma cruzi
3.5.9. Dirofilaria repens
| Pathogen | Antigen | References |
|---|---|---|
| SARS-CoV-2 | Spike, N | [4,68,69] |
| M. tuberculosis | Acetaldehyde dehydrogenase, transposase, and unknown proteins | [6] |
| HAV | VP1, VP3 | [70] |
| HBV | HBsAg | [72] |
| HCV | Core protein | [74] |
| HEV | ORF2 capsid, | [75] |
| H5N1 | neuraminidase, PB1-F2, H5 HA, M2 | [75] |
| HIV | gp120 C2, gp41, V3 | [78,80] |
| HTLV-1 | gp46 | [51] |
| Plasmodium falciparum | DBL3χ, PfEMP-1, erythrocyte membrane protein | [82] |
| Trypanosoma cruzi | Novel | [83] |
| Dirofilaria repens | Novel | [84] |
3.6. Use of Phagotomes in Vaccines
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bjorkman, P.J.; Burmeister, W.P. Structures of Two Classes of MHC Molecules Elucidated: Crucial Differences and Similarities. Curr. Opin. Struct. Biol. 1994, 4, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Rappuoli, R. Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Ferrin, T.E. Visualization Software for Molecular Assemblies. Curr. Opin. Struct. Biol. 2007, 17, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Ballmann, R.; Hotop, S.-K.; Bertoglio, F.; Steinke, S.; Heine, P.A.; Chaudhry, M.Z.; Jahn, D.; Pucker, B.; Baldanti, F.; Piralla, A.; et al. ORFeome Phage Display Reveals a Major Immunogenic Epitope on the S2 Subdomain of SARS-CoV-2 Spike Protein. Viruses 2022, 14, 1326. [Google Scholar] [CrossRef]
- Samrat, S.K.; Tharappel, A.M.; Li, Z.; Li, H. Prospect of SARS-CoV-2 Spike Protein: Potential Role in Vaccine and Therapeutic Development. Virus Res. 2020, 288, 198141. [Google Scholar] [CrossRef]
- Wang, L.; Deng, X.; Liu, H.; Zhao, L.; You, X.; Dai, P.; Wan, K.; Zeng, Y. The Mimic Epitopes of Mycobacterium Tuberculosis Screened by Phage Display Peptide Library Have Serodiagnostic Potential for Tuberculosis. Pathog. Dis. 2016, 74, ftw091. [Google Scholar] [CrossRef]
- Toride King, M.; Brooks, C.L. Epitope Mapping of Antibody-Antigen Interactions with X-Ray Crystallography. In Epitope Mapping Protocols; Rockberg, J., Nilvebrant, J., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1785, pp. 13–27. [Google Scholar]
- Bardelli, M.; Livoti, E.; Simonelli, L.; Pedotti, M.; Moraes, A.; Valente, A.P.; Varani, L. Epitope Mapping by Solution NMR Spectroscopy. J. Mol. Recognit. 2015, 28, 393–400. [Google Scholar] [CrossRef]
- Zheng, H.; Handing, K.B.; Zimmerman, M.D.; Shabalin, I.G.; Almo, S.C.; Minor, W. X-Ray Crystallography over the Past Decade for Novel Drug Discovery—Where Are We Heading Next? Expert Opin. Drug Discov. 2015, 10, 975–989. [Google Scholar] [CrossRef]
- Bianchi, M.; Turner, H.L.; Nogal, B.; Cottrell, C.A.; Oyen, D.; Pauthner, M.; Bastidas, R.; Nedellec, R.; McCoy, L.E.; Wilson, I.A.; et al. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 2018, 49, 288–300.e8. [Google Scholar] [CrossRef]
- Li, N.; Li, Z.; Fu, Y.; Cao, S. Cryo-EM Studies of Virus-Antibody Immune Complexes. Virol. Sin. 2020, 35, 1–13. [Google Scholar] [CrossRef]
- Amartely, H.; Iosub-Amir, A.; Friedler, A. Identifying Protein-Protein Interaction Sites Using Peptide Arrays. J. Vis. Exp. 2014, Volume 93, 52097. [Google Scholar] [CrossRef]
- Hernandez, D.P.; Dittmar, G. Peptide Array–Based Interactomics. Anal. Bioanal. Chem. 2021, 413, 5561–5566. [Google Scholar] [CrossRef]
- Vandervaart, J.P.; Inniss, N.L.; Ling-Hu, T.; Minasov, G.; Wiersum, G.; Rosas-Lemus, M.; Shuvalova, L.; Achenbach, C.J.; Hultquist, J.F.; Satchell, K.J.F.; et al. Serodominant SARS-CoV-2 Nucleocapsid Peptides Map to Unstructured Protein Regions. Microbiol. Spectr. 2023, 11, e00324-23. [Google Scholar] [CrossRef]
- Nagy, K.; McBride, R.; Head, S.R.; Ordoukhanian, P.; Law, M. Low-Cost Peptide Microarrays for Mapping Continuous Antibody Epitopes. In Peptide Microarrays; Cretich, M., Chiari, M., Eds.; Humana Press: New York, NY, USA, 2023; Volume 1352, pp. 63–81. [Google Scholar]
- Chen, C.-W.; Chang, C.-Y. Peptide Scanning-Assisted Identification of a Monoclonal Antibody-Recognized Linear B-Cell Epitope. J. Vis. Exp. 2017, 121, e55417. [Google Scholar] [CrossRef]
- Zheng, N.; Li, C.; Hou, H.; Chen, Y.; Zhang, A.; Han, S.; Wan, B.; Wu, Y.; He, H.; Wang, N.; et al. A Novel Linear B-Cell Epitope on the P54 Protein of African Swine Fever Virus Identified Using Monoclonal Antibodies. Viruses 2023, 15, 867. [Google Scholar] [CrossRef]
- Hagoss, Y.T.; Shen, D.; Zhang, Z.; Li, F.; Bu, Z.; Zhao, D. Novel Epitopes Mapping of African Swine Fever Virus CP312R Protein Using Monoclonal Antibodies. Viruses 2023, 15, 557. [Google Scholar] [CrossRef]
- Szymczak, L.C.; Kuo, H.-Y.; Mrksich, M. Peptide Arrays: Development and Application. Anal. Chem. 2018, 90, 266–282. [Google Scholar] [CrossRef]
- Mitro, N.; Gilardi, F.; Giudici, M.; Godio, C.; Scotti, E.; Crestani, M. Site-Directed Mutagenesis to Study the Role of Specific Amino Acids in the Ligand Binding Domain of PPARs. In Peroxisome Proliferator-Activated Receptors (PPARs); Badr, M., Youssef, J., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 952, pp. 137–144. [Google Scholar]
- Meng, T.; Bezstarosti, S.; Singh, U.; Yap, M.; Scott, L.; Petrosyan, N.; Quiroz, F.; van Eps, N.; Hui, E.K.; Suh, D.; et al. Site-directed Mutagenesis of HLA Molecules Reveals the Functional Epitope of a Human HLA-A1/A36-specific Monoclonal Antibody. HLA 2023, 101, 138–142. [Google Scholar] [CrossRef]
- Nonaka, Y.; Ogawa, T.; Shoji, H.; Nishi, N.; Kamitori, S.; Nakamura, T. Modulation of the Carbohydrate-Binding Specificity of Two Xenopus Proto-Type Galectins by Site-Directed Mutagenesis. Biochim. Biophys. Acta Proteins Proteomics 2021, 1869, 140684. [Google Scholar] [CrossRef]
- EL-Manzalawy, Y.; Dobbs, D.; Honavar, V.G. In Silico Prediction of Linear B-Cell Epitopes on Proteins. In Prediction of Protein Secondary Structure; Zhou, Y., Kloczkowski, A., Faraggi, E., Yang, Y., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1484, pp. 255–264. [Google Scholar]
- Sohail, M.S.; Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. In Silico T Cell Epitope Identification for SARS-CoV-2: Progress and Perspectives. Adv. Drug Deliv. Rev. 2021, 171, 29–47. [Google Scholar] [CrossRef]
- Ezzemani, W.; Windisch, M.P.; Altawalah, H.; Guessous, F.; Saile, R.; Benjelloun, S.; Kettani, A.; Ezzikouri, S. Design of a Multi-Epitope Zika Virus Vaccine Candidate—An in-Silico Study. J. Biomol. Struct. Dyn. 2023, 41, 3762–3771. [Google Scholar] [CrossRef] [PubMed]
- McCaffrey, P. Bioinformatic Techniques for Vaccine Development: Epitope Prediction and Structural Vaccinology. In Resources for Vaccine Development; Thomas, S., Ed.; Humana: New York, NY, USA, 2022; Volume 2412, pp. 413–423. [Google Scholar]
- Wang, L.-F.; Yu, M. Epitope Identification and Discovery Using Phage Display Libraries: Applications in Vaccine Development and Diagnostics. Curr. Drug Targets 2004, 5, 1–15. [Google Scholar] [CrossRef]
- Zhou, J.; Li, Y.; Liu, Z.; Qian, W.; Chen, Y.; Qi, Y.; Wang, A. Induction of Anti-Zearalenone Immune Response with Mimotopes Identified from a Phage Display Peptide Library. Toxicon 2021, 199, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Budania, S.; Dubey, A.; Singh, A. Trypanosoma Evansi RoTat 1.2 Variant Surface Antigen Mimotopes Selected by Panning of the Random Peptide Phage-display Library against Monoclonal Antibodies. J. Mol. Recognit. 2022, 35, e2984. [Google Scholar] [CrossRef] [PubMed]
- Palma, M. Perspectives on Passive Antibody Therapy and Peptide-Based Vaccines against Emerging Pathogens like SARS-CoV-2. GERMS 2021, 11, 287–305. [Google Scholar] [CrossRef]
- Palma, M. Aspects of Phage-Based Vaccines for Protein and Epitope Immunization. Vaccines 2023, 11, 436. [Google Scholar] [CrossRef]
- González-Mora, A.; Hernández-Pérez, J.; Iqbal, H.M.N.; Rito-Palomares, M.; Benavides, J. Bacteriophage-Based Vaccines: A Potent Approach for Antigen Delivery. Vaccines 2020, 8, 504. [Google Scholar] [CrossRef]
- Luzar, J.; Štrukelj, B.; Lunder, M. Phage Display Peptide Libraries in Molecular Allergology: From Epitope Mapping to Mimotope-Based Immunotherapy. Allergy 2016, 71, 1526–1532. [Google Scholar] [CrossRef]
- Apostolopoulos, V.; Bojarska, J.; Chai, T.-T.; Elnagdy, S.; Kaczmarek, K.; Matsoukas, J.; New, R.; Parang, K.; Lopez, O.P.; Parhiz, H.; et al. A Global Review on Short Peptides: Frontiers and Perspectives. Molecules 2021, 26, 430. [Google Scholar] [CrossRef]
- Joo, S.-H. Cyclic Peptides as Therapeutic Agents and Biochemical Tools. Biomol. Ther. 2012, 20, 19–26. [Google Scholar] [CrossRef]
- Kamstrup Sell, D.; Sloth, A.B.; Bakhshinejad, B.; Kjaer, A. A White Plaque, Associated with Genomic Deletion, Derived from M13KE-Based Peptide Library Is Enriched in a Target-Unrelated Manner during Phage Display Biopanning Due to Propagation Advantage. Int. J. Mol. Sci. 2022, 23, 3308. [Google Scholar] [CrossRef]
- Rentero Rebollo, I.; Heinis, C. Phage Selection of Bicyclic Peptides. Methods 2013, 60, 46–54. [Google Scholar] [CrossRef]
- Bosma, T.; Rink, R.; Moosmeier, M.A.; Moll, G.N. Genetically Encoded Libraries of Constrained Peptides. ChemBioChem 2019, 20, 1754–1758. [Google Scholar] [CrossRef]
- Skerra, A. Alternative Non-Antibody Scaffolds for Molecular Recognition. Curr. Opin. Biotechnol. 2007, 18, 295–304. [Google Scholar] [CrossRef]
- Irving, M.B.; Pan, O.; Scott, J.K. Random-Peptide Libraries and Antigen-Fragment Libraries for Epitope Mapping and the Development of Vaccines and Diagnostics. Curr. Opin. Chem. Biol. 2001, 5, 314–324. [Google Scholar] [CrossRef]
- Qi, H.; Lu, H.; Qiu, H.-J.; Petrenko, V.; Liu, A. Phagemid Vectors for Phage Display: Properties, Characteristics and Construction. J. Mol. Biol. 2012, 417, 129–143. [Google Scholar] [CrossRef]
- Palma, M. Phage Display Identification of Immunodominant Epitopes and Autoantibodies in Autoimmune Diseases. Curr. Biosci. 2021, 1, e02. [Google Scholar] [CrossRef]
- Stern, Z.; Stylianou, D.C.; Kostrikis, L.G. The Development of Inovirus-Associated Vector Vaccines Using Phage-Display Technologies. Expert Rev. Vaccines 2019, 18, 913–920. [Google Scholar] [CrossRef]
- Gershoni, J.M.; Roitburd-Berman, A.; Siman-Tov, D.D.; Tarnovitski Freund, N.; Weiss, Y. Epitope Mapping. BioDrugs 2007, 21, 145–156. [Google Scholar] [CrossRef]
- Maghsood, F.; Amiri, M.M.; Zarnani, A.-H.; Salimi, V.; Kardar, G.A.; Khoshnoodi, J.; Mobini, M.; Ahmadi Zare, H.; Ghaderi, A.; Jeddi-Tehrani, M.; et al. Epitope Mapping of Severe Acute Respiratory Syndrome Coronavirus 2 Neutralizing Receptor Binding Domain-Specific Monoclonal Antibodies. Front. Med. 2022, 9. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, B.; Ding, M.; Song, S.; Kang, Y.; Yu, Y.; Xu, M.; Xiang, T.; Gao, L.; Feng, Q.; et al. A Novel Vaccine Candidate Based on Chimeric Virus-like Particle Displaying Multiple Conserved Epitope Peptides Induced Neutralizing Antibodies against EBV Infection. Theranostics 2020, 10, 5704–5718. [Google Scholar] [CrossRef] [PubMed]
- Fishman, J.B.; Berg, E.A. Protein A and Protein G Purification of Antibodies. Cold Spring Harb. Protoc. 2019, 2019, pdb.prot099143. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, S.; Zhang, Y.; Wang, Y.; Zhu, Y.; Wang, B.; Chen, Z.-N. Purification of a Polyclonal Antibody against CD147 for ELISA Using Antigen-Immunoaffinity Chromatography. Mol. Med. Rep. 2017, 15, 4035–4040. [Google Scholar] [CrossRef] [PubMed]
- Valadon, P.; Scharff, M.D. Enhancement of ELISAs for Screening Peptides in Epitope Phage Display Libraries. J. Immunol. Methods 1996, 197, 171–179. [Google Scholar] [CrossRef]
- Taipa, M. Immunoassays: Biological Tools for High Throughput Screening and Characterisation of Combinatorial Libraries. Comb. Chem. High Throughput Screen. 2008, 11, 325–335. [Google Scholar] [CrossRef]
- McGuire, M.J.; Li, S.; Brown, K.C. Biopanning of Phage Displayed Peptide Libraries for the Isolation of Cell-Specific Ligands. In Biosensors and Biodetection; Rasooly, A., Herold, K.E., Eds.; Humana Press: New York, NY, USA, 2009; Volume 504, pp. 291–321. [Google Scholar]
- Kay, B.K.; Kasanov, J.; Yamabhai, M. Screening Phage-Displayed Combinatorial Peptide Libraries. Methods 2001, 24, 240–246. [Google Scholar] [CrossRef]
- Kim, H.; Ho, M. Isolation of Antibodies to Heparan Sulfate on Glypicans by Phage Display. Curr. Protoc. Protein Sci. 2018, 94, e66. [Google Scholar] [CrossRef]
- Mohd Ali, M.R.; Sum, J.S.; Aminuddin Baki, N.N.; Choong, Y.S.; Nor Amdan, N.A.; Amran, F.; Lim, T.S. Development of Monoclonal Antibodies against Recombinant LipL21 Protein of Pathogenic Leptospira through Phage Display Technology. Int. J. Biol. Macromol. 2021, 168, 289–300. [Google Scholar] [CrossRef]
- Yu, J.; Smith, G.P. [1] Affinity Maturation of Phage-Displayed Peptide Ligands. In Methods in Enzymelogy; Academic Press: Cambridge, MA, USA, 1996; pp. 3–27. [Google Scholar]
- Zhou, J.; Chen, J.; Peng, Y.; Xie, Y.; Xiao, Y. A Promising Tool in Serological Diagnosis: Current Research Progress of Antigenic Epitopes in Infectious Diseases. Pathogens 2022, 11, 1095. [Google Scholar] [CrossRef]
- Ren, H.J.; Liu, R.D.; Wang, Z.Q.; Cui, J. Construction and Use of a Trichinella Spiralis Phage Display Library to Identify the Interactions between Parasite and Host Enterocytes. Parasitol. Res. 2013, 112, 1857–1863. [Google Scholar] [CrossRef]
- Ma, M.; Liu, J.; Jin, S.; Wang, L. Development of Tumour Peptide Vaccines: From Universalization to Personalization. Scand. J. Immunol. 2020, 91, e12875. [Google Scholar] [CrossRef]
- Gan, H.K.; Burgess, A.W.; Clayton, A.H.A.; Scott, A.M. Targeting of a Conformationally Exposed, Tumor-Specific Epitope of EGFR as a Strategy for Cancer Therapy. Cancer Res. 2012, 72, 2924–2930. [Google Scholar] [CrossRef]
- Zhu, B.; Yang, J.; Van, R.; Yang, F.; Yu, Y.; Yu, A.; Ran, K.; Yin, K.; Liang, Y.; Shen, X.; et al. Epitope Alteration by Small Molecules and Applications in Drug Discovery. Chem. Sci. 2022, 13, 8104–8116. [Google Scholar] [CrossRef]
- Hosseinidoust, Z. Phage-Mediated Gene Therapy. Curr. Gene Ther. 2017, 17, 120–126. [Google Scholar] [CrossRef]
- Nakagami, H. Development of COVID-19 Vaccines Utilizing Gene Therapy Technology. Int. Immunol. 2021, 33, 521–527. [Google Scholar] [CrossRef]
- Lundstrom, K. Application of DNA Replicons in Gene Therapy and Vaccine Development. Pharmaceutics 2023, 15, 947. [Google Scholar] [CrossRef]
- Welsh, R.M.; Fujinami, R.S. Pathogenic Epitopes, Heterologous Immunity and Vaccine Design. Nat. Rev. Microbiol. 2007, 5, 555–563. [Google Scholar] [CrossRef]
- Zhang, Z.; Yao, F.; Lv, J.; Ding, Y.; Liu, X.; Zhang, L.; Ma, Z.; Zhou, P.; Wang, Y.; Guo, H.; et al. Identification of B-Cell Epitopes on Structural Proteins VP1 and VP2 of Senecavirus A and Development of a Multi-Epitope Recombinant Protein Vaccine. Virology 2023, 582, 48–56. [Google Scholar] [CrossRef]
- Vishweshwaraiah, Y.L.; Hnath, B.; Rackley, B.; Wang, J.; Gontu, A.; Chandler, M.; Afonin, K.A.; Kuchipudi, S.V.; Christensen, N.; Yennawar, N.H.; et al. Adaptation-Proof SARS-CoV-2 Vaccine Design. Adv. Funct. Mater. 2022, 32, 2206055. [Google Scholar] [CrossRef]
- Guo, J.-Y.; Liu, I.-J.; Lin, H.-T.; Wang, M.-J.; Chang, Y.-L.; Lin, S.-C.; Liao, M.-Y.; Hsu, W.-C.; Lin, Y.-L.; Liao, J.C.; et al. Identification of COVID-19 B-Cell Epitopes with Phage-Displayed Peptide Library. J. Biomed. Sci. 2021, 28, 43. [Google Scholar] [CrossRef]
- Staquicini, D.I.; Tang, F.H.F.; Markosian, C.; Yao, V.J.; Staquicini, F.I.; Dodero-Rojas, E.; Contessoto, V.G.; Davis, D.; O’Brien, P.; Habib, N.; et al. Design and Proof of Concept for Targeted Phage-Based COVID-19 Vaccination Strategies with a Streamlined Cold-Free Supply Chain. Proc. Natl. Acad. Sci. USA 2021, 118, e2105739118. [Google Scholar] [CrossRef]
- Kumar, G.; Sterrett, S.; Hall, L.; Tabengwa, E.; Honjo, K.; Larimer, M.; Davis, R.S.; Goepfert, P.A.; Larimer, B.M. Comprehensive Mapping of SARS-CoV-2 Peptide Epitopes for Development of a Highly Sensitive Serological Test for Total and Neutralizing Antibodies. Protein Eng. Des. Sel. 2022, 35, gzab033. [Google Scholar] [CrossRef] [PubMed]
- Larralde, O.G.; Martinez, R.; Camacho, F.; Amin, N.; Aguilar, A.; Talavera, A.; Stott, D.I.; Perez, E.M. Identification of Hepatitis A Virus Mimotopes by Phage Display, Antigenicity and Immunogenicity. J. Virol. Methods 2007, 140, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Burritt, J.B.; Bond, C.W.; Doss, K.W.; Jesaitis, A.J. Filamentous Phage Display of Oligopeptide Libraries. Anal. Biochem. 1996, 238, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Wan, Y.; Li, D.G.; Tang, Y.; Zhou, W. A Mimotope of Pre-S2 Region of Surface Antigen of Viral Hepatitis B Screened by Phage Display. Cell Res. 2001, 11, 203–208. [Google Scholar] [CrossRef]
- Folgori, A.; Tafi, R.; Meola, A.; Felici, F.; Galfré, G.; Cortese, R.; Monaci, P.; Nicosia, A. A General Strategy to Identify Mimotopes of Pathological Antigens Using Only Random Peptide Libraries and Human Sera. EMBO J. 1994, 13, 2236–2243. [Google Scholar] [CrossRef]
- Tafi, R.; Bandi, R.; Prezzi, C.; Mondelli, M.U.; Cortese, R.; Monaci, P.; Nicosia, A. Identification of HCV Core Mimotopes: Improved Methods for the Selection and Use of Disease-Related Phage-Displayed Peptides. Biol. Chem. 1997, 378, 495–502. [Google Scholar] [CrossRef]
- Larralde, O.; Petrik, J. Phage-Displayed Peptides That Mimic Epitopes of Hepatitis E Virus Capsid. Med. Microbiol. Immunol. 2017, 206, 301–309. [Google Scholar] [CrossRef]
- Khurana, S.; Suguitan, A.L.; Rivera, Y.; Simmons, C.P.; Lanzavecchia, A.; Sallusto, F.; Manischewitz, J.; King, L.R.; Subbarao, K.; Golding, H. Antigenic Fingerprinting of H5N1 Avian Influenza Using Convalescent Sera and Monoclonal Antibodies Reveals Potential Vaccine and Diagnostic Targets. PLoS Med. 2009, 6, e1000049. [Google Scholar] [CrossRef]
- Khurana, S.; Wu, J.; Verma, N.; Verma, S.; Raghunandan, R.; Manischewitz, J.; King, L.R.; Kpamegan, E.; Pincus, S.; Smith, G.; et al. H5N1 Virus-Like Particle Vaccine Elicits Cross-Reactive Neutralizing Antibodies That Preferentially Bind to the Oligomeric Form of Influenza Virus Hemagglutinin in Humans. J. Virol. 2011, 85, 10945–10954. [Google Scholar] [CrossRef]
- Scala, G.; Chen, X.; Liu, W.; Telles, J.N.; Cohen, O.J.; Vaccarezza, M.; Igarashi, T.; Fauci, A.S. Selection of HIV-Specific Immunogenic Epitopes by Screening Random Peptide Libraries with HIV-1-Positive Sera. J. Immunol. 1999, 162, 6155–6161. [Google Scholar] [CrossRef]
- Gazarian, K.G.; Palacios-Rodríguez, Y.; Gazarian, T.G.; Huerta, L. HIV-1 V3 Loop Crown Epitope-Focused Mimotope Selection by Patient Serum from Random Phage Display Libraries: Implications for the Epitope Structural Features. Mol. Immunol. 2013, 54, 148–156. [Google Scholar] [CrossRef]
- Palacios-Rodríguez, Y.; Gazarian, T.; Rowley, M.; Majluf-Cruz, A.; Gazarian, K. Collection of Phage–Peptide Probes for HIV-1 Immunodominant Loop-Epitope. J. Microbiol. Methods 2007, 68, 225–235. [Google Scholar] [CrossRef]
- Machado, L.F.A.; Filho, L.R.G.; Santos, F.A.A.; Siravenha, L.Q.; Silva, A.N.M.R.; Queiroz, M.A.F.; Vallinoto, A.C.R.; Ishak, M.O.G.; Ishak, R. Bioprospection and Selection of Peptides by Phage Display as Novel Epitope-Based Diagnostic Probes for Serological Detection of HTLV-1 and Use in Future Vaccines. Front. Med. 2022, 9, 884738. [Google Scholar] [CrossRef]
- Eda, S.; Sherman, I.W. Selection of Peptides Recognized by Human Antibodies against the Surface of Plasmodium Falciparum -Infected Erythrocytes. Parasitology 2005, 130, 1–11. [Google Scholar] [CrossRef]
- Teixeira, A.A.R.; Carnero, L.R.; Kuramoto, A.; Tang, F.H.F.; Gomes, C.H.; Pereira, N.B.; de Oliveira, L.C.; Garrini, R.; Monteiro, J.S.; Setubal, J.C.; et al. A Refined Genome Phage Display Methodology Delineates the Human Antibody Response in Patients with Chagas Disease. iScience 2021, 24, 102540. [Google Scholar] [CrossRef]
- Pękacz, M.; Basałaj, K.; Kalinowska, A.; Klockiewicz, M.; Stopka, D.; Bąska, P.; Długosz, E.; Karabowicz, J.; Młocicki, D.; Wiśniewski, M.; et al. Selection of New Diagnostic Markers for Dirofilaria Repens Infections with the Use of Phage Display Technology. Sci. Rep. 2022, 12, 2288. [Google Scholar] [CrossRef]
- Roehnisch, T.; Then, C.; Nagel, W.; Blumenthal, C.; Braciak, T.; Donzeau, M.; Böhm, T.; Flaig, M.; Bourquin, C.; Oduncu, F.S. Phage Idiotype Vaccination: First Phase I/II Clinical Trial in Patients with Multiple Myeloma. J. Transl. Med. 2014, 12, 119. [Google Scholar] [CrossRef]
- Johnson, A.K.; Jones, R.L.; Kraneburg, C.J.; Cochran, A.M.; Samoylov, A.M.; Wright, J.C.; Hutchinson, C.; Picut, C.; Cattley, R.C.; Martin, D.R.; et al. Phage Constructs Targeting Gonadotropin-Releasing Hormone for Fertility Control: Evaluation in Cats. J. Feline Med. Surg. 2020, 22, 685–695. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, M. Epitopes and Mimotopes Identification Using Phage Display for Vaccine Development against Infectious Pathogens. Vaccines 2023, 11, 1176. https://doi.org/10.3390/vaccines11071176
Palma M. Epitopes and Mimotopes Identification Using Phage Display for Vaccine Development against Infectious Pathogens. Vaccines. 2023; 11(7):1176. https://doi.org/10.3390/vaccines11071176
Chicago/Turabian StylePalma, Marco. 2023. "Epitopes and Mimotopes Identification Using Phage Display for Vaccine Development against Infectious Pathogens" Vaccines 11, no. 7: 1176. https://doi.org/10.3390/vaccines11071176
APA StylePalma, M. (2023). Epitopes and Mimotopes Identification Using Phage Display for Vaccine Development against Infectious Pathogens. Vaccines, 11(7), 1176. https://doi.org/10.3390/vaccines11071176







