Use of an ETEC Proteome Microarray to Evaluate Cross-Reactivity of ETVAX® Vaccine-Induced IgG Antibodies in Zambian Children
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Lab Analysis
2.2.1. Microarray Creation
2.2.2. Sample Analysis
2.2.3. Data Analysis
2.2.4. Statistical Analysis
3. Results
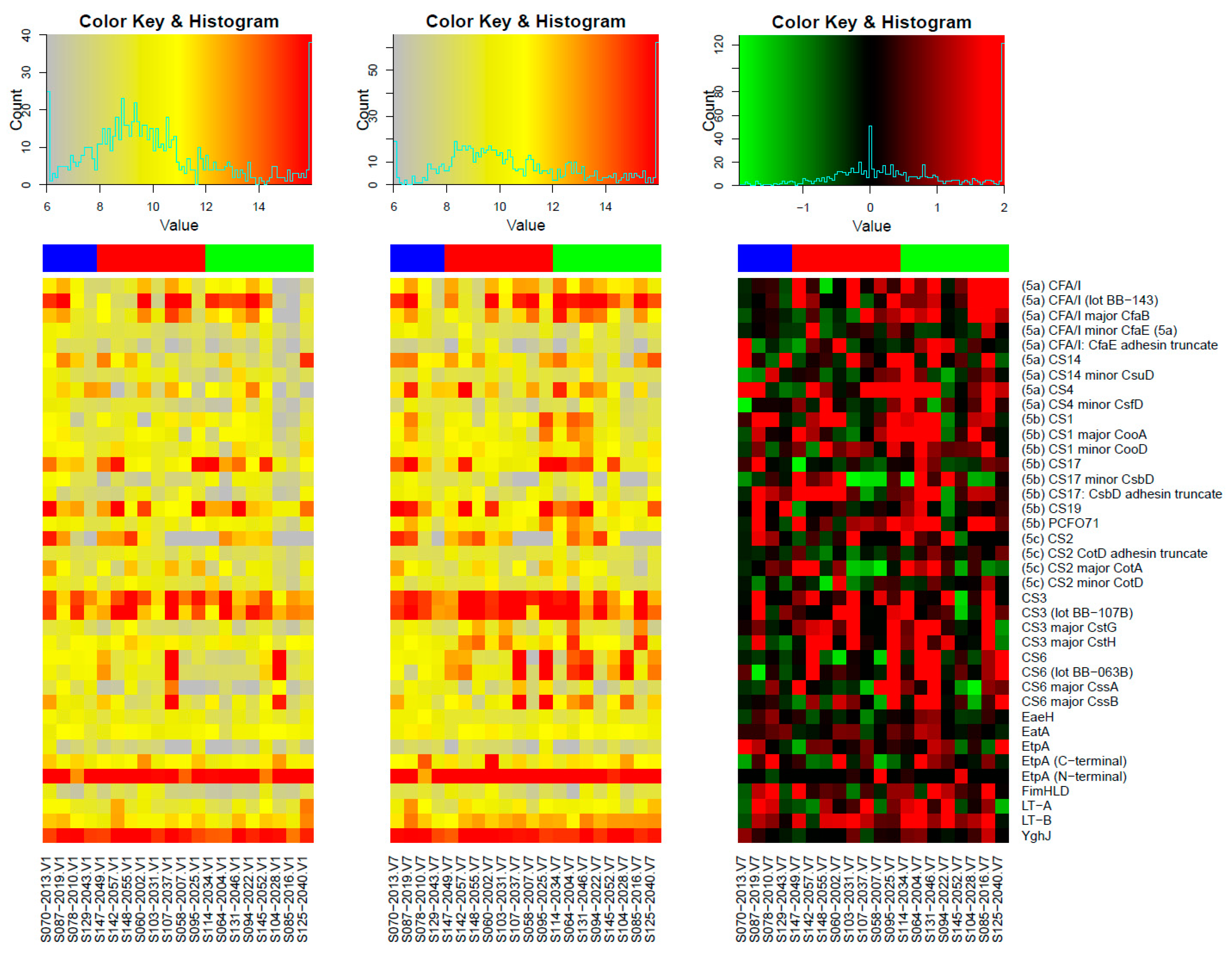
3.1. Responses to Purified Proteins
Microarray Responses to ETVAX® Antigens and Non-ETVAX® Antigens
3.2. Microarray Responses to Other ETEC Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khalil, I.A.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.; Atherly, D.E.; Brewer, T.G.; Engmann, C.M.; Houpt, E.R.; Kang, G.; et al. Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Hosangadi, D.; Smith, P.G.; Giersing, B.K. Considerations for using ETEC and Shigella disease burden estimates to guide vaccine development strategy. Vaccine 2019, 37, 7372–7380. [Google Scholar] [CrossRef] [PubMed]
- Kotloff, K.L.; Platts-Mills, J.A.; Nasrin, D.; Roose, A.; Blackwelder, W.C.; Levine, M.M. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 2017, 35, 6783–6789. Available online: https://pubmed.ncbi.nlm.nih.gov/28765005/ (accessed on 12 October 2021).
- Kotloff, K.L. The Burden and Etiology of Diarrheal Illness in Developing Countries. Pediatr. Clin. N. Am. 2017, 64, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Immunization, Vaccines and Biologicals. Available online: https://www.who.int/teams/immunization-vaccines-and-biologicals/diseases/enterotoxigenic-escherichia-coli-(etec) (accessed on 8 August 2022).
- Khalil, I.; Walker, R.; Porter, C.K.; Muhib, F.; Chilengi, R.; Cravioto, A.; Guerrant, R.; Svennerholm, A.-M.; Qadri, F.; Baqar, S.; et al. Enterotoxigenic Escherichia coli (ETEC) vaccines: Priority activities to enable product development, licensure, and global access. Vaccine 2021, 39, 4266–4277. [Google Scholar] [CrossRef] [PubMed]
- Chisenga, C.C.; Bosomprah, S.; Laban, N.M.; Kazimbaya, K.M.; Mwaba, J.; Simuyandi, M.; Chilengi, R. vAetiology of Diarrhoea in Children Under Five in Zambia Detected Using Luminex xTAG Gastrointestinal Pathogen Panel. Pediatr. Infect. Dis. 2018, 3, 8. [Google Scholar] [CrossRef]
- Qadri, F.; Svennerholm, A.M.; Faruque, A.S.G.; Sack, R.B. Enterotoxigenic Escherichia coli in Developing Countries: Epidemiology, Microbiology, Clinical Features, Treatment, and Prevention. Clin. Microbiol. Rev. 2005, 18, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Sahl, J.W.; Sistrunk, J.R.; Baby, N.I.; Begum, Y.; Luo, Q.; Sheikh, A.; Qadri, F.; Fleckenstein, J.M.; Rasko, D.A. Insights into enterotoxigenic Escherichia coli diversity in Bangladesh utilizing genomic epidemiology. Sci. Rep. 2017, 7, 3402. [Google Scholar] [CrossRef]
- Chakraborty, S.; Randall, A.; Vickers, T.J.; Molina, D.; Harro, C.D.; DeNearing, B.; Brubaker, J.; Sack, D.A.; Bourgeois, A.L.; Felgner, P.L.; et al. Interrogation of a live-attenuated enterotoxigenic Escherichia coli vaccine highlights features unique to wild-type infection. NPJ Vaccines 2019, 4, 37. [Google Scholar] [CrossRef]
- Zegeye, E.D.; Govasli, M.L.; Sommerfelt, H.; Puntervoll, P. Development of an enterotoxigenic Escherichia coli vaccine based on the heat-stable toxin. Hum. Vaccines Immunother. 2018, 15, 1379–1388. [Google Scholar] [CrossRef]
- Vidal, R.M.; Muhsen, K.; Tennant, S.M.; Svennerholm, A.-M.; Sow, S.O.; Sur, D.; Zaidi, A.K.M.; Faruque, A.S.G.; Saha, D.; Adegbola, R.; et al. Colonization factors among enterotoxigenic Escherichia coli isolates from children with moderate-to-severe diarrhea and from matched controls in the Global Enteric Multicenter Study (GEMS). PLoS Negl. Trop. Dis. 2019, 13, e0007037. [Google Scholar] [CrossRef]
- Mortezaei, N.; Epler, C.R.; Shao, P.P.; Shirdel, M.; Singh, B.; McVeigh, A.; Uhlin, B.E.; Savarino, S.J.; Andersson, M.; Bullitt, E. Structure and function of Enterotoxigenic Escherichia coli fimbriae from differing assembly pathways. Mol. Microbiol. 2015, 95, 116. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shahabudin, S.; Farid, S.; Lee, L.H.; McVeigh, A.L.; Maciel, M.; Poole, S.T.; Broadman, M.; Prouty, M.G.; Savarino, S.J. Cross-Reactivity, Epitope Mapping, and Potency of Monoclonal Antibodies to Class 5 Fimbrial Tip Adhesins of Enterotoxigenic Escherichia coli. Infect. Immun. 2020, 88, e00246-20. [Google Scholar] [CrossRef]
- Anantha, R.P.; McVeigh, A.L.; Lee, L.H.; Agnew, M.K.; Cassels, F.J.; Scott, D.A.; Whittam, T.S.; Savarino, S.J. Evolutionary and Functional Relationships of Colonization Factor Antigen I and Other Class 5 Adhesive Fimbriae of Enterotoxigenic Escherichia coli. Infect. Immun. 2004, 72, 7190–7201. [Google Scholar] [CrossRef] [PubMed]
- Kipkirui, E.; Koech, M.; Ombogo, A.; Kirera, R.; Ndonye, J.; Kipkemoi, N.; Kirui, M.; Philip, C.; Roth, A.; Flynn, A.; et al. Molecular characterization of enterotoxigenic Escherichia coli toxins and colonization factors in children under five years with acute diarrhea attending Kisii Teaching and Referral Hospital, Kenya. Trop. Dis. Travel Med. Vaccines 2021, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Simuyandi, M.; Chilengi, R.; Connor, S.B.; Voeglein, J.B.; Laban, N.M.; Mwila-Kazimbaya, K.; Chisenga, C.C.; Mwaba, J.; Sack, D.A.; Chakraborty, S. Enterotoxigenic Escherichia coli Toxins and Colonization Factors among Zambian Children Presenting with Moderate to Severe Diarrhea to Selected Health Facilities. Arch. Microbiol. Immunol. 2019, 3, 173–184. [Google Scholar] [CrossRef]
- Leach, S.; Lundgren, A.; Carlin, N.; Löfstrand, M.; Svennerholm, A.-M. Cross-reactivity and avidity of antibody responses induced in humans by the oral inactivated multivalent enterotoxigenicEscherichia coli (ETEC) vaccine ETVAX. Vaccine 2017, 35, 3966–3973. [Google Scholar] [CrossRef] [PubMed]
- von Mentzer, A.; Tobias, J.; Wiklund, G.; Nordqvist, S.; Aslett, M.; Dougan, G.; Sjöling, Å.; Svennerholm, A.M. Identification and characterization of the novel colonization factor CS30 based on whole genome sequencing in enterotoxigenic Escherichia coli (ETEC). Sci Rep. 2017, 7, 12514. [Google Scholar] [CrossRef]
- Chakraborty, S.; Harro, C.; DeNearing, B.; Brubaker, J.; Connor, S.; Maier, N.; Dally, L.; Flores, J.; Bourgeois, A.L.; Walker, R.; et al. Impact of lower challenge doses of enterotoxigenic Escherichia coli on clinical outcome, intestinal colonization and immune responses in adult volunteers. PLOS Negl. Trop. Dis. 2018, 12, e0006442. [Google Scholar] [CrossRef]
- Harris, J.A.; Roy, K.; Woo-Rasberry, V.; Hamilton, D.J.; Kansal, R.; Qadri, F.; Fleckenstein, J.M. Directed Evaluation of Enterotoxigenic Escherichia coli Autotransporter Proteins as Putative Vaccine Candidates. PLoS Negl. Trop. Dis. 2011, 5, e1428. [Google Scholar] [CrossRef]
- Montero, D.; Orellana, P.; Gutiérrez, D.; Araya, D.; Salazar, J.C.; Prado, V.; Oñate, Á.; del Canto, F.; Vidal, R. Immunoproteomic Analysis to Identify Shiga Toxin-Producing Escherichia coli Outer Membrane Proteins Expressed during Human Infection. Infect. Immun. 2014, 82, 4767–4777. [Google Scholar] [CrossRef] [PubMed]
- Fleckenstein, J.M. Confronting Challenges to Enterotoxigenic Escherichia coli Vaccine Development. Front. Trop. Dis. 2021, 2, 709907. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Zhang, W. Development of effective vaccines for enterotoxigenic Escherichia coli. Lancet Infect. Dis. 2020, 20, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sack, D.A. Current Progress in Developing Subunit Vaccines against Enterotoxigenic Escherichia coli-Associated Diarrhea. Clin. Vaccine Immunol. 2015, 22, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Tobias, J.; Lebens, M.; Bölin, I.; Wiklund, G.; Svennerholm, A.-M. Construction of non-toxic Escherichia coli and Vibrio cholerae strains expressing high and immunogenic levels of enterotoxigenic E. coli colonization factor I fimbriae. Vaccine 2008, 26, 743–752. [Google Scholar] [CrossRef]
- Tobias, J.; Svennerholm, A.-M.; Carlin, N.I.; Lebens, M.; Holmgren, J. Construction of a non-toxigenic Escherichia coli oral vaccine strain expressing large amounts of CS6 and inducing strong intestinal and serum anti-CS6 antibody responses in mice. Vaccine 2011, 29, 8863–8869. [Google Scholar] [CrossRef]
- Clements, J.D.; Norton, E.B. The Mucosal Vaccine Adjuvant LT(R192G/L211A) or dmLT. Msphere 2018, 3, e00215-18. [Google Scholar] [CrossRef]
- Svennerholm, A.-M.; Lundgren, A.; Leach, S.; Akhtar, M.; Qadri, F. Mucosal Immune Responses Against an Oral Enterotoxigenic Escherichia coli Vaccine Evaluated in Clinical Trials. J. Infect. Dis. 2021, 224, S821–S828. [Google Scholar] [CrossRef]
- Chen, K.; Magri, G.; Grasset, E.K.; Cerutti, A. Rethinking mucosal antibody responses: IgM, IgG and IgD join IgA. Nat. Rev. Immunol. 2020, 20, 427–441. Available online: https://www.nature.com/articles/s41577-019-0261-1 (accessed on 10 January 2022). [CrossRef]
- Roy, K.; Bartels, S.; Qadri, F.; Fleckenstein, J.M. Enterotoxigenic Escherichia coli Elicits Immune Responses to Multiple Surface Proteins. Infect. Immun. 2010, 78, 3027–3035. [Google Scholar] [CrossRef]
- Chakraborty, S.; Randall, A.; Vickers, T.J.; Molina, D.; Harro, C.D.; DeNearing, B.; Brubaker, J.; Sack, D.A.; Bourgeois, A.L.; Felgner, P.L.; et al. Human Experimental Challenge with Enterotoxigenic Escherichia coli Elicits Immune Responses to Canonical and Novel Antigens Relevant to Vaccine Development. J. Infect. Dis. 2018, 218, 1436–1446. [Google Scholar] [CrossRef]
- Giuntini, S.; Stoppato, M.; Sedic, M.; Ejemel, M.; Pondish, J.R.; Wisheart, D.; Schiller, Z.A.; Thomas, W.D.; Barry, E.M.; Cavacini, L.A.; et al. Identification and Characterization of Human Monoclonal Antibodies for Immunoprophylaxis against Enterotoxigenic Escherichia coli Infection. Infect. Immun. 2018, 86, e00355-18. Available online: https://journals.asm.org/doi/abs/10.1128/IAI.00355-18 (accessed on 12 December 2021). [CrossRef]
- Niewiesk, S. Maternal Antibodies: Clinical Significance, Mechanism of Interference with Immune Responses, and Possible Vaccination Strategies. Front. Immunol. 2014, 5, 446. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.N.; Tu, L.T.P.; Anders, K.L.; Hieu, N.T.; Vi, L.L.; Chau, N.V.V.; Duong, V.T.; Chau, T.T.H.; Tuyen, H.T.; Nga, T.V.T.; et al. The transfer and decay of maternal antibody against Shigella sonnei in a longitudinal cohort of Vietnamese infants. Vaccine 2016, 34, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Chisenga, C.C.; Bosomprah, S.; Simuyandi, M.; Mwila-Kazimbaya, K.; Chilyabanyama, O.N.; Laban, N.M.; Bialik, A.; Asato, V.; Meron-Sudai, S.; Frankel, G.; et al. Shigella-specific antibodies in the first year of life among Zambian infants: A longitudinal cohort study. PLoS ONE 2021, 16, e0252222. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.r-project.org/ (accessed on 25 March 2022).
- Benjaminit, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Charan, J.; Kantharia, N.D. How to calculate sample size in animal studies? J. Pharmacol. Pharmacother. 2013, 4, 303–306. Available online: http://www.ncbi.nlm.nih.gov/pubmed/24250214 (accessed on 29 July 2019). [CrossRef]
- Qadri, F.; Saha, A.; Ahmed, T.; Al Tarique, A.; Begum, Y.A.; Svennerholm, A.-M. Disease Burden Due to Enterotoxigenic Escherichia coli in the First 2 Years of Life in an Urban Community in Bangladesh. Infect. Immun. 2007, 75, 3961–3968. [Google Scholar] [CrossRef]
- Khampanisong, P.; Pauly, M.; Nouanthong, P.; Vickers, M.A.; Virachith, S.; Xaydalasouk, K.; Black, A.P.; Muller, C.P.; Hübschen, J.M. Waning of Maternal Antibodies against Measles Suggests a Large Window of Susceptibility in Infants in Lao People’s Democratic Republic. Pathogens 2021, 10, 1316. [Google Scholar] [CrossRef]
- Ares, M.A.; Abundes-Gallegos, J.; Rodríguez-Valverde, D.; Panunzi, L.G.; Jiménez-Galicia, C.; Jarillo-Quijada, M.D.; Cedillo, M.L.; Alcántar-Curiel, M.D.; Torres, J.; Girón, J.A.; et al. The Coli Surface Antigen CS3 of Enterotoxigenic Escherichia coli Is Differentially Regulated by H-NS, CRP, and CpxRA Global Regulators. Front. Microbiol. 2019, 10, 1685. [Google Scholar] [CrossRef]
- Poole, S.T., Jr.; Maciel, M.; Dinadayala, P.; Dori, K.E.; McVeigh, A.L.; Liu, Y.; Barry, E.; Grassel, C.; Prouty, M.G.; Renauld-Mongénie, G.; et al. Biochemical and Immunological Evaluation of Recombinant CS6-Derived Subunit Enterotoxigenic Escherichia coli Vaccine Candidates. Infect. Immun. 2019, 87, e00788-18. [Google Scholar] [CrossRef]
- Norton, E.B.; Branco, L.M.; Clements, J.D. Evaluating the A-Subunit of the Heat-Labile Toxin (LT) As an Immunogen and a Protective Antigen Against Enterotoxigenic Escherichia coli (ETEC). PLoS ONE 2015, 10, e0136302. [Google Scholar] [CrossRef] [PubMed]
- Norton, E.B.; Lawson, L.; Freytag, L.C.; Clements, J.D. Characterization of a Mutant Escherichia coli Heat-Labile Toxin, LT(R192G/L211A), as a Safe and Effective Oral Adjuvant. Clin. Vaccine Immunol. 2011, 18, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Qadri, F.; Akhtar, M.; Bhuiyan, T.R.; Chowdhury, M.I.; Ahmed, T.; Rafique, T.A.; Khan, A.; Rahman, S.I.A.; Khanam, F.; Lundgren, A.; et al. Safety and immunogenicity of the oral, inactivated, enterotoxigenic Escherichia coli vaccine ETVAX in Bangladeshi children and infants: A double-blind, randomised, placebo-controlled phase 1/2 trial. Lancet Infect. Dis. 2020, 20, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Qadri, F.; Kansal, R.; Rasko, D.A.; Sheikh, A.; Fleckenstein, J.M. Conservation and Immunogenicity of Novel Antigens in Diverse Isolates of Enterotoxigenic Escherichia coli. PLOS Negl. Trop. Dis. 2015, 9, e0003446. [Google Scholar] [CrossRef]
- Kumar, P.; Luo, Q.; Vickers, T.J.; Sheikh, A.; Lewis, W.G.; Fleckenstein, J. EatA, an Immunogenic Protective Antigen of Enterotoxigenic Escherichia coli, Degrades Intestinal Mucin. Infect. Immun. 2014, 82, 500–508. [Google Scholar] [CrossRef]
- Fleckenstein, J.; Sheikh, A.; Qadri, F. Novel Antigens for enterotoxigenic Escherichia coli (ETEC) Vaccines. Expert Rev. Vaccines 2014, 13, 631. [Google Scholar] [CrossRef]
- Dent, A.E.; Malhotra, I.; Wang, X.; Babineau, D.; Yeo, K.T.; Anderson, T.; Kimmel, R.J.; Angov, E.; Lanar, D.E.; Narum, D.; et al. Contrasting Patterns of Serologic and Functional Antibody Dynamics to Plasmodium falciparum Antigens in a Kenyan Birth Cohort. Clin. Vaccine Immunol. 2016, 23, 104–116. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mubanga, C.; Simuyandi, M.; Mwape, K.; Chibesa, K.; Chisenga, C.; Chilyabanyama, O.N.; Randall, A.; Liang, X.; Glashoff, R.H.; Chilengi, R. Use of an ETEC Proteome Microarray to Evaluate Cross-Reactivity of ETVAX® Vaccine-Induced IgG Antibodies in Zambian Children. Vaccines 2023, 11, 939. https://doi.org/10.3390/vaccines11050939
Mubanga C, Simuyandi M, Mwape K, Chibesa K, Chisenga C, Chilyabanyama ON, Randall A, Liang X, Glashoff RH, Chilengi R. Use of an ETEC Proteome Microarray to Evaluate Cross-Reactivity of ETVAX® Vaccine-Induced IgG Antibodies in Zambian Children. Vaccines. 2023; 11(5):939. https://doi.org/10.3390/vaccines11050939
Chicago/Turabian StyleMubanga, Cynthia, Michelo Simuyandi, Kapambwe Mwape, Kennedy Chibesa, Caroline Chisenga, Obvious Nchimunya Chilyabanyama, Arlo Randall, Xiaowu Liang, Richard H. Glashoff, and Roma Chilengi. 2023. "Use of an ETEC Proteome Microarray to Evaluate Cross-Reactivity of ETVAX® Vaccine-Induced IgG Antibodies in Zambian Children" Vaccines 11, no. 5: 939. https://doi.org/10.3390/vaccines11050939
APA StyleMubanga, C., Simuyandi, M., Mwape, K., Chibesa, K., Chisenga, C., Chilyabanyama, O. N., Randall, A., Liang, X., Glashoff, R. H., & Chilengi, R. (2023). Use of an ETEC Proteome Microarray to Evaluate Cross-Reactivity of ETVAX® Vaccine-Induced IgG Antibodies in Zambian Children. Vaccines, 11(5), 939. https://doi.org/10.3390/vaccines11050939




