Potent NTD-Targeting Neutralizing Antibodies against SARS-CoV-2 Selected from a Synthetic Immune System
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Expression and Purification of Antigen and Antibody
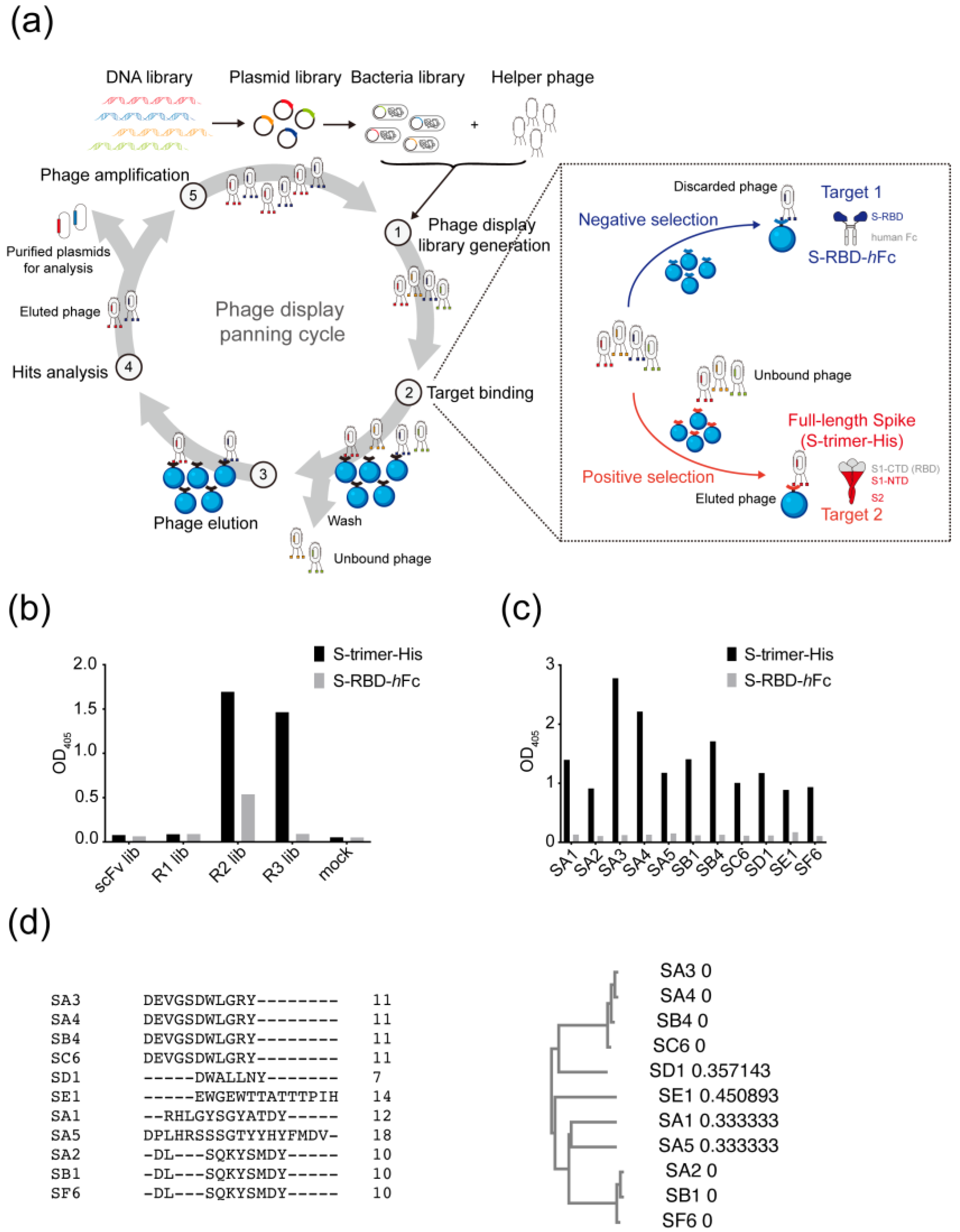
2.3. Phage Panning
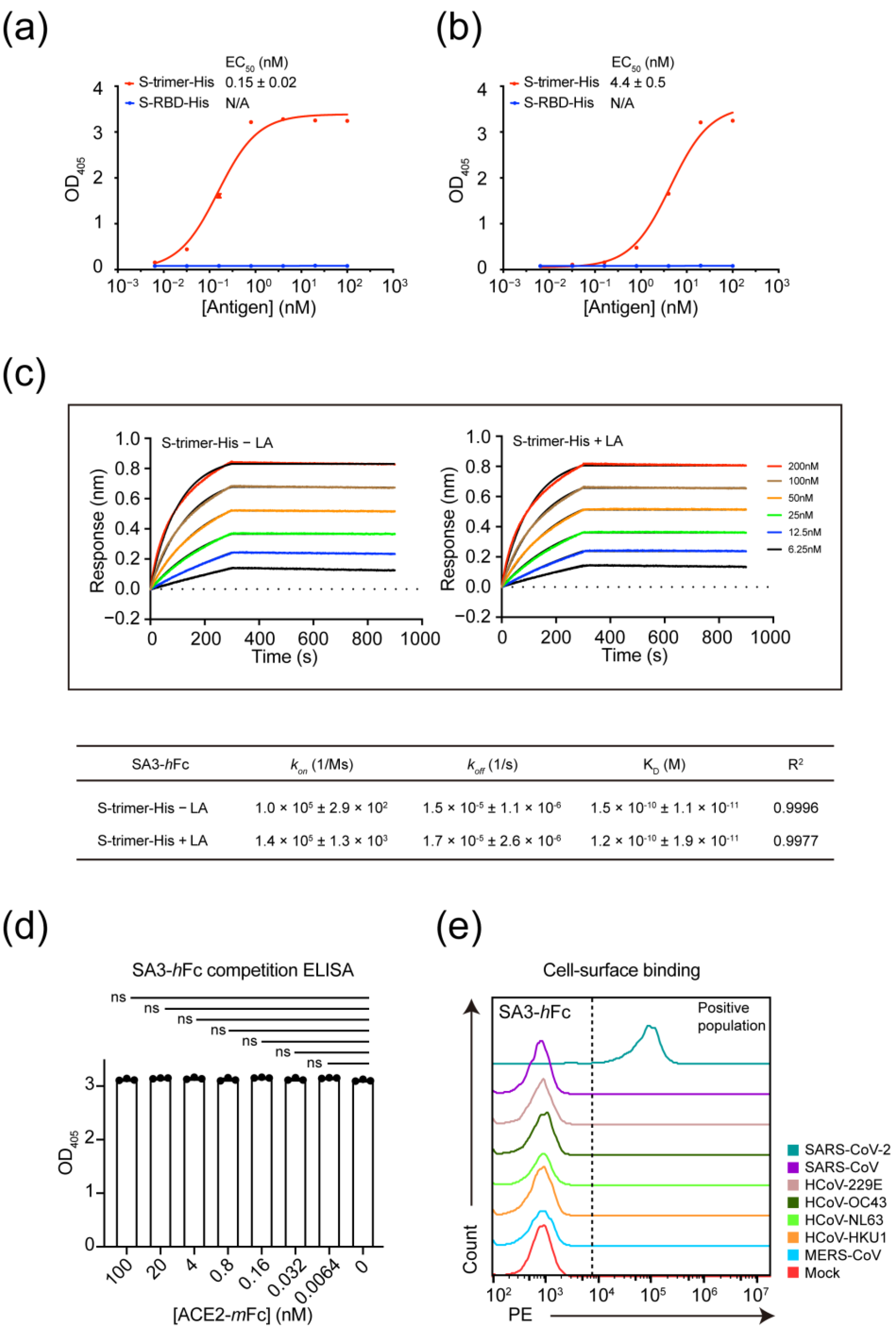
2.4. ELISA
2.5. Affinity Determination by Biolayer Interferometry (BLI)
2.6. Antibody Recognition of Cell Surface-Displayed Spikes by FACS
2.7. Size-Exclusion-High-Performance Liquid Chromatography
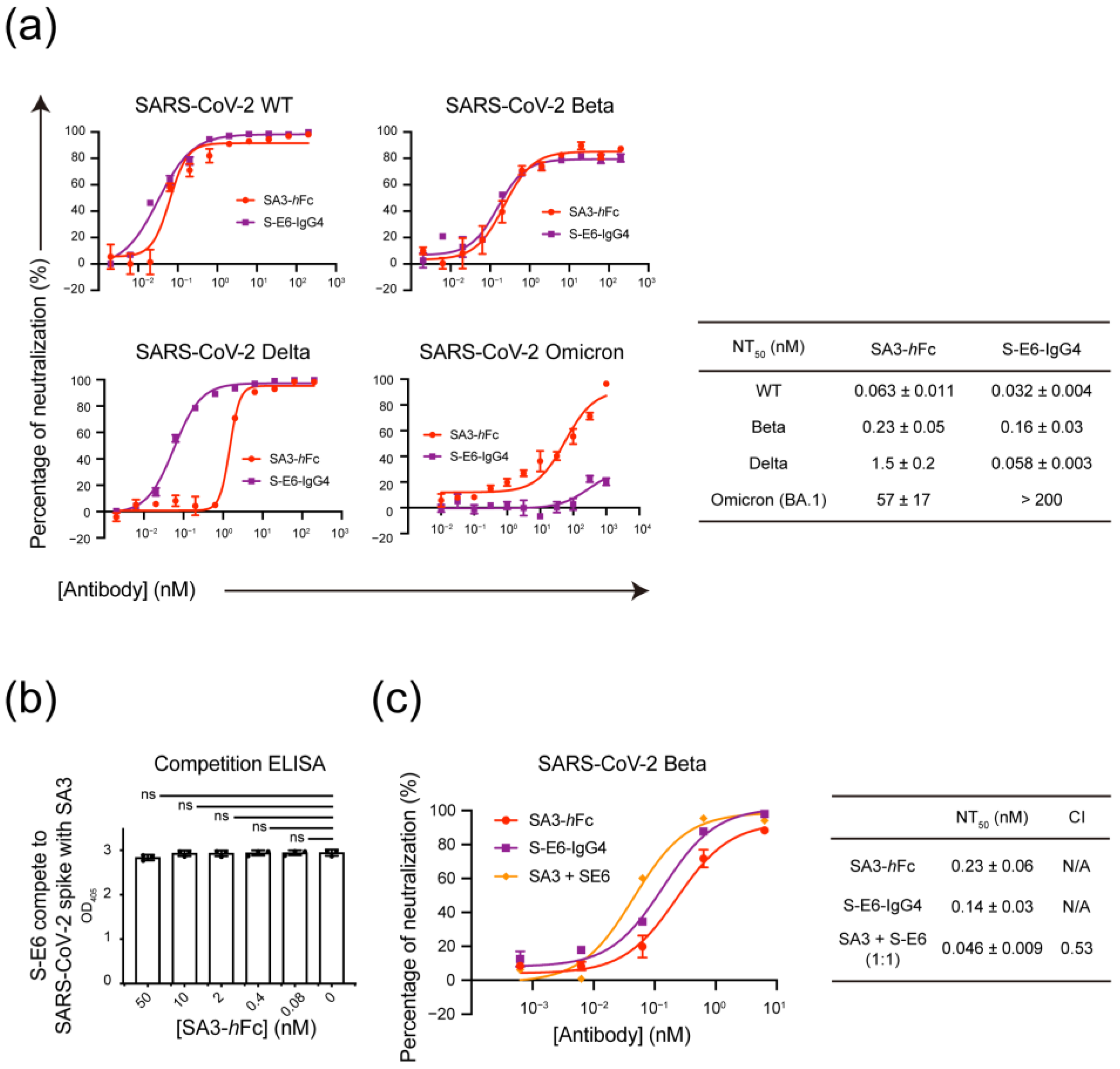
2.8. Pseudovirus-Based Neutralization Assay
2.9. Chemical Crosslinking and Mass Spectrometry
2.10. Quantification and Statistical Analysis
3. Results
3.1. Selection of Antibodies Targeting the Non-RBD Regions of SARS-CoV-2 Spike Protein
3.2. Kinetic Characterization and Mode of Interaction for Non-RBD Targeting Antibody SA3
3.3. Epitope Mapping of SA3
3.4. Mutually Non-Exclusive Binding and Synergistic Neutralization of SA3 and S-E6 against SARS-CoV-2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhou, Y.; Liu, S.; Kou, Z.; Li, W.; Farzan, M.; Jiang, S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004, 324, 773–781. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e899. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Hoffmann, M.; Arora, P.; Gross, R.; Seidel, A.; Hornich, B.F.; Hahn, A.S.; Kruger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e2312. [Google Scholar] [CrossRef]
- Ponde, R.A.A. Physicochemical effect of the N501Y, E484K/Q, K417N/T, L452R and T478K mutations on the SARS-CoV-2 spike protein RBD and its influence on agent fitness and on attributes developed by emerging variants of concern. Virology 2022, 572, 44–54. [Google Scholar] [CrossRef]
- Errico, J.M.; Adams, L.J.; Fremont, D.H. Antibody-mediated immunity to SARS-CoV-2 spike. Adv. Immunol. 2022, 154, 696370. [Google Scholar] [CrossRef]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e1021. [Google Scholar] [CrossRef]
- Kreer, C.; Zehner, M.; Weber, T.; Ercanoglu, M.S.; Gieselmann, L.; Rohde, C.; Halwe, S.; Korenkov, M.; Schommers, P.; Vanshylla, K.; et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell 2020, 182, 1663–1673. [Google Scholar] [CrossRef]
- Tong, P.; Gautam, A.; Windsor, I.W.; Travers, M.; Chen, Y.; Garcia, N.; Whiteman, N.B.; McKay, L.G.A.; Storm, N.; Malsick, L.E.; et al. Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. Cell 2021, 184, 4969–4980.e4915. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e1220. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e2316. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Lerner, R.A. Combinatorial antibody libraries: New advances, new immunological insights. Nat. Rev. Immunol. 2016, 16, 498–508. [Google Scholar] [CrossRef]
- Lerner, R.A. Manufacturing immunity to disease in a test tube: The magic bullet realized. Angew. Chem. Int. Ed. Engl. 2006, 45, 8106–8125. [Google Scholar] [CrossRef]
- Qiang, M.; Ma, P.; Li, Y.; Liu, H.; Harding, A.; Min, C.; Wang, F.; Liu, L.; Yuan, M.; Ji, Q.; et al. Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago. Adv. Sci. 2022, 9, e2102181. [Google Scholar] [CrossRef]
- Gao, C.; Brümmer, O.; Mao, S.; Janda, K.D. Selection of Human Metalloantibodies from a Combinatorial Phage Single-Chain Antibody Library. J. Am. Chem. Soc. 1999, 121, 6517–6518. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Ren, P.; Zhang, C.; Tang, J.; Yu, Z.; Zhu, X.; Fan, K.; Li, G.; Zhu, W.; Sang, W.; et al. Avidity-Based Selection of Tissue-Specific CAR-T Cells from a Combinatorial Cellular Library of CARs. Adv. Sci. 2021, 8, 2003091. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Lossl, P.; Scheltema, R.; Viner, R.; Heck, A.J.R. Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 2017, 8, 15473. [Google Scholar] [CrossRef] [PubMed]
- Toelzer, C.; Gupta, K.; Yadav, S.; Borucu, U.; Davidson, A.; Williamson, M.K.; Shoemark, D.; Garzoni, F.; Staufer, O.; Milligan, R.; et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 2020, 370, 725–730. [Google Scholar] [CrossRef]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef]
- McCallum, M.; Czudnochowski, N.; Rosen, L.E.; Zepeda, S.K.; Bowen, J.E.; Walls, A.C.; Hauser, K.; Joshi, A.; Stewart, C.; Dillen, J.R.; et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 2022, 375, 864–868. [Google Scholar] [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Cerutti, G.; Guo, Y.; Zhou, T.; Gorman, J.; Lee, M.; Rapp, M.; Reddem, E.R.; Yu, J.; Bahna, F.; Bimela, J.; et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 2021, 29, 819–833.e817. [Google Scholar] [CrossRef]
- Tao, P.; Kuang, Y.; Li, Y.; Li, W.; Gao, Z.; Liu, L.; Qiang, M.; Zha, Z.; Fan, K.; Ma, P.; et al. Selection of a Full Agonist Combinatorial Antibody that Rescues Leptin Deficiency In Vivo. Adv. Sci. 2020, 7, 2000818. [Google Scholar] [CrossRef]
- Zost, S.J.; Gilchuk, P.; Chen, R.E.; Case, J.B.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; Sutton, R.E.; Suryadevara, N.; Chen, E.C.; et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020, 26, 1422–1427. [Google Scholar] [CrossRef]
- Seydoux, E.; Homad, L.J.; MacCamy, A.J.; Parks, K.R.; Hurlburt, N.K.; Jennewein, M.F.; Akins, N.R.; Stuart, A.B.; Wan, Y.H.; Feng, J.; et al. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, Z.; Muecksch, F.; Cho, A.; Gaebler, C.; Hoffmann, H.H.; Ramos, V.; Zong, S.; Cipolla, M.; Johnson, B.; Schmidt, F.; et al. Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-CoV-2 spike proteins. Immunity 2022, 55, 998–1012.e1018. [Google Scholar] [CrossRef] [PubMed]
- Vanshylla, K.; Fan, C.; Wunsch, M.; Poopalasingam, N.; Meijers, M.; Kreer, C.; Kleipass, F.; Ruchnewitz, D.; Ercanoglu, M.S.; Gruell, H.; et al. Discovery of ultrapotent broadly neutralizing antibodies from SARS-CoV-2 elite neutralizers. Cell Host Microbe 2022, 30, 69–82.e10. [Google Scholar] [CrossRef]
- Seydoux, E.; Homad, L.J.; MacCamy, A.J.; Parks, K.R.; Hurlburt, N.K.; Jennewein, M.F.; Akins, N.R.; Stuart, A.B.; Wan, Y.H.; Feng, J.; et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity 2020, 53, 98–105.e105. [Google Scholar] [CrossRef] [PubMed]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef]
- Suryadevara, N.; Shrihari, S.; Gilchuk, P.; VanBlargan, L.A.; Binshtein, E.; Zost, S.J.; Nargi, R.S.; Sutton, R.E.; Winkler, E.S.; Chen, E.C.; et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 2021, 184, 2316–2331.e2315. [Google Scholar] [CrossRef]
- Murdaca, G.; Noberasco, G.; Olobardi, D.; Lunardi, C.; Maule, M.; Delfino, L.; Triggiani, M.; Cardamone, C.; Benfaremo, D.; Moroncini, G.; et al. Current Take on Systemic Sclerosis Patients’ Vaccination Recommendations. Vaccines 2021, 9, 1426. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Wang, F.; Li, Y.; Yan, L.; Liu, L.; Zhu, W.; Ma, P.; Shi, X.; Yang, G. Potent NTD-Targeting Neutralizing Antibodies against SARS-CoV-2 Selected from a Synthetic Immune System. Vaccines 2023, 11, 771. https://doi.org/10.3390/vaccines11040771
Li W, Wang F, Li Y, Yan L, Liu L, Zhu W, Ma P, Shi X, Yang G. Potent NTD-Targeting Neutralizing Antibodies against SARS-CoV-2 Selected from a Synthetic Immune System. Vaccines. 2023; 11(4):771. https://doi.org/10.3390/vaccines11040771
Chicago/Turabian StyleLi, Wenping, Fulian Wang, Yu Li, Lei Yan, Lili Liu, Wei Zhu, Peixiang Ma, Xiaojie Shi, and Guang Yang. 2023. "Potent NTD-Targeting Neutralizing Antibodies against SARS-CoV-2 Selected from a Synthetic Immune System" Vaccines 11, no. 4: 771. https://doi.org/10.3390/vaccines11040771
APA StyleLi, W., Wang, F., Li, Y., Yan, L., Liu, L., Zhu, W., Ma, P., Shi, X., & Yang, G. (2023). Potent NTD-Targeting Neutralizing Antibodies against SARS-CoV-2 Selected from a Synthetic Immune System. Vaccines, 11(4), 771. https://doi.org/10.3390/vaccines11040771





